Structural basis for effector transmembrane domain recognition by type VI secretion system chaperones
Figures
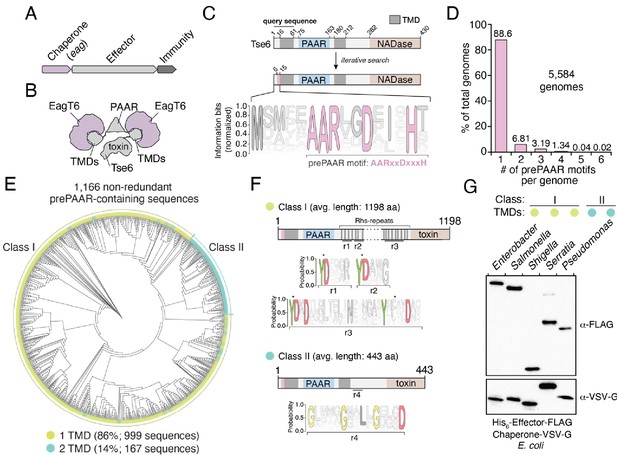
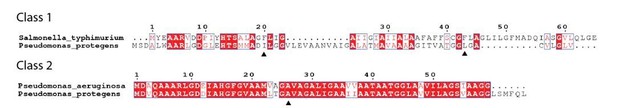
The prePAAR motif is conserved across multiple bacterial genera and is found in T6SS effectors that interact with Eag chaperones.
(A) Genomic arrangement of T6SS chaperone-effector-immunity genes for characterized effector-associated gene family members (eag; shown in purple), which encode DUF1795 domain-containing chaperones. (B) Schematic depicting Eag chaperone interactions with the transmembrane domain (TMD) regions of the model chaperone-effector pair EagT6-Tse6. (C) Protein architecture and sequence logo for the prePAAR motif found in the N-terminus of Tse6. An alignment of 2,054 sequences was generated using the 61 N-terminal residues of Tse6 as the search query. The relative frequency of each residue and information content in bits was calculated at every position of the sequence and then normalized by the sum of each position’s information bits. Transparency is used to indicate probability of a residue appearing at a specific position. Residues colored in pink correspond to the prePAAR motif: AARxxDxxxH. (D) Genomes from genera of Proteobacteria known to contain functional T6SSs (Burkholderia, Escherichia, Enterobacter, Pseudomonas, Salmonella, Serratia, Shigella, Yersinia) were screened for unique prePAAR effectors. Percentage of total genomes that contained 1 to 6 prePAAR motifs is indicated. (E) Phylogenetic distribution of 1,166 non-redundant prePAAR-containing effectors identified in B. TM prediction algorithms were used to quantify the number of TM regions in each effector. The two classes that emerged are labeled in green (class I; 1 TM region-containing effectors) and blue (class II; 2 TM region-containing effectors). Branch lengths indicates evolutionary distances. (F) Effector sequences within class I or class II were aligned, and a sequence logo was generated based on the relative frequency of each residue at each position to identify characteristic motifs of both classes. Four different regions (r1–r4) after the PAAR and TM regions were found to harbor conserved residues. Class I effectors contain YD repeat regions (r1-3) characteristic of Rhs proteins, whereas a GxxxxGxxLxGxxxD motif (r4) was identified in class II effectors. (G) Western blot analysis of five effector-chaperone pairs that were selected from the indicated genera, based on the analysis in B. Each pair was co-expressed in E. coli and co-purified using nickel affinity chromatography. The class and number of TM regions from each pair are indicated. Locus tags for each pair (e, effector; c, chaperone) are as follows: Enterobacter (e: ECL_01567, c: ECL_01566), Shigella (e: SF0266, c: SF3490), Salmonella (e: SL1344_0286, c: SL1344_0285), Serratia (e: Spro_3017, c: Spro_3016), Pseudomonas (e: PA0093, c: PA0094). Note that the Rhs component of the class I prePAAR effector SF0266 is encoded by the downstream open-reading frame SF0267 (see Figure 1—figure supplement 1C for details).
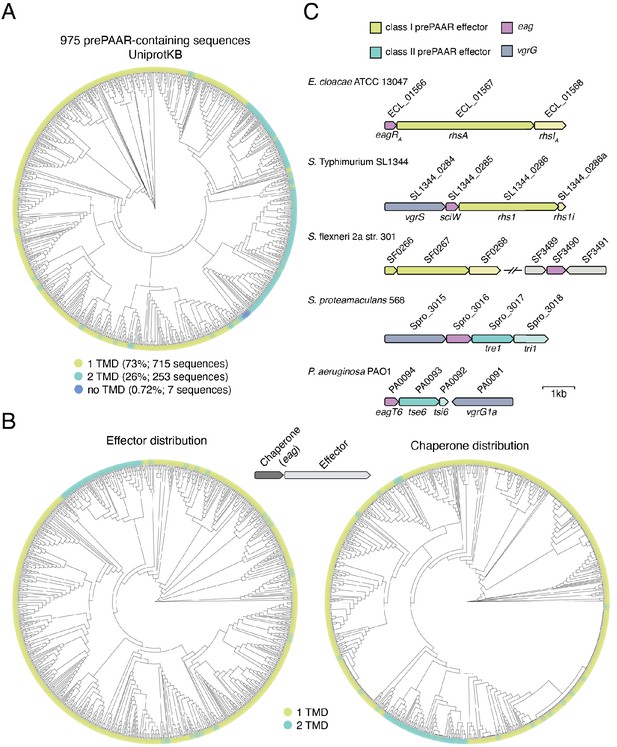
prePAAR effectors contain a fixed number of transmembrane domains.
(A) Phylogenetic distribution of 975 prePAAR-containing proteins identified in the UniProtKB database using the N-terminus of Tse6 (Tse6NT) as a search query (see Materials and Methods). The TM helix predictors TMHMM and Phobius (Krogh et al., 2001; Käll et al., 2007) were used to quantify the number of TMDs in each protein (green, 1 TMD; blue, 2 TMDs). (B) Similar analysis as Figure 1E, except that only prePAAR-containing effectors with an adjacent eag gene are depicted (left). The adjacently encoded eag chaperone sequences for each prePAAR effector were then used to build a second tree to depicting their distribution and association with an effector class (right). The eag chaperones were labelled with their neighboring effector’s TMD prediction. All branch length represents evolutionary distance. (C) Genomic arrangement of the five chaperone-effector pairs used for the co-purification experiment shown in Figure 1G. Shading was used to differentiate effector (dark) from potential immunity (light) genes. Locus tags and previously established names for each open reading frame are indicated above and below the gene diagram, respectively. Scale bar indicates 1 kilobase pair.
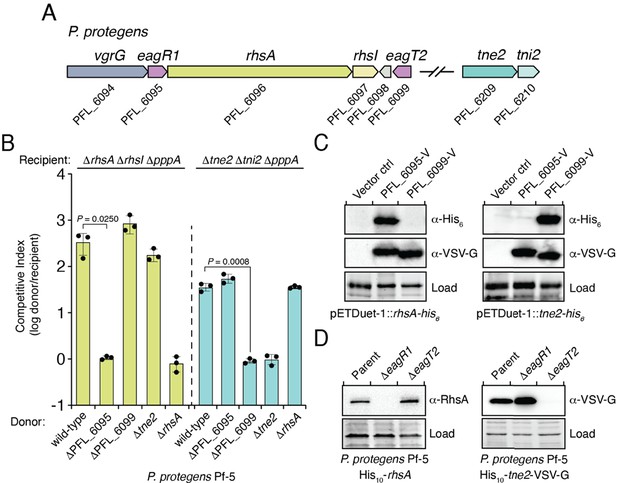
Eag chaperones are specific for their cognate prePAAR effector and are necessary for effector stability in vivo.
(A) Genomic context of two prePAAR-containing effector-immunity pairs from P. protegens Pf-5. RhsA is a class I effector (shown in green) and Tne2 is a class II effector (shown in blue). Shading is used to differentiate effector (dark) and immunity genes (light). Predicted eag genes are shown in purple. (B) Outcome of intraspecific growth competition assays between the indicated P. protegens donor and recipient strains. Donor strains were competed with recipient strains lacking rhsA-rhsI (green) or tne2-tni2 (blue). Both recipients are lacking pppA to stimulate type VI secretion. Data are mean ± s.d. for n = 3 biological replicates and are representative of two independent experiments; p values shown are from two-tailed, unpaired t-tests. (C) Western blot analysis of E. coli cell lysates from cells expressing the indicated effectors (RhsA or Tne2) and either empty vector, PFL_6095 V or PFL_6099 V. (D) Affinity-tagged RhsA or Tne2 were purified from cell fractions of the indicated P. protegens strains and visualized using western blot analysis. Deletion constructs for each eag gene were introduced into each of the indicated parent backgrounds. A non-specific band present in the SDS-PAGE gel was used as a loading control. (C–D) Data are representative of two independent experiments.
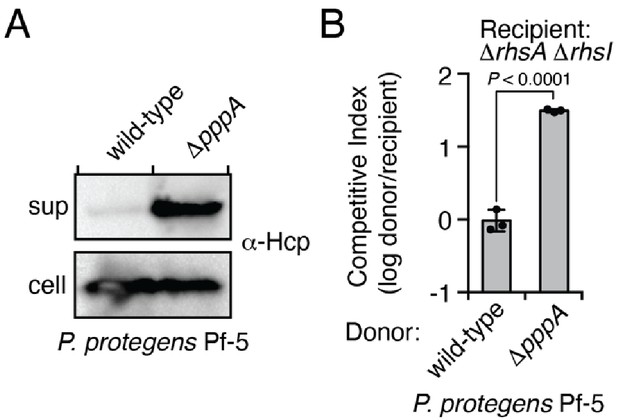
The type VI secretion system of P. protegens Pf-5 is repressed by the threonine phosphorylation pathway.
(A) Western blot of supernatant (sup) and cell fractions of the indicated P. protegens Pf-5 strains grown to OD 0.8. An Hcp (PFL_6089)-specific antibody was used to assess T6SS activity. (B) Intraspecific growth competition assay of the indicated donor P. protegens strains against a recipient susceptible to intoxication by the class I prePAAR effector RhsA. Data are mean ± s.d. for n = 3 biological replicates; p value shown is from a two-tailed, unpaired t-test.
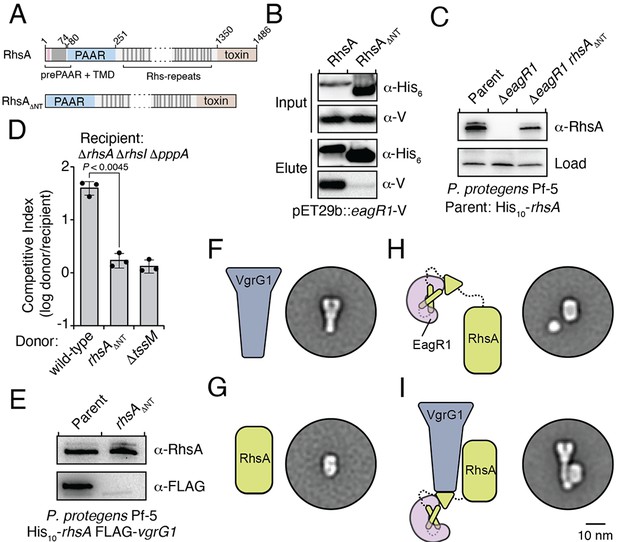
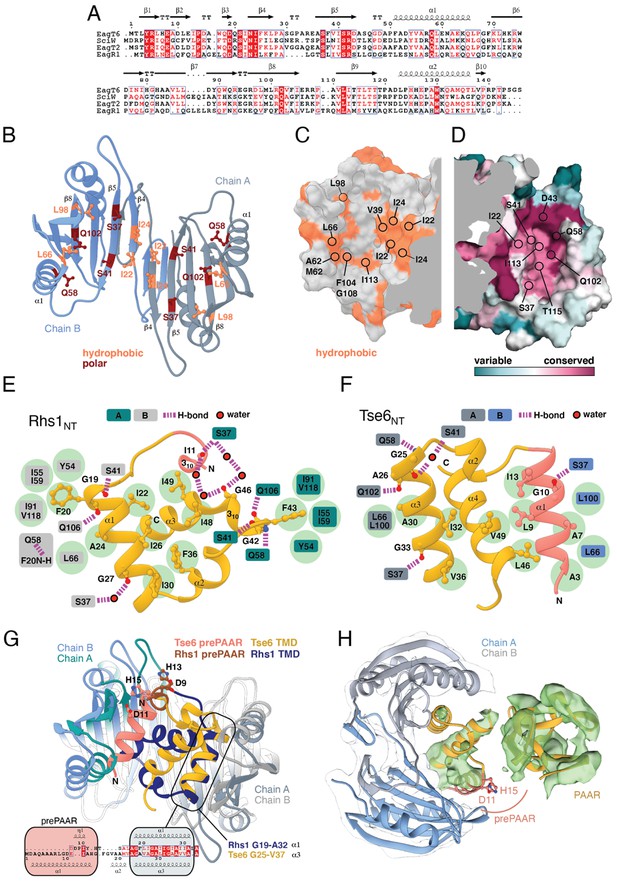
An Eag chaperone promotes the stability of its cognate class I prePAAR effector by interacting with its prePAAR and TMD-containing N-terminus.
(A) Domain architecture of P. protegens RhsA and a truncated variant lacking its prePAAR and TMD-containing N-terminus (RhsA∆NT). (B) EagR1 interacts with the N-terminus of RhsA. His6-tagged RhsA or RhsA∆NT and co-expressed with EagR1 in E. coli, purified using affinity chromatography and detected by western blot. (C) Affinity purification of chromosomally His10-tagged RhsA or RhsA∆NT from cell fractions of the indicated P. protegens strains. The parent strain expresses chromosomally encoded His10-tagged RhsA. The loading control is a non-specific band on the blot. (D) Outcome of growth competition assays between the indicated donor and recipient strains of P. protegens. Data are mean ± s.d. for n = 3 biological replicates; p value shown is from a two-tailed, unpaired t-test. (E) Affinity purification of His10-RhsA or His10-RhsA∆NT from a P. protegens Pf-5 strain containing a chromosomally encoded FLAG epitope tag fused to vgrG1. FLAG-tagged VgrG1 was detected by western blot. (F–I) Representative negative-stain EM class averages for purified VgrG1 (F), RhsA∆NT (G), EagR1-RhsA complex (H) and EagR1-RhsA-VgrG complex (I). Scale bar represents 10 nm for all images. All proteins were expressed and purified from E. coli. (B–C, E) Data are representative of two independent experiments.
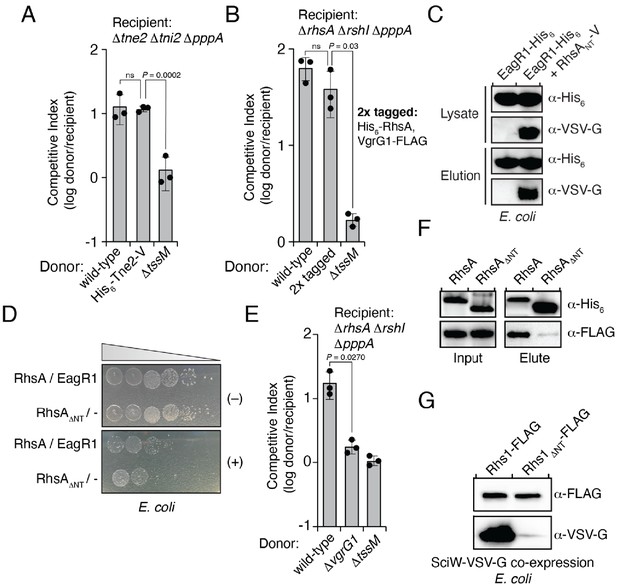
RhsA interacts with EagR1 and requires VgrG1 for delivery into target cells.
(A and B) Growth competition assays between the indicated P. protegens donor strains and either Tne2 (A) or RhsA (B) susceptible recipients. (C) Western blot of lysate and pull-down elution fractions of His6-tagged EagR1 co-expressed with an empty vector or RhsANT-VSV-G (residues 1–74) in E. coli. (D) Growth of E. coli co-expressing inducible plasmids harboring RhsA and EagR1 or RhsA∆NT with an empty vector. Overnight cultures were plated on media containing (+) or lacking (-) inducers and were imaged after 24 hr of growth. (E) Competition assay of the indicated P. protegens donor strains against a recipient susceptible to RhsA. (F) Affinity pull-down of His6-tagged RhsA or RhsA∆NT co-expressed with VgrG-FLAG and EagR1-VSV-G in E. coli. Samples were analysed by western blot using the indicated antibodies. (G) Western blot of affinity pull-down elution fractions of His6-tagged Rhs1 or Rhs1∆NT co-expressed with VSV-G tagged SciW. (A–B, E) Data are mean ± s.d. for n = 3 biological replicates; p values shown are from a two-tailed, unpaired t-test. (C–D, F–G) Data are representative of two independent experiments.
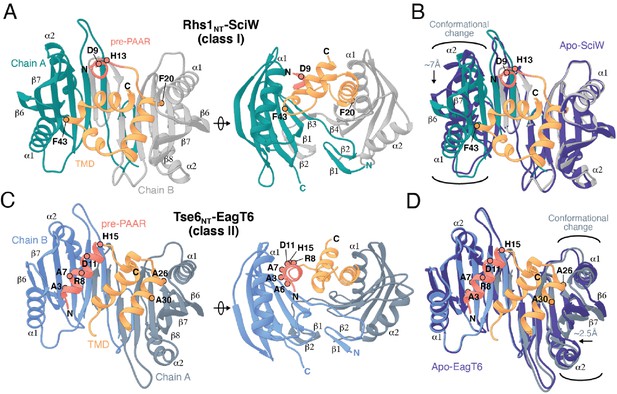
Co-crystal structures of the N-terminus of class I and class II prePAAR effectors in complex with their cognate Eag chaperones.
(A) An X-ray crystal structure of the Eag chaperone SciW bound to the N-terminus of Salmonella Typhimurium class I prePAAR effector Rhs1 (Rhs1NT, residues 8–57 are modeled) shown in two views related by a ~ 90° rotation. (B) Structural overlay of the apo-SciW structure with SciW-Rhs1NT complex demonstrates that a considerable conformational change in SciW occurs upon effector binding. (C) An X-ray crystal structure of the Eag chaperone EagT6 bound to the N-terminus of Tse6 (Tse6NT, residues 1–38 and 41–58 are modeled) shown in two views related by a ~ 90° rotation. (D) Structural overlay of the apo-EagT6 structure (PDB 1TU1) with the EagT6-Tse6NT complex shows a minor conformational change in EagT6 occurring upon effector binding. Eag chaperones are colored by chain, N-terminal transmembrane domains (TMDs) are colored in orange, the pre-PAAR motif in red, and apo chaperone structure in dark blue. Positions of residues of interest in the effector N-terminal regions are labeled.
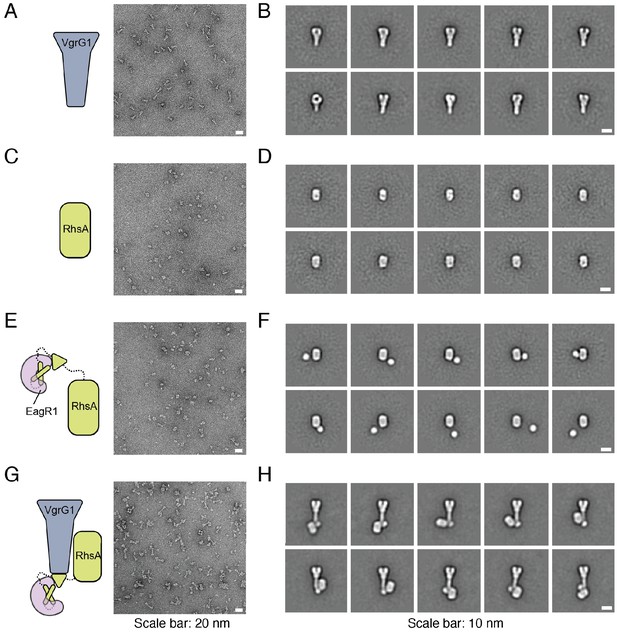
RhsA, EagR1, and VgrG1 form a ternary complex in vitro.
Unprocessed micrographs (A, C, E, G) and representative 2-D class averages (B, D, F, H) of negatively stained VgrG1 (A, B), RhsA∆NT (C, D), EagR1-RhsA complex (E, F) and EagR1-RhsA-VgrG1 complex (G, H). Scale bar represents 20 nm for unprocessed micrographs and 10 nm for class averages.
Eag chaperones interact with effector TMDs by mimicking interhelical interactions of alpha helical membrane proteins.
(A) Alignment of Eag chaperones that interact with class I (SciW, EagR1) or class II (EagT6 and EagT2) prePAAR effectors plotted with secondary structure elements. (B) Residues making intimate molecular contacts with their respective TMDs that are conserved among SciW, EagR1, EagT6, and EagT2 are shown. Hydrophobic contacts are colored in light orange and polar contacts in deep red. Residue numbers are based on EagT6. (C and D) The conserved hydrophobic molecular surface of the chaperones is shown in light orange (C) and their molecular surface residue conservation is shown as determined by the Consurf server (D) (Ashkenazy et al., 2016). Conserved residues making contacts with the TMDs in both co-crystal structures are shown. (E) Molecular contact map of Rhs1NT (residues 1–59) and SciW. prePAAR is shown in pink and the TMD regions in gold. Amino acids making contacts with the conserved residues of the Eag chaperones are shown by side chain/and or by main chain atoms (red for oxygen and blue for nitrogen). Residues in the Eag chaperone are highlighted by color of chain A or B. Polar (H-bond) contacts are drawn with a purple dashed line and are made with the side chain of the listed Eag residue. Outlined red circles indicate a water molecule. Light green circles indicate van der Waals interactions and hydrophobic interactions. The central group of hydrophobic residues without a listed chaperone residue all pack into the Eag hydrophobic face in Figure 4G (EagT6 I22/24 and V39). (F) Molecular contact map of Tse6NT (residues 1–61) and EagT6. Schematic is the same as panel B. Q102 in EagT6 corresponds to Q106 in SciW. (G) Structural alignment of SciW-Rhs1NT and EagT6-Tse6NT co-crystal structures using the structurally conserved TM helix as a reference. Eag chain coloring is the same as Figure 4. Rhs1NT is colored in dark blue with a brown prePAAR and Tse6NT in gold with a pink prePAAR. The conserved solvent accessible prePAAR residues D9/11 and H13/15 are shown in ball and stick model. Inset sequence alignment reflects the structurally aligned residues of Rhs1NT (top) and Tse6NT (bottom) as calculated by UCSF Chimera (Pettersen et al., 2004). Secondary structural elements are labeled. (H) Docking of the EagT6-TMD crystal structure from Figure 4C into the previously obtained cryo-EM density map of the EagT6-Tse6-EF-Tu-Tsi6-VgrG1a complex (EMD-0135). Cryo-EM density corresponding to EagT6 is depicted in transparent gray and Tse6-TMD and Tse6-PAAR in green; prePAAR residues are shown in pink. Note that Tse6-TMD was docked independent of EagT6 into the Tse6 density. One of three possible orientations for the PAAR domain is shown.
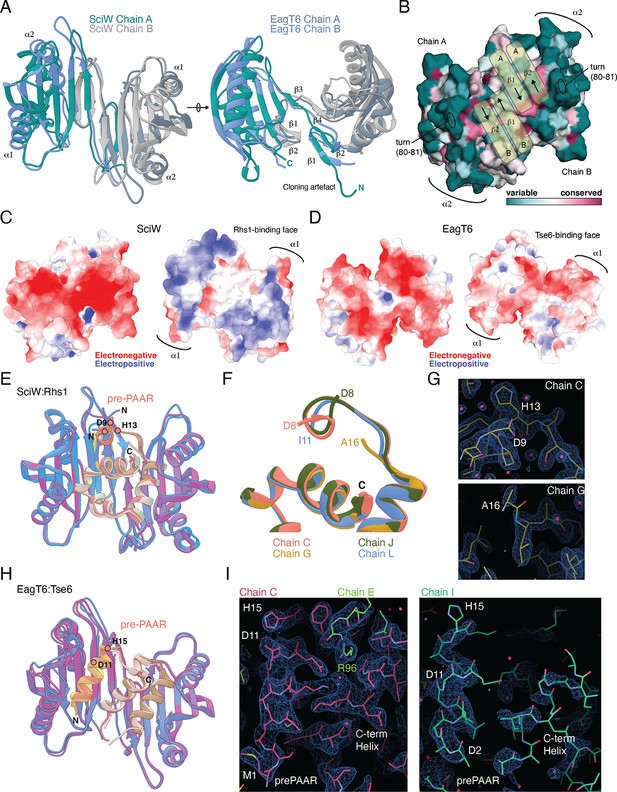
Structural comparison of Eag chaperones and effector complexes.
(A) Structural comparison of apo-SciW and apo-EagT6. Two views are shown related by an ~90° rotation. Each chaperone is colored by chain as in Figure 4. (B) Conserved surface residues as determined by the Consurf server. The view is a 180° rotation of panel A from Figure 4. The domain-swap created by the beta-strands from chain A and chain B are labeled and shown with yellow bar overlays. (C) Electrostatic surface potential of apo-SciW. The back (left, same surface as panel B) and Rhs1 binding surfaces (right) are shown. (D) Electrostatic surface potential of apo-SciW. The convex (left, same surface as panel B) and concave (Tse6 binding) surfaces (right) are shown. (E) Structural overlay of the four SciW-Rhs1NT complexes in the asymmetric unit of the crystal structure. The modeled prePAAR and C-terminus of Rhs1 are indicated and colored by chain. (F) View of the Rhs1 prePAAR region of each complex in the crystal structure. The N-terminal residue for each chain is listed. (G) Electron density maps of SciW-Rhs1NT Chain C and Chain G contoured at 1.4 rmsd (0.6816e/Å3). (H) Structural overlay of the three EagT6-Tse6NT complexes in the asymmetric unit of the crystal structure. The modeled prePAAR and C-terminus of Tse6 are indicated and colored by chain. (I) Electron density maps of EagT6-Tse6NT Chain C and Chain I contoured at 1.2 rmsd (0.0344e/Å3). The prePAAR and modelled C-terminal helix of the TMD region are labeled. A crystal packing artefact from Chain E including residue R96 that locks the prePAAR-TMD into place is shown. Electrostatic surface potentials were calculated by the adaptive-Poisson-Boltzmann server. Potentials are colored from −5 to 5 kT/e at pH 7.0. Images were created using UCSF Chimera, Coot, and Pymol.
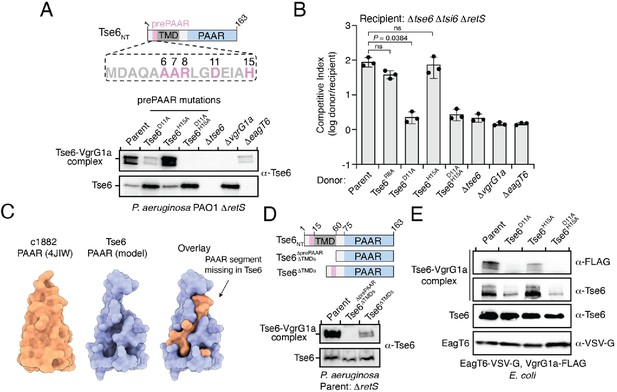
prePAAR is required for PAAR domain interaction with the VgrG spike.
(A) Western blot analysis of Tse6 from cell fractions of the indicated P. aeruginosa strains. Low-molecular-weight band indicates Tse6 alone whereas high-molecular-weight band indicates Tse6-VgrG1a complex. The parental strain contains a ∆retS deletion to transcriptionally activate the T6SS (Goodman et al., 2004). Schematic shows the N-terminal construct of Tse6 (Tse6NT), prePAAR is indicated in pink. (B) Outcome of growth competition assay between the indicated P. aeruginosa donor and recipient strains. The parent strain is P. aeruginosa ∆retS. Data are mean ± s.d. for n = 3 biological replicates; p value shown is from a two-tailed, unpaired t-test; ns indicates data that are not significantly different. (C) Structural comparison of the c1882 PAAR protein from E. coli (PDB: 4JIW) with a model of the PAAR domain of Tse6 generated using Phyre2 (Kelley et al., 2015). The overlay shows the additional N-terminal segment present in c1882 that is absent in Tse6. (D) Western blot of cell fractions of the indicated P. aeruginosa strains. The identity of the low-molecular weight and high-molecular weight bands are the same as described in A. The parent strain contains a retS deletion. Schematic shows mutants of Tse6 natively expressed in P. aeruginosa. Only the N-terminus of Tse6 is shown for simplicity. prePAAR is indicated in pink. Constructs lacking TMD1 also lack TMD2. (E) Western blot of elution samples from affinity pull-down with the indicated His6-tagged Tse6 variants co-purified with VgrG1a-FLAG and EagT6-VSV-G in E. coli. (A, D–E) Data are representative of two independent experiments.
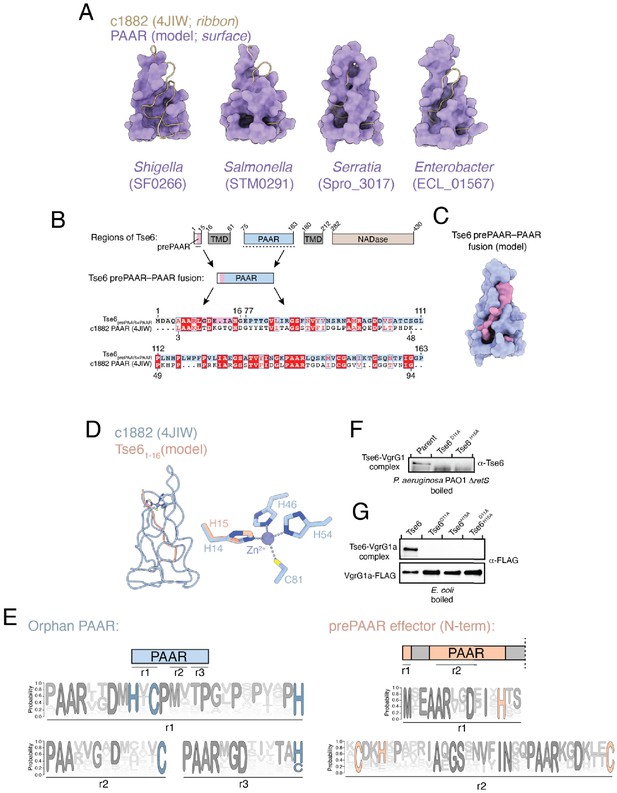
The PAAR domain of prePAAR effectors lacks a critical N-terminal segment.
(A) Surface representation of structural models of the PAAR domain from each of the indicated prePAAR effector proteins (purple) overlaid with a ribbon representation of the c1882 PAAR protein crystal structure (beige). Structural alignments were performed using ChimeraX. (B and C) Schematic showing the residue boundaries of the different regions of Tse6. The prePAAR (pink) and PAAR (blue) sequences were artificially fused to generate Tse6prePAAR+PAAR and used to generate an alignment with c1882 (B) and a structural model (C). Pink space-filling representation indicates the region of the model comprised of prePAAR. (D) Structural overlay of the prePAAR segment (peach) from the artificially fused Tse6prePAAR+PAAR sequence in Figure 6D with the entire c1882 PAAR protein (blue). The zoom shows the Zn2+-coordinating residues of c1882 and the overlap of H15 from Tse6’s prePAAR with H14 of c1882. (E) Sequence logos developed from multiple sequence alignments of 564 orphan PAAR sequences and the N-terminus of 1765 prePAAR-containing effectors. Sequence logos were developed for different regions (r1, r2, r3) in each construct that were contained for Zn2+-coordinating residues histidine and cysteine. (F) Same samples from Figure 6A, except samples were boiled before being subject to electrophoresis. (G) Same samples from 6D, except samples were boiled before being subject to electrophoresis.
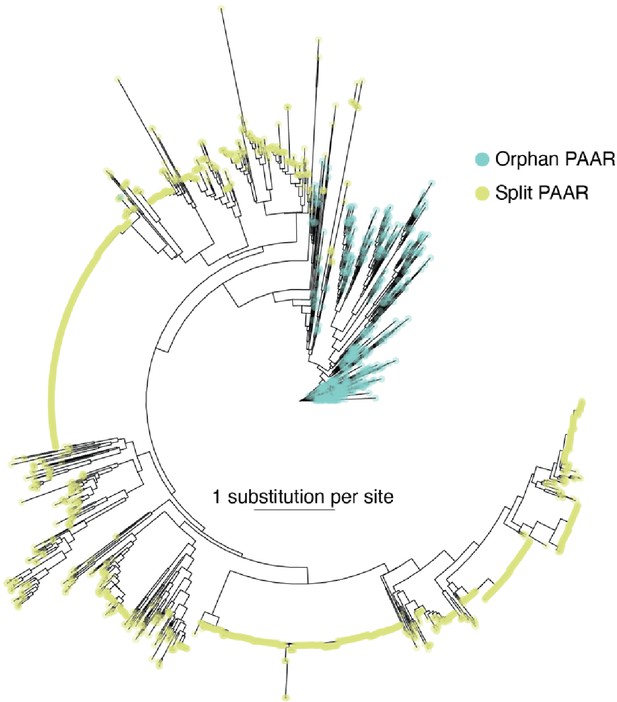
Orphan PAARs are ancestral to split PAARs.
Phylogenetic distribution of 564 orphan PAAR sequences (blue) and 1765 split PAAR (green) sequences. The scale bar indicates the substitutions per base.
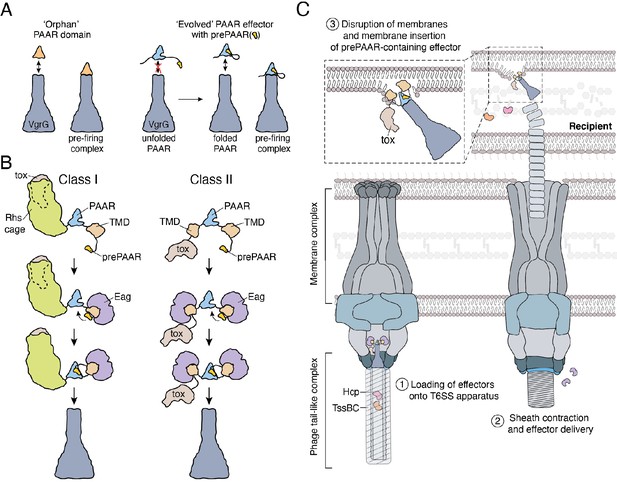
Model depicting the role of Eag chaperones and prePAAR in type VI secretion.
(A) PAAR proteins exist with or without prePAAR domains. Those that lack prePAAR (orphan), can interact with VgrG and form a functional T6SS spike complex without any additional factors. By contrast, prePAAR-containing effectors contain multiple domains (evolved) and likely require the prePAAR motif for proper folding of the PAAR domain and thus, loading onto the T6SS apparatus. (B) prePAAR-containing effectors can be divided into two classes: class I effectors have a single TMD and contain a C-terminal toxin domain that is likely housed within a Rhs cage whereas class II effectors contain two TMDs and do not possess a Rhs cage. TMD-chaperone and prePAAR-PAAR interactions are required for effector stability and VgrG interaction, respectively, for both classes of prePAAR effectors. (C) Depiction of a prePAAR-containing effector being exported by the T6SS into recipient cells. Inset shows the hydrophobic TMDs of a class II prePAAR effector disrupting the inner membrane of the target bacterium to allow entry of the effector toxin domain into the cytoplasm.
Tables
X-ray data collection and refinement statistics.
| SciW (native) | SciW (Iodide) | SciW-Rhs11-59 | EagT6-Tse61-61 | |
|---|---|---|---|---|
| Data Collection | ||||
| Wavelength (Å) | 1.5418 | 1.5418 | 0.97895 | 1.5418 |
| Space group | P212121 | P212121 | P3121 | P32 |
| Cell dimensions | ||||
| a, b, c (Å) | 55.27 75.1 76.6 | 55.6 75.3 76.4 | 105.3 105.3 248.4 | 68.9 68.9 173.1 |
| α, β, γ (°) | 90 90 90 | 90 90 90 | 90 90 120 | 90 90 120 |
| Resolution (Å) | 29.03–1.75 | 19.63–2.21 | 91.20–1.90 | 28.22–2.55 |
| (1.82–1.75) | (2.33–2.21) | (1.98–1.90) | (2.65–2.55) | |
| Unique reflections | 32309 (3162)* | 29933 (4888) | 126298 (12473) | 29267 (2832) |
| CC(1/2) | 99.8 (89.1) | 99.6 (81.4) | 99.9 (53.9) | 99.6 (52.8) |
| Rmerge (%)† | 6.2 (91.3) | 6.1 (44.7) | 5.7 (34.6) | 15.5 (179.8) |
| I/σI | 14.2 (1.9) | 8.0 (1.8) | 11.6 (1.26) | 7.27 (0.92) |
| Completeness (%) | 99.5 (98.8) | 96.0 (97.9) | 99.9 (99.9) | 99.3 (96.9) |
| Redundancy | 7.0 (6.8) | 2.0 (1.9) | 9.9 (9.7) | 4.9 (4.8) |
| Refinement | ||||
| Rwork/Rfree (%)‡ | 19.8/22.6 | 18.7/21.4 | 22.9/26.6 | |
| Average B-factors (Å2) | 46.1 | 42.9 | 71.7 | |
| Protein | 45.1 | 42.5 | 72.1 | |
| Ligands | 60.8 | 123.4 | ||
| Water | 53.9 | 42.2 | 59.3 | |
| No. atoms | ||||
| Protein | 2331 | 10492 | 7827 | |
| Ligands | 10 | 60 | ||
| Water | 256 | 1119 | 248 | |
| Rms deviations | ||||
| Bond lengths (Å) | 0.003 | 0.005 | 0.004 | |
| Bond angles (°) | 0.67 | 0.68 | 0.73 | |
| Ramachandran plot (%)§ | ||||
| Total favored | 99.65 | 99.24 | 98.26 | |
| Total allowed | 0.35 | 0.68 | 1.74 | |
| PDB code | 6XRB | 6XRR | 6XRF |
-
*Values in parentheses correspond to the highest resolution shell.
†Rmerge = Σ Σ |I(k) - < I > |/ Σ I(k) where I(k) and <I > represent the diffraction intensity values of the individual measurements and the corresponding mean values. The summation is over all unique measurements.
-
‡Rwork = Σ ||Fobs| - k|Fcalc||/|Fobs| where Fobs and Fcalc are the observed and calculated structure factors, respectively. Rfree is the sum extended over a subset of reflections excluded from all stages of the refinement.
§As calculated using MOLPROBITY (Chen et al., 2010).
Additional files
-
Supplementary file 1
List of prePAAR motif-containing proteins identified in the UniProtKB Database .
The document contains two separate sheets. List A corresponds to 2054 prePAAR-containing sequences that were identified through an iterative search of the UniprotKB using Tse6NT. List B corresponds to 975 sequences collected following filtering of list A (see Materials and Methods for details).
- https://cdn.elifesciences.org/articles/62816/elife-62816-supp1-v3.xlsx
-
Supplementary file 2
List of prePAAR motif-containing proteins from assembled genomes of all species belonging to the genera Burkholderia, Escherichia, Enterobacter, Pseudomonas, Salmonella, Serratia, Shigella, and Yersinia.
The document contains two separate sheets. List C corresponds to 6101 prePAAR-containing sequences that were identified through an iterative search of the UniprotKB using Tse6NT. List D corresponds to 1166 sequences collected following filtering of list C (see Materials and methods for details).
- https://cdn.elifesciences.org/articles/62816/elife-62816-supp2-v3.xlsx
-
Supplementary file 3
Summary of the number of genomes and effector sequences used in our informatics analyses.
This document contains three separate sheets. The ‘UniprotKB-effectors’ sheet shows the quantity of initial prePAAR-containing sequences that were identified in our search and the number of sequences that were used following filtering and removal of redundancy (plotted in the cladogram in Figure 1—figure supplement 1A). The numbers in bold indicate the number of sequences in Supplementary file 1. The ‘eight genera - genomes’ sheet corresponds to the number of genomes from the eight genera (Burkholderia, Escherichia, Enterobacter, Pseudomonas, Salmonella, Serratia, Shigella, and Yersinia) that contained one prePAAR-containing sequence and the number that remained following filtering and removal of redundancy. The ‘8-genera – effectors’ sheet corresponds to initial and final numbers of prePAAR-containing sequences that were identified in the eight genera listed above. The final number of sequences in this sheet were used to construct the cladogram in Figure 1E. The numbers in bold indicate the numbers of sequences in the lists in Supplementary file 2.
- https://cdn.elifesciences.org/articles/62816/elife-62816-supp3-v3.xlsx
-
Supplementary file 4
Strains used in this study.
- https://cdn.elifesciences.org/articles/62816/elife-62816-supp4-v3.docx
-
Supplementary file 5
Plasmids used in this study.
- https://cdn.elifesciences.org/articles/62816/elife-62816-supp5-v3.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/62816/elife-62816-transrepform-v3.pdf