An interbacterial DNA deaminase toxin directly mutagenizes surviving target populations
Figures
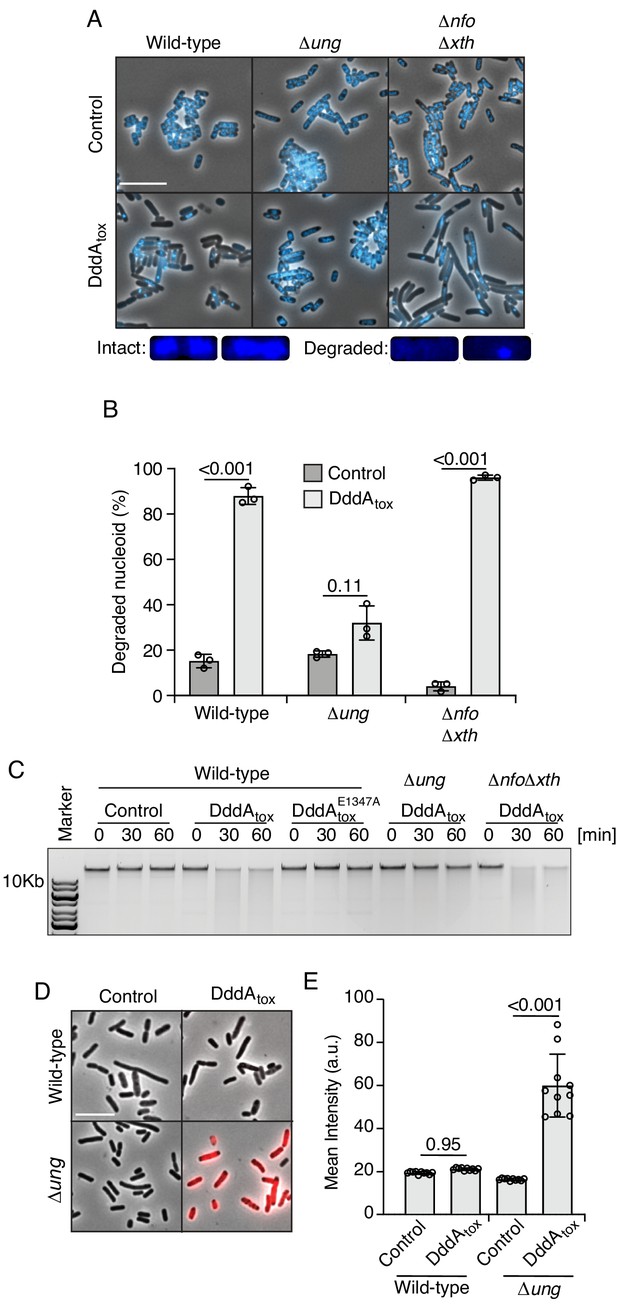
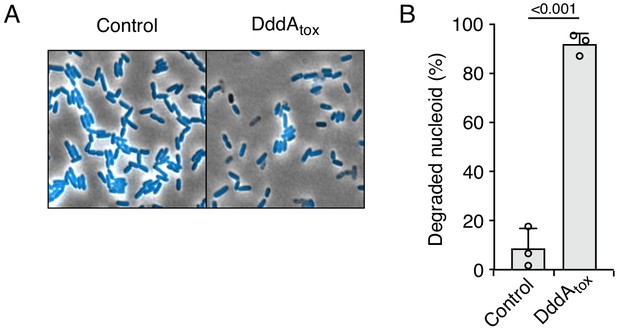
Double-stranded DNA deaminase A (DddA) expression leads to nucleoid degradation in E. coli wild-type cells and uracil accumulation in E. coli Δung.
(A) Fluorescence microscopy of the indicated E. coli strains expressing DddAtox or carrying an empty vector (Control). DAPI staining (DNA) is shown in cyan. Top: Representative micrographs for each condition, scale bar = 10 μm. Bottom: representative images of cells with intact or degraded nucleoids. (B) Quantification of nucleoid state in cells shown in A (n ≈100–200 cells per condition). (C) Agarose gel electrophoresis analysis of total genomic DNA isolated from the indicated E. coli strains expressing DddAtox, DddAtoxE1347A, or carrying an empty vector (Control) after induction for the time period shown. (D) Fluorescence microscopy indicating genomic uracil incorporation (red) of E. coli strains expressing DddAtox or carrying an empty vector (Control), scale bar = 10 μm. (E) Quantification of uracil labeling signal from cells shown in D (n ≈50 cells per condition). Values and error bars reflect mean ± s.d. of n = 2 independent biological replicates. p-Values derive from unpaired two-tailed t-test.
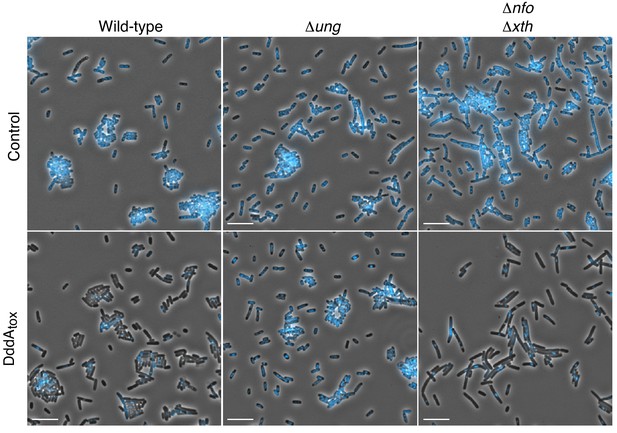
Expression of DddAtox leads to nucleoid degradation in E. coli.
Full field view of fluorescence micrograph shown in Figure 1 (A) depicting E. coli strains expressing DddAtox or carrying an empty vector (Control). DAPI staining (DNA) is shown in cyan. Scale bar = 10 µM.
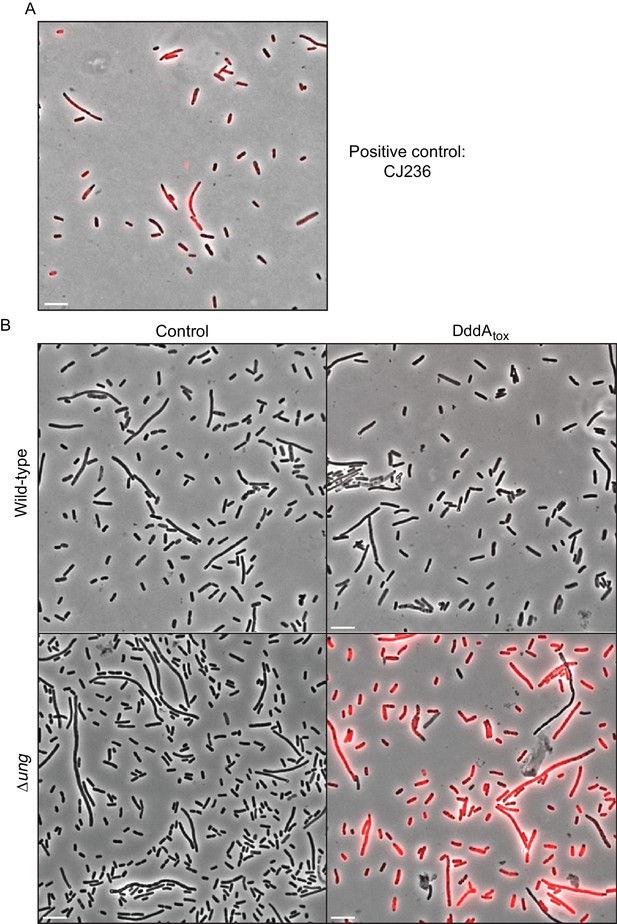
DddAtox induction leads to genomic uracil accumulation in E. coli.
(A,B) Fluorescence microscopy indicating genomic uracil incorporation (red) in the noted E. coli strains. Scale bar = 10 µM. (A) Staining of a positive control strain for uracil incorporation, E. coli CJ236 [dut−, ung−]. This strain accumulates ~500 times more uracil than wild type. (B) Complete fields of view used to generate fluorescence micrographs depicted in Figure 1D.
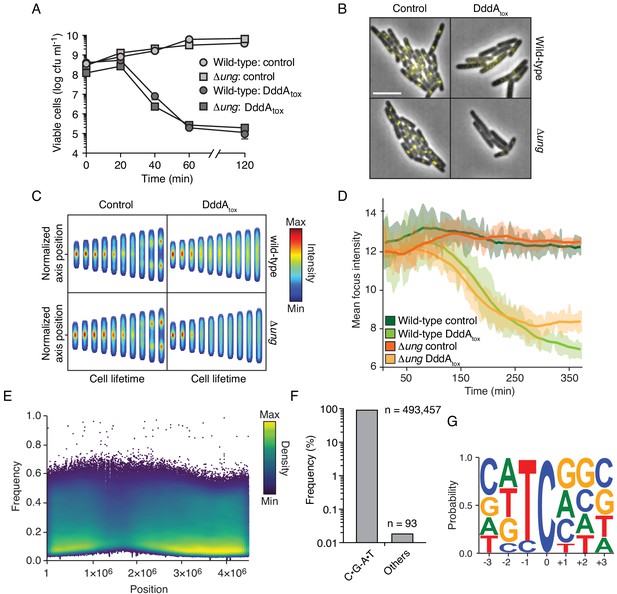
Intoxication by DddA leads to DNA replication arrest and widespread uracil incorporation across the genome.
(A) Viable cells (colony-forming units, cfu) of the indicated E. coli strains recovered following induction of DddAtox or empty vector (Control) for the time period shown. Values represent mean ± s.d. of n = 3 technical replicate and data are representative of three independent experiments. (B) Representative images from time-lapse microscopy of DnaN-YPet-expressing strains 6 hr post-induction of DddAtox, scale bar = 5 μm. (C) Cell tower representation of averaged localized fluorescence intensity of DnaN-YPet-expressing strains shown in B over the course of cell lifetimes. (n ≈ 20–300 cells per condition at start of experiment). (D) Mean focus intensity of each frame during time-lapse microscopy of DnaN-YPet-expressing strains over the course of 6 hr (n ≈ 20–300 cells per condition at start of experiment). (E) Representation of single-nucleotide variants (SNVs) by chromosomal position, frequency, and density in E. coli Δung following 60 min expression of DddAtox. (F) Frequency of the indicated substitutions among the SNVs shown in E. (G) Probability sequence logo of the region flanking mutated cytosines among the SNVs shown in (E).
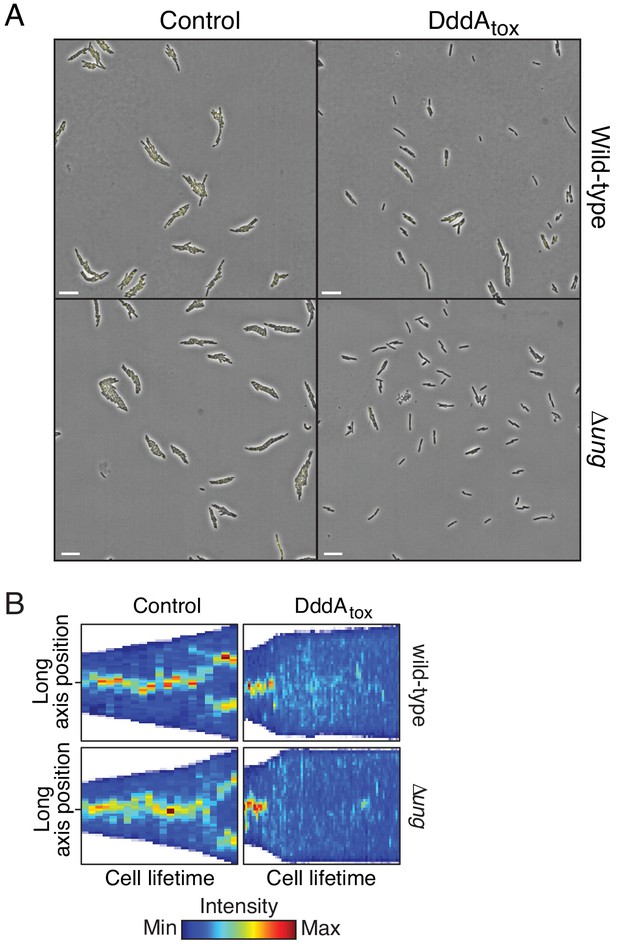
Expression of DddAtox in E. coli leads to replication arrest but does not mutagenize RNA.
(A) Full field of view of fluorescence micrographs shown in Figure 2B. Scale bar = 10 µM. (B) Kymograph of representative cells from time-lapse microscopy of DnaN-YPet expressing strains during a 6 hr timelapse post-induction of DddAtox or empty vector (Control), scale bar = 5 μm.
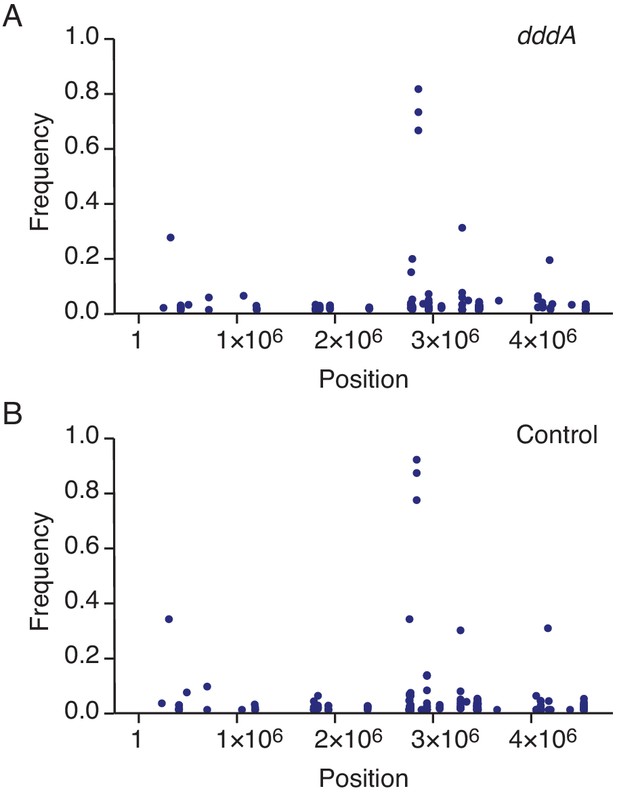
Expression of DddAtox does not mutagenize RNA.
Representation by chromosomal position, frequency, and density of single-nucleotide variants (SNVs) detected with RNA-seq analysis of total cDNA collected from E. coli 60 min after induction of DddAtox (A) or the empty vector (Control) (B). The high-frequency SNVs detected near position 3 × 106 are the result of reads from highly expressed genes mapping incorrectly to homologous but not identical regions of the genome.
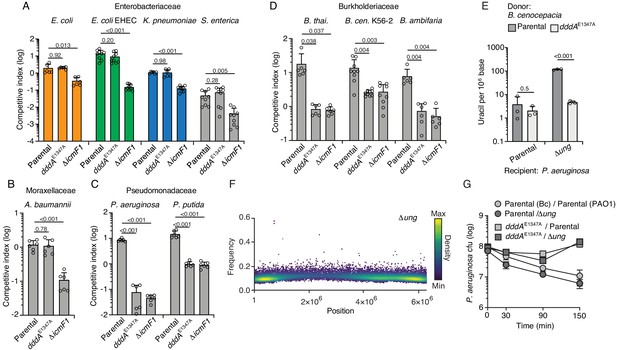
Double-stranded DNA deaminase A (DddA) delivery via the T6SS during interbacterial competition results in varying outcomes among different recipient species.
(A–D) Competitiveness of the B. cenocepcia strains indicated at bottom against selected Enterobacteriaceae (colors relate to Figure 4) (A), Moraxellaceae (B), Pseudomonadaceae (C), or Burkholderiaceae (D). Pairs of organisms were cocultured on a solid surface for 6 hr. (E) Mass spectrometric quantification of uracil in genomic DNA obtained from 1 hr co-cultures of the indicated strains of P. aeruginosa with B. cenocepacia wild-type or dddAE1347A. Values and error bars reflect mean ± s.d. of n = 3 independent biological replicates. p-Values derive from unpaired two-tailed t-test. (F) Single-nucleotide variants (SNVs) detected in populations of P. aeruginosa Δung after 1 hr growth in competition with B. cenocepacia. SNVs are plotted according to their chromosomal position and frequency and colored according to their relative density. (G) Cell viability of the indicated P. aeruginosa strains after 1 hr growth in competition with B. cenocepacia wild-type or dddAE1347A. Values and error bars reflect mean ± s.d. of n = 2 independent biological replicates.
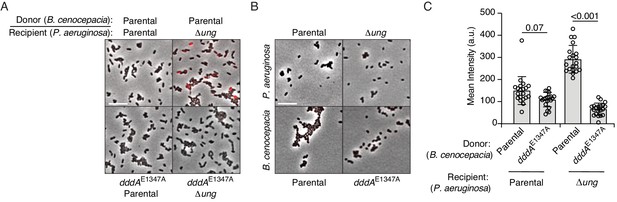
P. aeruginosa accumulates genomic uracil during competition with B. cenocepacia.
(A–B) Fluorescence microscopy indicating uracil incorporation (red) into genomic DNA of cells recovered after one hour of growth from cocultures of P. aeruginosa wild-type or Δung cells and the indicated strains of B. cenocepacia (A) or each of the strains used in (A) grown in monoculture (B). (C) Quantification of uracil labeling signal from cells shown in A (n = ~50 cells per condition). Values and error bars reflect mean ± s.d. of n = 2 independent biological replicates. p-Values derive from unpaired two-tailed t-test. (D) Mutation frequency as measured by spontaneous rifampicin resistance frequency in P. aeruginosa Δung recovered after 1 hr growth in competition with the indicated strain of B. cenocepacia. Values and error bars reflect mean ± s.d. of n = 2 independent biological replicates with n = 3 technical replicates each. p-Values derive from unpaired two-tailed t-test.
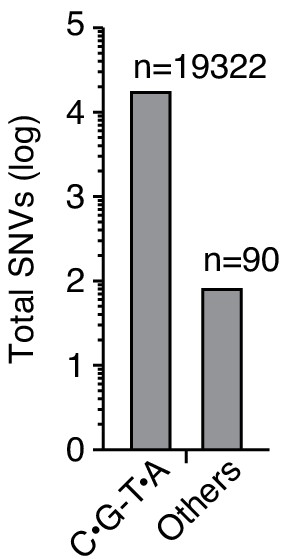
Delivery of double-stranded DNA deaminase A (DddA) to P. aeruginosa Δung during interbacteria competition with B. cenocepacia results in the accumulation of C•G-to-T•A transitions.
Quantification of total single-nucleotide variants (SNVs) detected in P. aeruginosa Δung after intoxication by B. cenocepacia, as shown in Figure 3F.
Expression of DddAtox in P. aeruginosa results in nucleoid degradation.
(A) Representative micrographs of P. aeruginosa expressing DddAtox or carrying an empty vector (Control). DAPI staining (DNA) is shown in cyan, scale bar = 10 μm. (B) Quantification of nucleoid state in cells shown in A (n = ~100–200 cells per condition). Values and error bars reflect mean ± s.d. of n = 3 independent biological replicates. p-Values derive from unpaired two-tailed t-test.
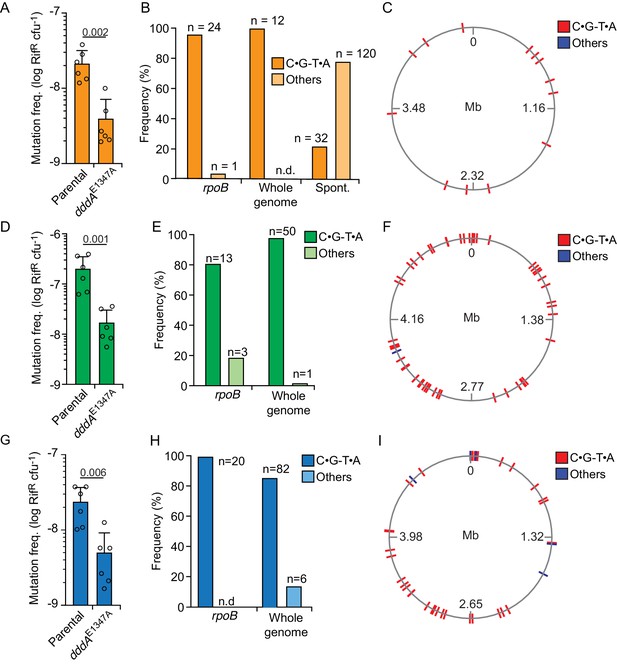
Delivery of double-stranded DNA deaminase A (DddA) by B. cenocepacia induces mutagenesis in a subset of resistant recipient species.
(A, D, and G) Mutation frequency as measured by spontaneous rifampicin resistance frequency in clones of different species recovered from growth in competition with B. cenocepacia wild-type or B. cenocepacia dddAE1347A (n = 2). (B, E, and H). Distribution of different mutation types detected in rpoB or the whole genome of rifampicin-resistant clones of different species recovered after growth in competition with wild-type B. cenocepacia. (B) Pattern of spontaneous mutation types observed in rpoB of E.coli derived from Garibyan et al., 2003. (C, F, and I). Genome distribution of different mutation types detected by whole-genome sequencing of rifampicin-resistant clones recovered after competition with B. cenocepacia. Species targeted include E. coli (A–C), E. coli EHEC (D–F), and K. pneumoniae (G–I). Colors relate to Figure 3. Values and error bars reflect mean ± s.d. of n = 2 independent biological replicates with n = 3 technical replicates each. p-values derive from unpaired two-tailed t-test. Spont. Spontaneous.
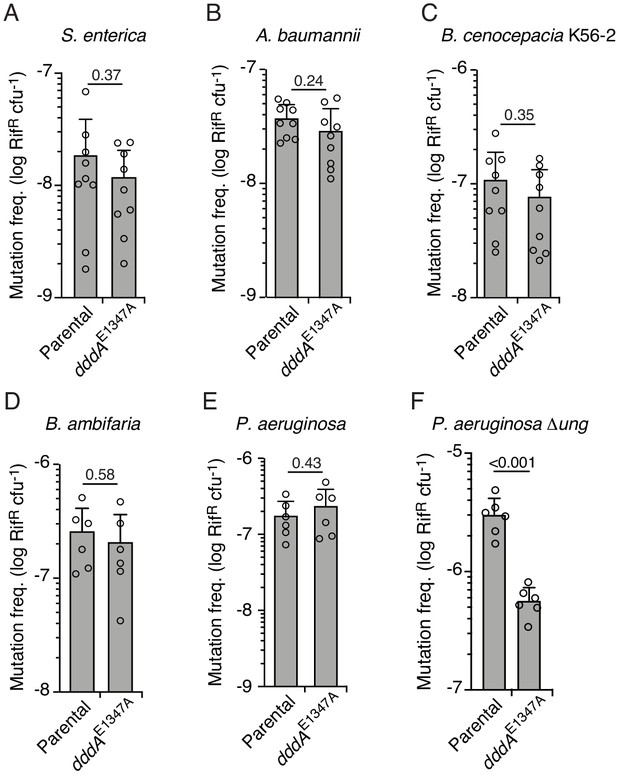
Double-stranded DNA deaminase A (DddA)-mediated intoxication is not mutagenic during competition between B. cenocepacia and certain recipient species.
(A–F) Mutation frequency as measured by spontaneous rifampicin resistance frequency in clones of different species recovered from growth in competition with B. cenocepacia wild-type or B. cenocepacia dddAE1347A (n = 2). Values and error bars reflect mean ± s.d. of n = 2 independent biological replicates with n = 3 technical replicates each. p-values derive from unpaired two-tailed t-test.
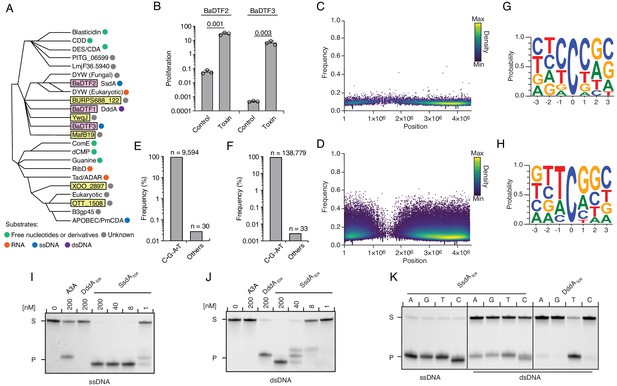
Predicted deaminase toxins from BaDTF2 and BaDTF3 clades exhibit mutagenic activity and a BaDTF2 representative targets ssDNA.
(A) Dendogram indicated evolutionary history of clades within the deaminase superfamily their predicted substrates (colored dots), modified from Iyer et al., 2011. Predicted toxins with unknown substrates, yellow boxes; deaminases toxins with defined biochemical activity, pink boxes. (B) Toxicity of representative BaDTF2 and BaDTF3 toxins as indicated by the proliferation (fold change in colony-forming unit [cfu] recovered) of E. coli after 1 hr expressing the toxins or the empty vector (Control). Values represent means ± s.d., and p-values derive from unpaired two-tailed t-test. (C and D) Representation of single-nucleotide variants (SNVs) by chromosomal position, frequency, and density in E. coli Δung after 1 hr expression of representative BaDTF2 (C) or BaDTF3 (D) toxins. (E and F). Distribution of different nucleotides substitutions among SNVs detected in E. coli Δung expressing representative BaDTF2 (E) or BaDTF3 (F) toxins. (G and H) Probability sequence logo of the region flanking mutated cytosines from E. coli Δung intoxicated with representative BaDTF2 (G) or BaDTF3 (H) toxins. (I and J). In vitro cytidine deamination assays for BaDTF2 toxin single-strand DNA deaminase toxin A (SsdA) using a single-stranded (I) or double-stranded (J) FAM-labeled DNA substrate 'S' with cytidines in the contexts CC, TC, AC, and GC. Cytidine deamination leads to products 'P' with increased mobility. A3A, APOBEC3A (control for activity on ssDNA). DddAtox was used as a control for activity toward dsDNA. K. In vitro cytidine deamination assays for SsdAtox or DddAtox using a single-stranded or double-stranded FAM-labeled DNA substrate with a single cytidine in the context indicated at top. Gels shown in I-K are representative of two replicates. Data in B represent means ± s.d., p-values derive from unpaired two-tailed t-test (n = 3).
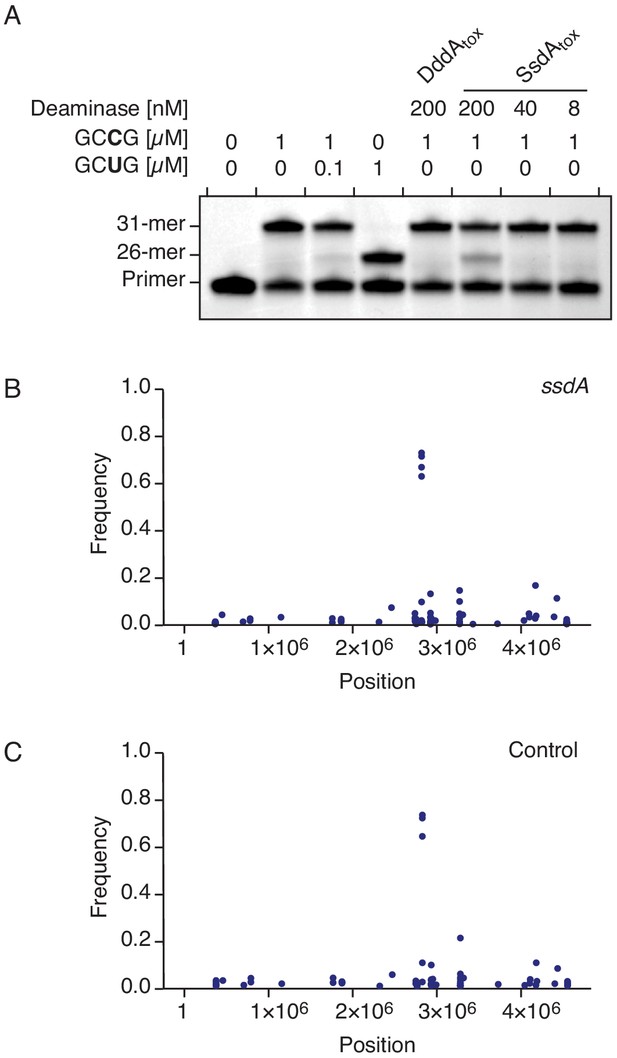
SsdAtox exhibits cytidine deaminase activity toward RNA in vitro, but not in vivo.
(A) Poisoned primer extension assay to detect deamination of cytidine in single-stranded RNA substrates. Deamination activity generates a 26-mer product. Image is representative of n = 2 independent replicates. (B–C) Representation of single-nucleotide variants (SNVs) by chromosomal position, frequency, and density detected with RNA-seq analysis of total cDNA collected from E. coli 60 min after induction of SsdAtox (B) or an empty vector (Control) (C). The high-frequency SNVs detected near position 3 × 106 are the result of reads from highly expressed genes mapping incorrectly to homologous but not identical regions of the genome.
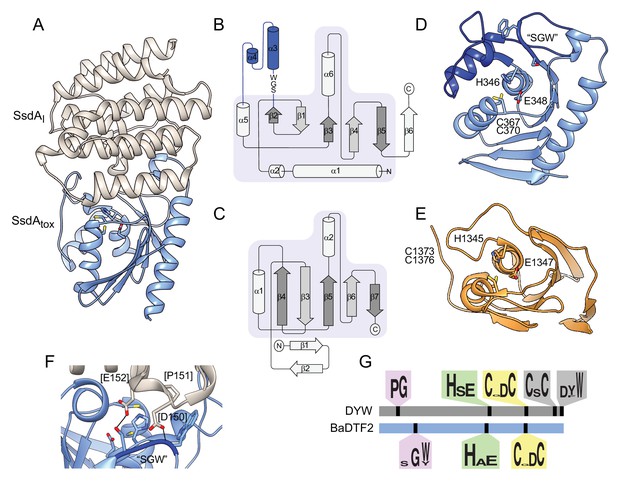
The structure of BaDF2 member single-strand DNA deaminase toxin A (SsdA) bears little resemblance to double-stranded DNA deaminase A (DddA) and reveals motifs differentiating toxins from RNA-targeting DYW proteins.
(A) Ribbon diagram depiction of the SsdAtox-SsdAI structure. (B,C) Secondary structure diagrams for SsdAtox and the core fold of deaminase superfamily proteins (Iyer et al., 2011). The SWG motif conserved in BaDTF2 toxins and the accompanying α-helical insertion (blue) are indicated in B. (D) Active site view of SsdAtox. Catalytic and zinc-coordinating residues, SGW motif and α-helical insertion (blue) are indicated. (E) Active site view of DddAtox indicating catalytic and zinc-coordinating residues. (F) Zoom-in view of the contact site between SsdAI and the active site of SsdAtox. (G) Conserved motifs identified in DYW and BaDTF2 proteins.
Videos
Double-stranded DNA deaminase A (DddA) inhibits E. coli growth.
Time lapse phase microscopy of E. coli expressing DddA during a 300-min period.
Double-stranded DNA deaminase A (DddA) arrests replication in E. coli growth.
Time lapse fluorescence microscopy of E. coli with YPet-labeled DnaN-expressing DddA during a 300-min period.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Escherichia coli) | DH5α | Thermo Fisher Scientific Cat#18258012 | F–φ80lacZΔM15 Δ (lacZYA-argF)U169 recA1 endA1 hsdR17(rK–, mK+) phoAsupE44 λ–thi-1 gyrA96relA1 | Used for cloning |
| Strain, strain background (Escherichia coli) | BL21 (DE3) | EMD Millipore Cat#69450 | F–ompT hsdSB(rB–, mB–)gal dcm (DE3) | Used for protein expression |
| Strain, strain background (Escherichia coli) | BL21 ung-151 | PMID:15096615 | BDSC:31777; FLYB:FBtp0001612; RRID:BDSC_31777 | Used for protein expression |
| Strain, strain background (Escherichia coli) | XK1502 | PMID:16923902 | F– ompT hsdSB (rB–, mB–) gal dcm ung-151 | Used for protein expression |
| Strain, strain background (Escherichia coli) | XK1502 Δung | PMID:32641830 | F− ΔlacU169 nalA Δung | Used for protein expression |
| Strain, strain background (Escherichia coli) | AB1157 | PMID:17248104 | F- thr-1 leuB6(Am) glnX44(AS) hisG4(Oc) rfbC1 rpsL31(strR) argE3(Oc) | |
| Strain, strain background (Escherichia coli) | AB1157 γpet-dnaN | PMID:28114307 | γpet-dnaN, KanR | |
| Strain, strain background (Escherichia coli) | CJ236 | PMID:24723723 | F+ung-1 relA1 dut-1 spoT1 thiE1 | |
| Strain, strain background (Burkholderia cenocepacia) | H111 | PMID:10713433 | Wild-type | |
| Strain, strain background (Burkholderia cenocepacia) | H111 ΔI35_RS01770 | PMID:32641830 | ΔI35_RS01770 | |
| Strain, strain background (Burkholderia cenocepacia) | H111 dddAE1347A | PMID:32641830 | dddAE1347A | |
| Strain, strain background (Pseudomonas aeruginosa) | PAO1 | PMID:10984043 | Wild-type | |
| Strain, strain background (Klebsiella pneumoniae) | MGH 78578 | PMID:11677609 | Wild-type | |
| Strain, strain background (Escherichia coli) | O157:H7 EDL933 | PMID:11206551 | ||
| Strain, strain background (Acinetobacter baumannii) | ATCC 17978 | PMID:18931120 | Wild-type | |
| Strain, strain background (Burkholderia thailandensis) | E264 | PMID:16336651 | Wild-type | |
| Strain, strain background (Burkholderia thailandensis) | F1 | PMID:23618999 | Wild-type | |
| Strain, strain background (Burkholderia cenocepacia) | K56-2 | PMID:29208119 | Wild-type | |
| Strain, strain background (Salmonella enterica) | Sv. Typhimurium 14028 s | PMID:19897643 | Wild-type | |
| Recombinant DNA reagent | pPSV39-CV (plasmid) | PMID:23954347 | For inducible expression of proteins in E. coli | |
| Recombinant DNA reagent | pScrhaB2-V (plasmid) | PMID:15925406 | For inducible expression of proteins in E. coli | |
| Recombinant DNA reagent | pEXG2 (plasmid) | PMID:15911752 | For generation of markless P. aeruginosa mutants | |
| Recombinant DNA reagent | pScrhaB2-V::ssdA (plasmid) | This study | To express ssdA | |
| Recombinant DNA reagent | pPSV39-CV::ssdAI (plasmid) | This study | To express ssdAI | |
| Recombinant DNA reagent | pScrhaB2-V::TequE (plasmid) | This study | To express BadTF3 | |
| Recombinant DNA reagent | pPSV39-CV::TequE (plasmid) | This study | To express BadTF3-Imm | |
| Recombinant DNA reagent | pETDuet-1 mcs1::ssdA-his6 mcs1::ssdAI (plasmid) | This study | To express ssdA-ssdAI | |
| Recombinant DNA reagent | pexG2_Δung (plasmid) | This study | To delete ung in Pseudomonas aeruginosa | |
| Sequence-based reagent | ungDel-1 | This study | 5’-CAAGCTTCTGCAGGTCGACTCTAGAGGTATGGAGTTGTCCTTCGG | |
| Sequence-based reagent | ungDel-2 | This study | 5-AGAGGTCCGGATCGGTCATGGAACCCCC | |
| Sequence-based reagent | ungDel-3 | This study | 5’-CATGACCGATCCGGACCTCTGAAGGCCGC | |
| Sequence-based reagent | ungDel-4 | This study | GGAAATTAATTAAGGTACCGAATTCCCGCGCCGGTGGACTGGC | |
| Sequence-based reagent | PAO1-ung-F | This study | 5’-CCGGGGAGTACTTCTCGTTC | |
| Sequence-based reagent | PAO1-ung-R | This study | 5’-GGCGTTCCAGTACCTGCTC | |
| Sequence-based reagent | GA_duet_PsyrE1-F | This study | 5’-ACCATCATCACCACAGCCAGGATCCGAAGGTCTCAAATATTGCG | |
| Sequence-based reagent | GA_duet_PsyrE1-R | This study | 5’-CTTAAGCATTATGCGGCCGCTCATTCCGACCTCATAATTG | |
| Sequence-based reagent | GA_duet_PsyrI1-F | This study | 5’-TATAAGAAGGAGATATACATATGAATAACAAAAGTAAAGTATTGATTGAAAAGC | |
| Sequence-based reagent | GA_duet_PsyrI1-R | This study | 5’-GCCGGCCGATATCCAATTGAGATCTTCACACAACTTGCGGCAC | |
| Sequence-based reagent | GA_pRhB_PsyrE1-F | This study | 5’-TGAAATTCAGCAGGATCACATATGAAGGTCTCAAATATTGCG | |
| Sequence-based reagent | GA_pRhB_PsyrE1-R | This study | 5’-TCATTTCAATATCTGTATATCTAGATTCCGACCTCATAATTGTTTC | |
| Sequence-based reagent | GA_p39_PsyrI1-F | This study | 5’-ACAATTTCAGAATTCGAGCTCACGGGAGGAAAGATGAATAACAAAAGTAAAGTATTGATTGAAAAGC | |
| Sequence-based reagent | GA_p39_PsyrI1-R | This study | 5’-TCATTTCAATATCTGTATATCTAGATCACACAACTTGCGGCAC | |
| Sequence-based reagent | rpoB-F | This study | 5’-GGAAAACCAGTTCCGCGTTG | |
| Sequence-based reagent | rpoB-R | This study | 5’-TCCAAGTTGGAGTTCGCCTG | |
| Sequence-based reagent | nfo_del-F | This study | 5’-CATTACCGTTTTCCTCCAGCGGGTTTAACAGGAGTCCTCGCATGAAATACGTGTAGGCTGGAGCTGCTTC | |
| Sequence-based reagent | nfo_del-R | This study | 5’-CCGTAAAATTGCAAGGATCTCCTTTTCCCGGTTATTCATCTTCAGGCTACCATATGAATATCCTCCTTAG | |
| Sequence-based reagent | nfo_dt-F | This study | 5’-GCTGATGGCACTGGTACTGT | |
| Sequence-based reagent | nfo_dt-R | This study | 5’-CCTTTAATCCGGCCTTTGCG | |
| Sequence-based reagent | xthA_del-F | This study | 5’-TACCATCCACGCACTCTTTATCTGAATAAATGGCAGCGACTATGAAATTTGTGTAGGCTGGAGCTGCTTC | |
| Sequence-based reagent | xthA_del-R | This study | 5’-TTAATTCTCCTGACCCAGTTTGAGCCAGGAGAGCTGCTAAATTAGCGGCGCATATGAATATCCTCCTTAG | |
| Sequence-based reagent | xthA_dt-F | This study | 5’-TACGTTTGCGATGTGGGTGA | |
| Sequence-based reagent | xthA_dt-R | This study | 5’-ATAACAAAGGACGGCAGGCA | |
| Sequence-based reagent | EL142_RS06975[tox]-gBlock | This study | 5’-TGAAATTCAGCAGGATCACATATGCTTTTAGGTGGACTTAACAACTACCAATACGCCCCAAATCCAGTCGAATGGGTCGATCCTTTGGGTTGGAAATTCTCCAATGGCAAGCGTCGTCCGCCCCACAAGGCAACGGTTACCGTCACAGACAAGAACGGAGTGGTCAAACACAAATCCAATTTGGTGTCAGGAAATATGACAGAAGCCGAAAAAAAACTGGGTTTCCCGAACAACTCTTTGGCAACACATACCGAGAATCGTGCAACGCGCTTAATTGACCTGAATCAAGGTGATACCATGTTAATTGAAGGACAGTATCGCCCGTGCCCACGCTGTAAGGGTGCGATGCGTGTTAAGGCAGAGGAATCTGGGGCTAAGGTTATCTACACCTGGCCCGAAGACGGTGACTTGAAGAAGCGCGAGTGGGAAGGAACCCCCTGTGATAAAAAGTCTAGATATACAGATATTGAAATGA | |
| Sequence-based reagent | EL142_RS06970 -gBlock | This study | 5’-GCCCCAAGGGGTTATGCTAAAGCTTTCTTAATTATCGAACAGGAATTTCAGATGATTCTCGACTTCAAGAATGTCAATCTTTTCAATCAAGCGAAGACGAACGTAGTTATTAGGCTCAATTAAAAGATCGACACAATTTGGTTCGGACAGTAAATAGCCGGACAAGCCATAGGGGTTAAAATCAAGGAACAGATTAGACTCCAGATAGATGTCTCCCTCGGGGATACCACCGCTGCGTTCCGTTGTTGAACGTGTAATGATGAAATACTGCTCAGGCACTTCATAGCTATCAGATGCAAACGAAAGGATGGCGCAATAATCCTCAATCGCAAACTTCACTTCTTGAATCACGATGTTACTCAGCATCTTTCCTCCCGTGAGCTCGAATTCTGAAATTGT | |
| Sequence-based reagent | RNA-GGCCGG | This study | 5’-GAGGCCGGAAGUGGAUGUGGAUAAGAUGGAG | |
| Sequence-based reagent | RNA-GGCUGG | This study | 5’-GAGGCUGGAAGUGGAUGUGGAUAAGAUGGAG | |
| Sequence-based reagent | PPE-Oligonucleotide | This study | 5’FAM-CTCCATCTTATCCACATCCACT | |
| Sequence-based reagent | DNA-ATGCGCCA | This study | 5’FAM-AAAAAAAAAAAAAA ATGCGCCAAAAAAAAAAAAAAA | |
| Sequence-based reagent | revDNA-ATGCGCCA | This study | 5’-TTTTTTTTTTTTTTTGGCGCATTTTTTTTTTTTTTT | |
| Sequence-based reagent | DNA-GCG | This study | 5’-FAM-AAAAAAAAAAAAAAAA GCGAAAAAAAAAAAAAAAAA | |
| Sequence-based reagent | DNA-CCG | This study | 5’-FAM-AAAAAAAAAAAAAAAA CCGAAAAAAAAAAAAAAAAA | |
| Sequence-based reagent | DNA-TCG | This study | 5’-FAM-AAAAAAAAAAAAAAAA TCGAAAAAAAAAAAAAAAAA | |
| Sequence-based reagent | DNA-ACG | This study | 5’-FAM-AAAAAAAAAAAAAAAA ACGAAAAAAAAAAAAAAAAA | |
| Sequence-based reagent | revDNA-GCG | This study | 5’-TTTTTTTTTTTTTTTTTCGCTTTTTTTTTTTTTTTT | |
| Sequence-based reagent | revDNA-CCG | This study | 5’-TTTTTTTTTTTTTTTTTCGGTTTTTTTTTTTTTTTT | |
| Sequence-based reagent | revDNA-TCG | This study | 5’-TTTTTTTTTTTTTTTTTCGATTTTTTTTTTTTTTTT | |
| Sequence-based reagent | revDNA-ACG | This study | 5’-TTTTTTTTTTTTTTTTTCGTTTTTTTTTTTTTTTTT | |
| Gene (Pseudomonas syringae) | ssdA | GeneBank | PSYRH_RS14050 | |
| Gene (Pseudomonas syringae) | ssdAI | GeneBank | PSYRH_RS1404 | |
| Gene (Taylorella equigenitalis) | BadTF3 | GeneBank | EL142_RS06975 | |
| Gene (Taylorella equigenitalis) | BadTF3-Immunity | GeneBank | EL142_RS06970 |
Additional files
-
Supplementary file 1
Summary of single-nucleotide variants (SNPs) identified within target cells after co-culture with B. cenocepacia.
- https://cdn.elifesciences.org/articles/62967/elife-62967-supp1-v3.xlsx
-
Supplementary file 2
X-ray data collection and refinement statistics.
- https://cdn.elifesciences.org/articles/62967/elife-62967-supp2-v3.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/62967/elife-62967-transrepform-v3.pdf





