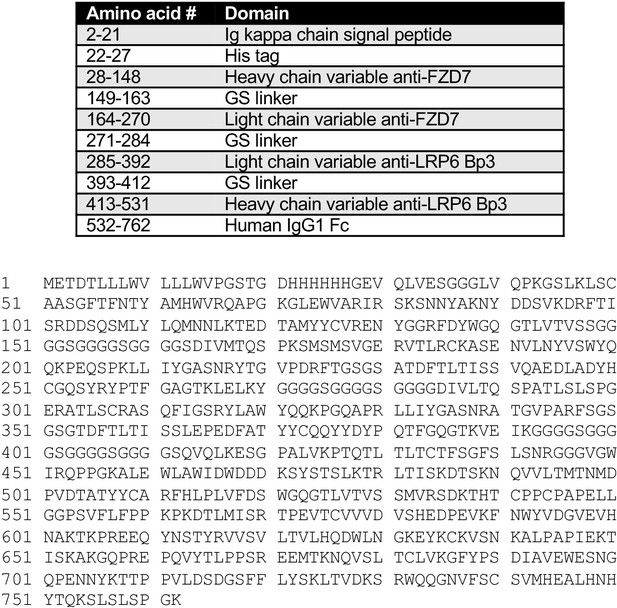
Selective activation of FZD7 promotes mesendodermal differentiation of human pluripotent stem cells
Figures
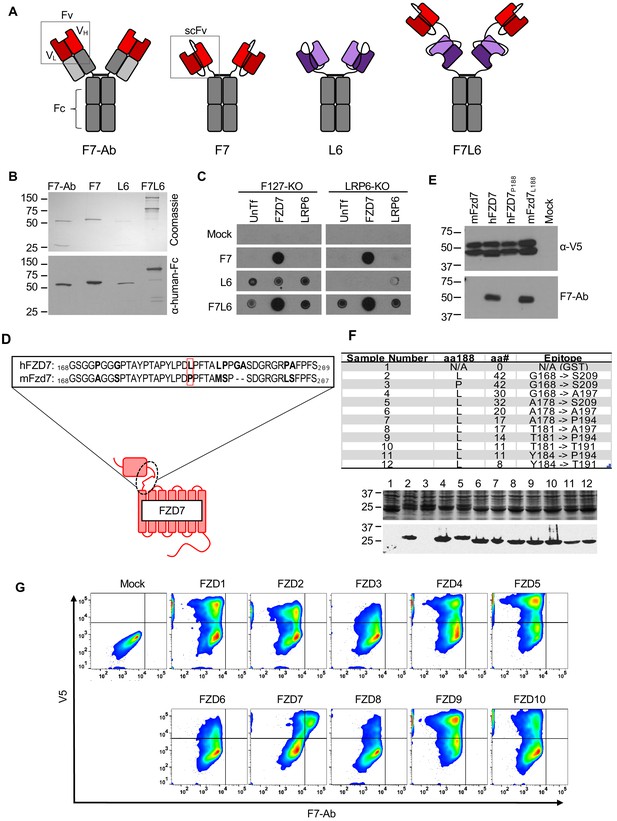
Design, specificity, and expression of F7L6.
(A) Schematic of FZD7- and LRP6-specific binders. Red blocks depict the variable light (VL) and heavy (VH) antibody domains that recognize FZD7 and were fused to form the single-chain variable fragment (scFv) in F7 and F7L6. Purple blocks depict the scFv that recognizes LRP6 and is used in L6 and F7L6. (B) Expression of FZD7- and LRP-specific binders. Transgenes encoding F7-Ab, F7, L6, and F7L6 were stably transduced in CHO cells. Recombinant proteins were harvested and purified from conditioned media and detected by Coomassie Blue staining (upper) and anti-human-Fc immunoblot (lower). (C) Binding specificity of F7, L6, and F7L6. HEK293T carrying mutations in FZD1, 2, and 7 (F127-KO) or in LRP6 (LRP6-KO) were transfected with FZD7 and LRP6, respectively, and whole cell lysates were probed in a dot blot format with conditioned media containing F7, L6, or F7L6. As a negative control (Mock), blots were incubated with CM from untransfected CHO cells. (D) Schematic of FZD7 and amino acid alignment of the extracellular ‘neck’ region of hFZD7 and mFzd7. The dashed oval indicates the neck region. The red box in the amino acid sequences indicates amino acid position 188. (E) HEK293T cells were transiently transfected with the indicated V5-tagged transgenes, and cell lysates were probed with either F7-Ab or V5 antibody (α-V5). Mock = untransfected cells. (F) Mapping F7-Ab epitope to an eight amino acid sequence. Bacterial lysates containing fusion proteins between GST and the FZD7 peptide sequences indicated in the table were detected by Coomassie staining (top) or by immunoblotting with F7-Ab (bottom). Abbreviations in table: aa188, amino acid at position 188; aa#, number of amino acids in FZD7 peptide; L, leucine; P, proline. (G) F7-Ab is specific to human FZD7 and does not cross-react with the other nine FZDs (1-6, 8-10). F127-KO were transfected with expression constructs carrying the indicated human FZD cDNAs tagged with an intracellular V5 sequence. Non-permeabilized cells were stained with F7-Ab for cell-surface FZD expression, and then permeabilized and stained for V5 expression. All FZD receptors were expressed as revealed by anti-V5 antibody staining.
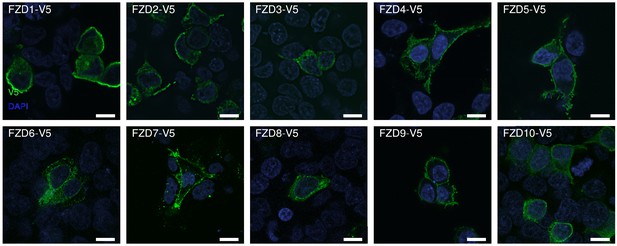
Immunofluorescent detection of V5-tagged FZD1-10 demonstrates cell surface localization.
Scale bar = 10 microns.
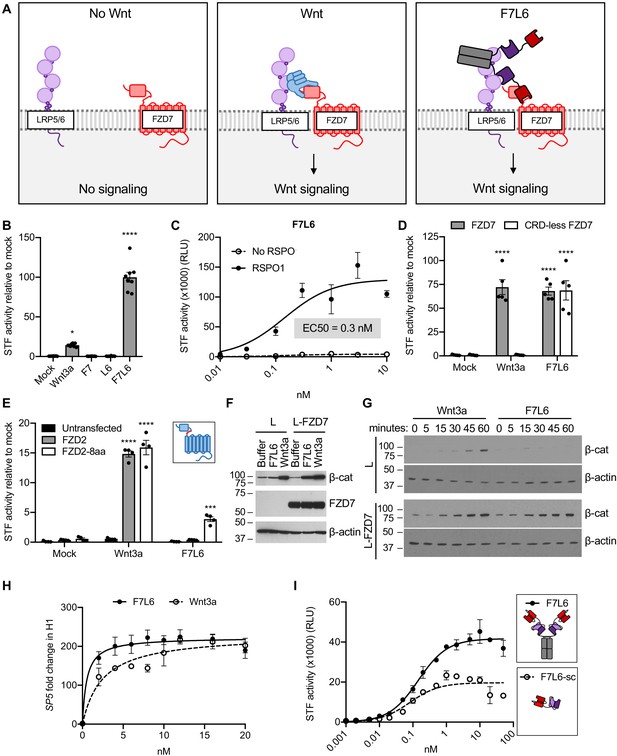
F7L6 activates Wnt/B-catenin signaling.
(A) Schematic of the proposed mechanism of action of F7L6 through the heterodimerization of FZD7 and LRP6 at the cell surface. (B) F7L6 activation of the WNT signaling pathway was evaluated using a luciferase-based WNT reporter (Super TOP-Flash, STF) assay. HEK293T stably transduced with the WNT reporter Super TOP-Flash (STF) were treated with indicated conditioned media for 24 hr and then assayed for luciferase activity. (C) F7L6 signaling activity is augmented by RSPO1. HEK293T:STF cells were treated with the indicated concentrations of F7L6 in the presence or absence of RSPO1 (100 ng/mL) for 24 hr and then assayed for luciferase activity (RLU = relative light units). (D) F7L6 activates signaling independently of the WNT-binding cysteine-rich domain (CRD). F127-KO cells carrying the STF reporter were transfected with expression plasmids carrying wildtype FZD7 or CRD-less FZD7, treated with Wnt3a or F7L6 for 24 hr and then assayed for luciferase activity. Inset illustrates FZD7 lacking the CRD. (E) F7L6 activates FZD2 tagged with the eight-amino acid epitope of FZD7. F127-KO cells carrying the STF reporter were transfected with expression plasmids carrying wildtype FZD2 or FZD2-8aa, treated with Wnt3a or F7L6 for 24 hr and then assayed for luciferase activity. Inset illustrates FZD2 (blue) with the eight-amino acid FZD7 tag (red). (F) F7L6 leads to β-catenin stabilization in mouse L-cells expressing human FZD7. Untransfected (L) or FZD7-expressing (L-FZD7) L-cells were treated with 10 nM F7L6 or Wnt3a for 3 hr. Cell lysates were immunoblotted for β-catenin and FZD7. Blotting for β-actin served as a loading control. (G) F7L6 leads to β-catenin stabilization in a time-dependent manner. L and L-FZD7 cells were treated with 10 nM Wnt3a or F7L6 for the indicated times, and cell lysates were immunoblotted for β-catenin. Blotting for β-actin served as a loading control. (H) F7L6 activates SP5 expression in hPS cells in a dose-dependent manner. H1/WA01 cells were treated with the indicated doses of F7L6 or Wnt3a for 24 hr. RNA was analyzed by qRT-PCR. Data represented as mean ± SEM for three technical replicates, with a nonlinear regression curve. All samples were normalized to the 0 nM (buffer) control. (I) Bivalent and tetravalent Wnt mimetics activate Wnt signaling. HEK293T:STF cells were treated with indicated concentrations of either F7L6 (tetravalent) or F7L6-sc (bivalent) in the presence of RSPO1 (100 ng/mL) for 24 hr and then assayed for luciferase activity (RLU = relative light units). For all statistical analyses: one-way ANOVA and Tukey’s multiple comparisons test: ****p≤0.0001, *p≤0.05.
-
Figure 2—source data 1
Raw data for STF and RT-qPCR assays shown in Figure 2.
- https://cdn.elifesciences.org/articles/63060/elife-63060-fig2-data1-v2.xlsx
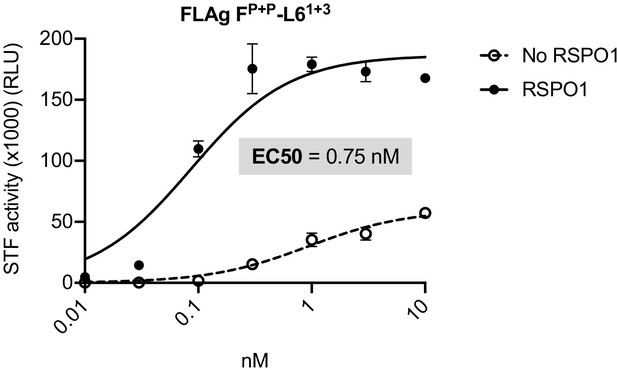
Dose response curves for a previously published Wnt mimetic, FLAgP+P-L61+3 (described in Tao et al., 2019) and Wnt3a.
HEK293T:STF cells were treated with the indicated concentrations of FLAgP+P-L61+3 or Wnt3a in the presence or absence of RSPO1 (100 ng/mL) for 24 hr and then assayed for luciferase activity (RLU = relative light units).
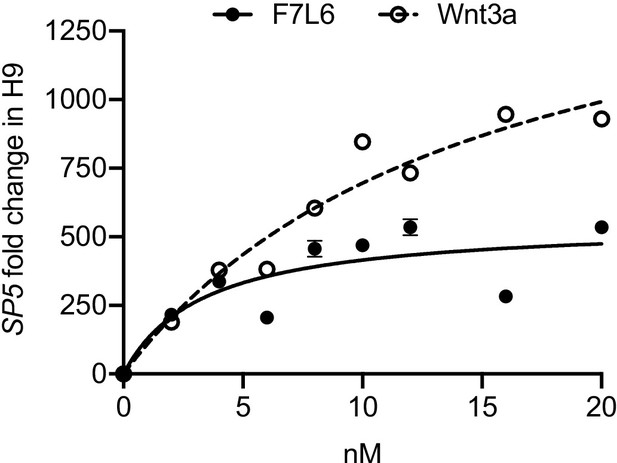
F7L6 activates Wnt/β-catenin signaling.
F7L6 activates Wnt target gene SP5 in human pluripotent stem cells in a dose-dependent manner. H9/WA09 cells were treated with the indicated doses of F7L6 or Wnt3a for 24 hr. RNA was analyzed by qRT-PCR. Data represented as mean ± SEM for three technical replicates, with a nonlinear regression curve. All samples were normalized to the 0 nM (buffer) control.
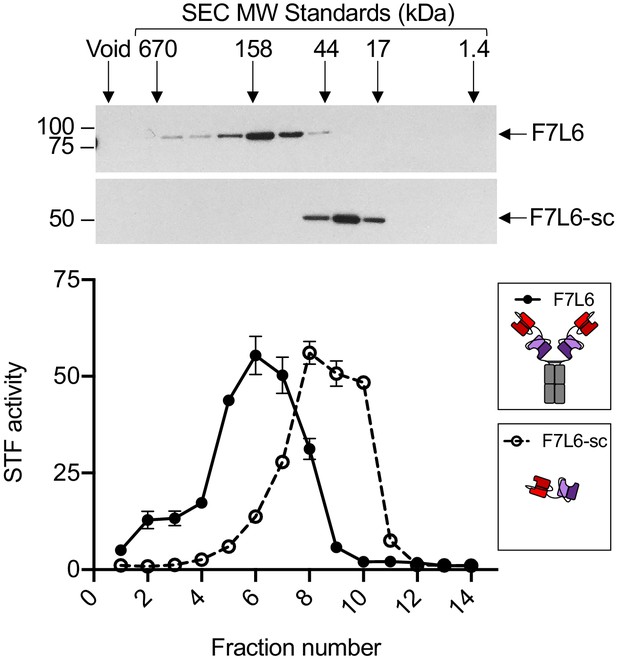
Size exclusion chromatography (SEC) of F7L6 and F7L6-sc.
Purified Wnt mimetics F7L6 and F7L6-sc were fractionated on a Superdex 200 column. Fractions were assayed by SDS-PAGE followed by Coomassie staining and by activation of the STF reporter in HEK293T cells.
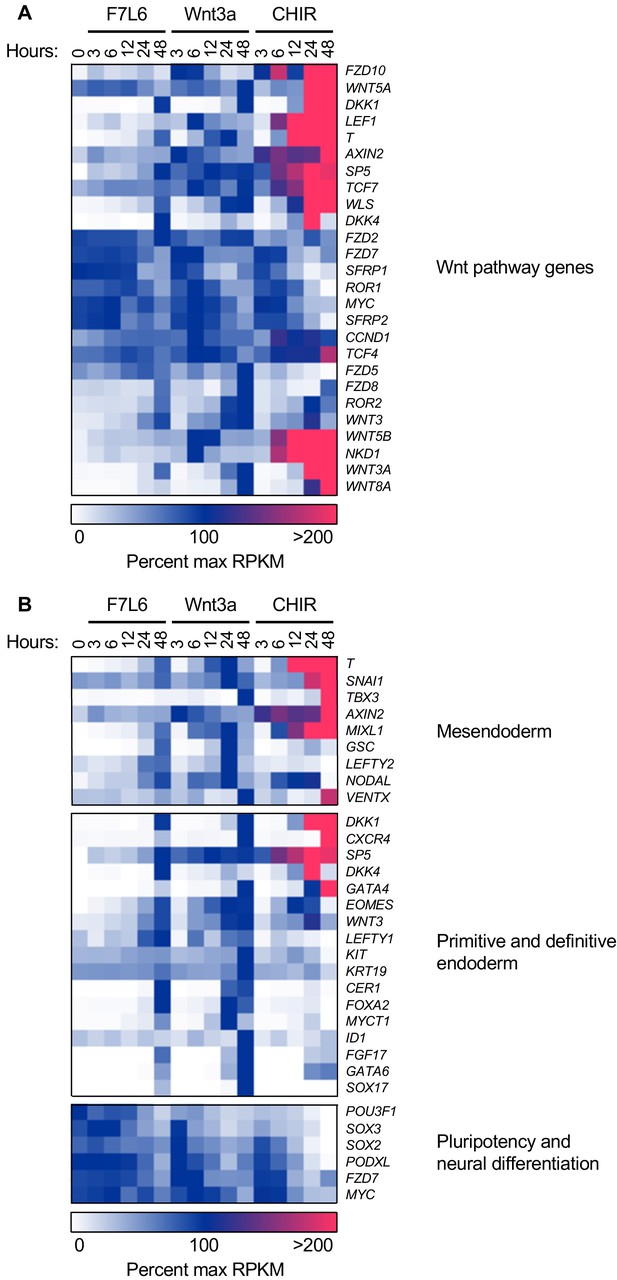
F7L6, Wnt3a, and CHIR differentially alter the transcriptome of human pluripotent stem (hPS) cells.
hPS cells (H1/WA01) were treated with 5 nM F7L6 or Wnt3a, or 250 nM CHIR98014 (CHIR) for the indicated hours. RNA was isolated and analyzed by RNA-seq. Significant differential gene expression was defined as a 1.75-fold increase or decrease in RPKM compared to the 0 hr (buffer) control. Expression is represented as percent maximum RPKM (0, white; 100, blue;≥100, pink). RPKM for each gene was normalized to the maximum RPKM across F7L6 and Wnt3a treatment groups. Supplementary Data 1 provides complete gene list. (A) Heat map of Wnt target genes changed in response to F7L6, Wnt3a, or CHIR. (B) Heat map of changed genes associated with mesendoderm and primitive/definitive endoderm differentiation, and pluripotency and neural differentiation. F7L6 promotes mesendodermal differentiation, similarly to Wnt3a.
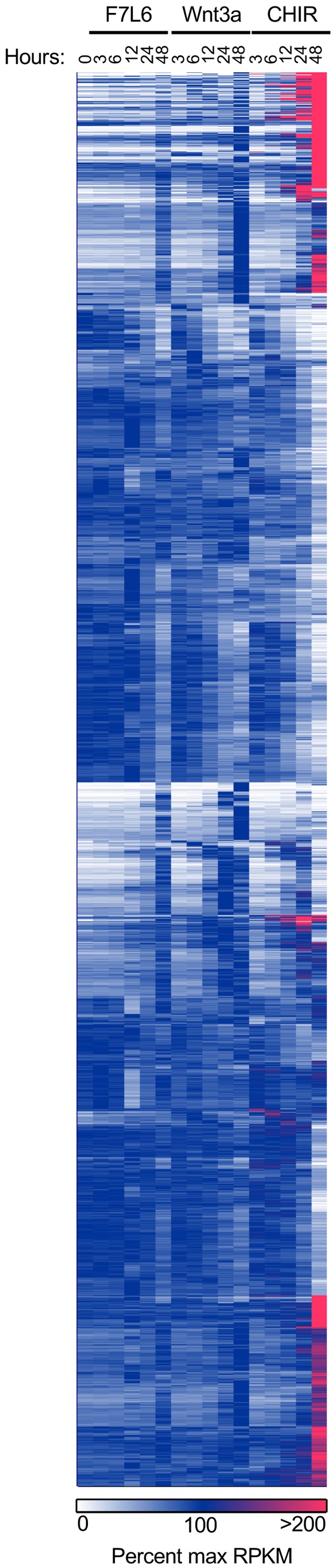
F7L6, Wnt3a, and CHIR differentially alter the transcriptome of human pluripotent stem (hPS) cells.
Heat map of 1814 significantly differentially expressed genes in response to F7L6, Wnt3a, and/or CHIR98014 (CHIR) treatments in hPS cells (H1/WA01). Cells were treated with 5 nM F7L6 or Wnt3a, or 250 nM CHIR98014 (CHIR) for the indicated hours, and RNA was isolated and analyzed by RNA-seq. Data are represented as percent maximum RPKM (0, white; 100, blue;≥100, pink). Supplementary file 1A provides the complete gene list.
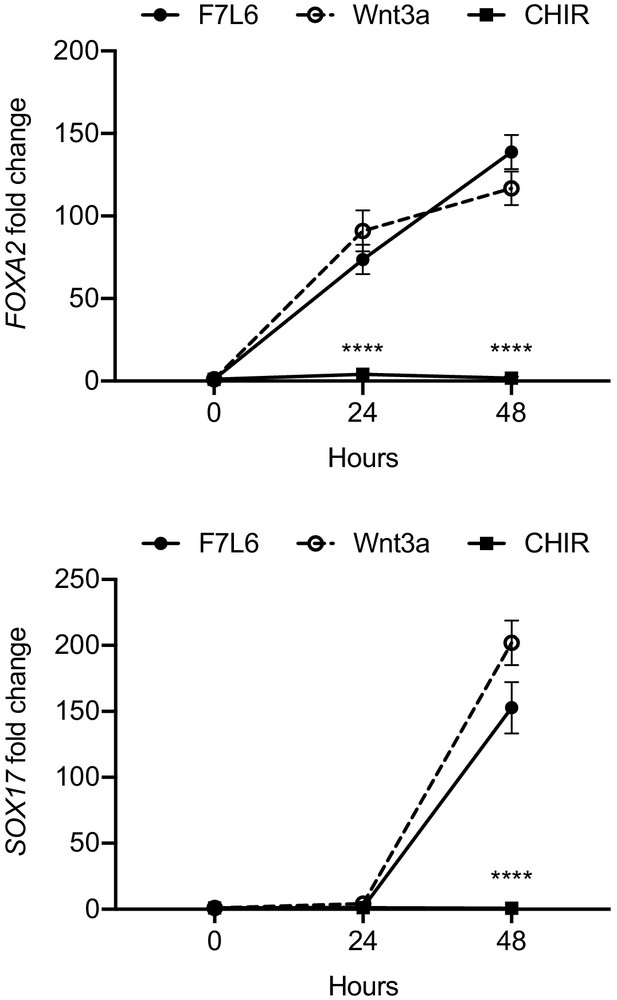
CHIR does not activate select definitive endoderm markers in undirected differentiation of human pluripotent stem cells.
H1/WA01 cells were treated with 5 nM F7L6 or Wnt3a, or 250 nM CHIR for the indicated hours. RNA was isolated and analyzed by RT-qPCR for FOXA2 and SOX17. Data represented as mean ± SEM for two independent experiments, three technical replicates each. All samples were normalized to the 0 hr (buffer) control. For statistical analyses: one-way ANOVA and Tukey’s multiple comparisons test for significance of CHIR against F7L6 and Wnt3a at each time point: ****p≤0.0001.
-
Figure 3—figure supplement 2—source data 1
Raw data for RT-qPCR assays shown in Figure 3—figure supplement 2.
- https://cdn.elifesciences.org/articles/63060/elife-63060-fig3-figsupp2-data1-v2.xlsx
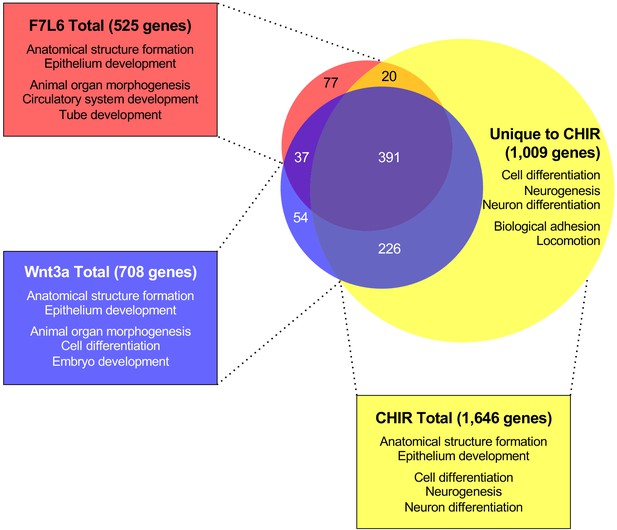
Gene Set Enrichment Analysis (GSEA) analyses of transcriptome changes induced by F7L6, Wnt3a, and CHIR.
Venn diagram and GSEA analyses of genes differentially expressed in response to F7L6, Wnt3a, and CHIR98014 treatments in hPS cells (H1/WA01). The top five GSEA gene set hits for each treatment group are listed by commonality among the groups and alphabetical order. Supplementary file 1 provides lists of gene names and Supplementary file 2 provides lists of gene set names.
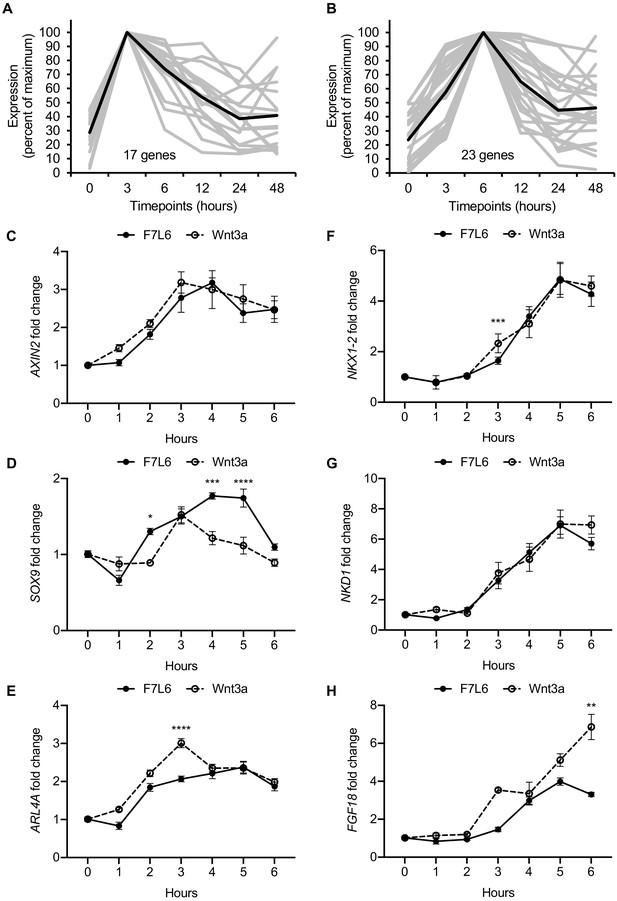
Early Wnt target genes activated by F7L6 and Wnt3a in hPS cells.
RNA-seq profiles of genes significantly activated at 3 hr (A) and 6 hr (B) in response to Wnt3a (5 nM) in H1/WA01 cells. Lists of genes are provided in Supplementary file 3 A and B. Validation of three target genes (AXIN2, SOX9, and ARL4A) maximally activated at 3 hr (C–E) and of three genes (NKX1-2, NKD1, and FGF18) maximally activated at 6 hr (F–G). H1 cells were treated with F7L6 or Wnt3a (each at 10 nM) for the indicated time and total RNA was analyzed by RT-qPCR. Gene expression was normalized to the expression of RPL13A. Data represented as mean ± SEM for two independent experiments, three technical replicates each. All samples were normalized to the 0 hr (buffer) control. For statistical analyses: one-way ANOVA and Tukey’s multiple comparisons test for significance between F7L6 and Wnt3a treatments at each time point: ****p≤0.0001, ***p≤0.001, **p≤0.01, *p≤0.05.
-
Figure 5—source data 1
Raw data for RT-qPCR assays shown in Figure 5.
- https://cdn.elifesciences.org/articles/63060/elife-63060-fig5-data1-v2.xlsx
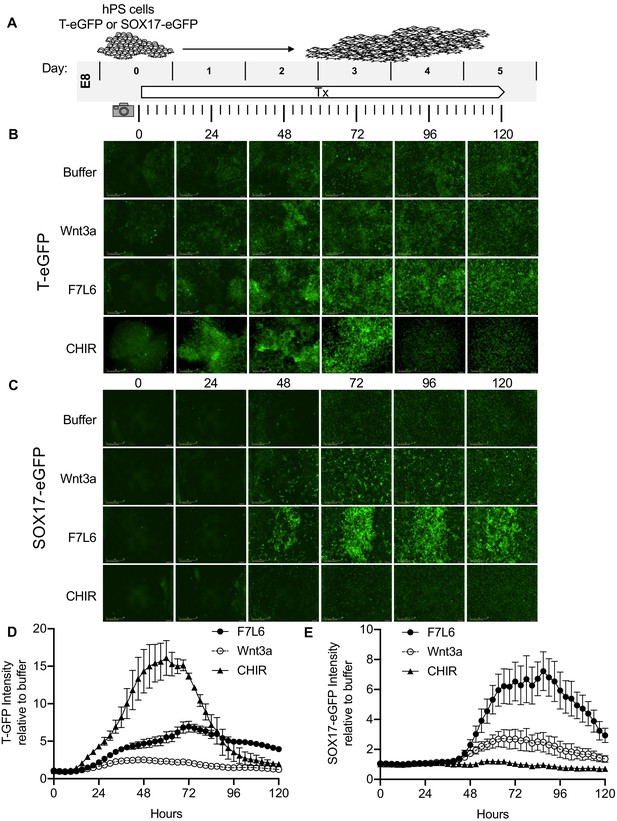
Activation of FZD7 with F7L6 promotes differentiation of hPS cells.
(A) Schematic of live cell imaging experiment. Abbreviations: E8, essential eight medium; Tx, treatment. H9/WA09 cells carrying a T-GFP (B, D) or a SOX17-eGFP (C, E) reporter gene were treated with the indicated compounds, and fluorescence was imaged every 3 hr for a total of 120 hr on an IncuCyte Life Cell Analysis System. Fluorescence was quantified by total green object integrated intensity (GCU x μm2/image).
Movies corresponding to Figure 6B,D.
H9 cells carrying a T-GFP reporter were treated with Buffer, Wnt3a, F7L6, or CHIR and imaged for 120 hr. Fluorescent images were captured every 3 hr and combined to generate a movie.
Movies corresponding to Figure 6C,E.
H9 cells carrying a SOX17-eGFP reporter were treated with Buffer, Wnt3a, F7L6, or CHIR and imaged for 120 hr. Fluorescent images were captured every 3 hr and combined to generate a movie.
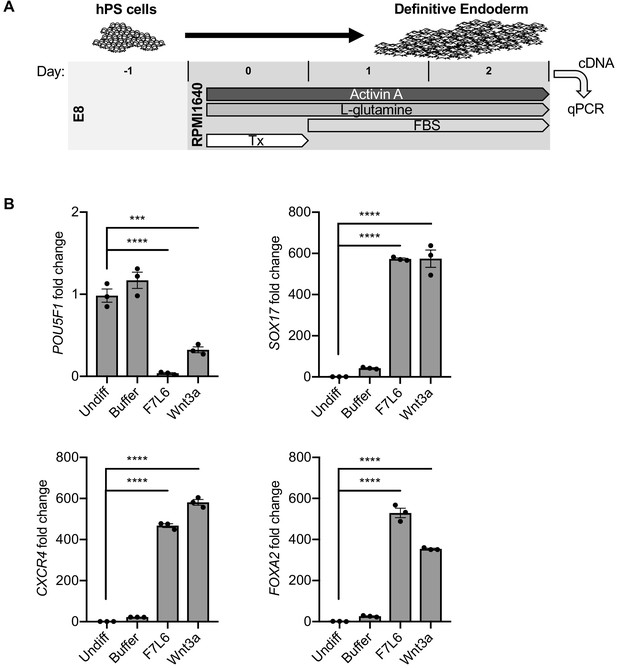
Activation of FZD7 with F7L6 promotes differentiation to definitive endoderm (DE).
(A) Schematic of DE differentiation protocol. Abbreviations: E8, essential eight medium; FBS, fetal bovine serum; Tx, treatment. (B) RT-qPCR analysis of the differentiation treated with the indicated compounds. Treatment of hPS cells (H9/WA01) with F7L6 or Wnt3a leads to downregulation of the pluripotency marker POU5F1 and upregulation of the DE markers CXCR4, SOX17 and FOXA2. Gene expression was normalized to the expression of RPL13A. All samples were normalized to undifferentiated (Undiff) samples. For all statistical analyses: one-way ANOVA and Tukey’s multiple comparisons test: ****p≤0.0001, ***p≤0.001.
-
Figure 7—source data 1
Raw data for RT-qPCR assays shown in Figure 7.
- https://cdn.elifesciences.org/articles/63060/elife-63060-fig7-data1-v2.xlsx
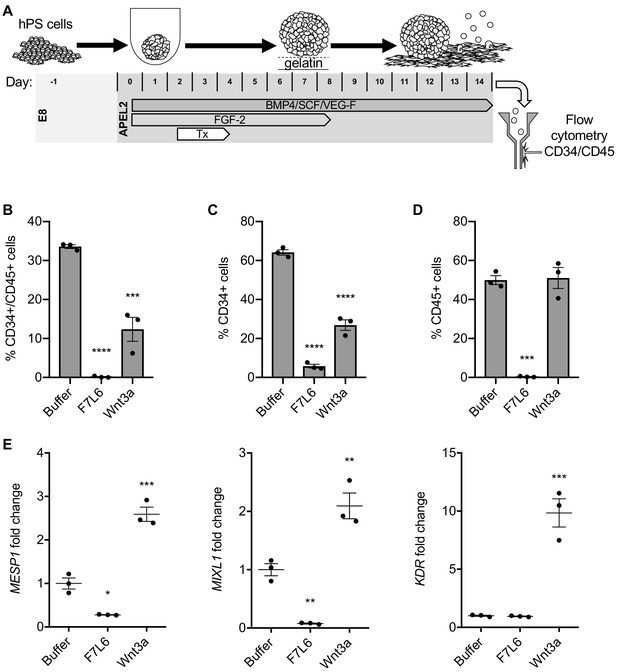
Activation of FZD7 with F7L6 blocks differentiation to hematopoietic stem and progenitor cells.
(A) Schematic of the APEL hematopoietic stem/progenitor cell (HSPC) differentiation protocol. HPS cells were treated (Tx) with either Wnt3a or F7L6 from Days 2 to 4 of the 14 day differentiation protocol. On day 14, cells were analyzed by flow cytometry for the cell surface markers CD34 and CD45. Quantitation of flow cytometry of CD34/CD45 double positive cells (B), CD34 single positive cells (C), and CD45 single positive cells (D). (E) RT-qPCR analysis of MESP1 and MIXL1 at day 4 and of KDR at day 11 of differentiation. For statistical analyses: one-way ANOVA and Tukey’s multiple comparisons test: ***p≤0.001, **p≤0.01, *p0.05, ns, not significant.
-
Figure 8—source data 1
Raw data for flow cytometry and RT-qPCR assays shown in Figure 8.
- https://cdn.elifesciences.org/articles/63060/elife-63060-fig8-data1-v2.xlsx
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Cricetulus griseus) | CHO | ATCC | CCL-61 | RRID:CVCL_0213 |
| Cell line (Homo sapiens) | HEK293 | ATCC | CRL-1573 | RRID:CVCL_0045 |
| Cell line (Homo sapiens) | HEK293T-F127-KO | Prof. M. Boutros, Heidelberg University, Germany | Voloshanenko et al., 2017 | |
| Cell line (Homo sapiens) | HEK293T-F124578-KO | Prof. M. Boutros, Heidelberg University, Germany | Voloshanenko et al., 2017 | |
| Cell line (Mus musculus) | L1 | ATCC | CRL-2648 | RRID:CVCL_4536 |
| Cell line (Homo sapiens) | WA01 (H1) | WiCell Research Institute | NIH Registration Number: 0043 | Male, RRID:CVCL_9771 |
| Cell line (Homo sapiens) | WA09 (H9) | WiCell Research Institute | NIH Registration Number: 0062 | Female RRID:CVCL_9773 |
| Cell line (Homo sapiens) | H9 SOX17:GFP | Prof. S. Kim, Stanford University, USA | Wang et al., 2011 | |
| Cell line (Homo sapiens) | H9 T-GFP | Prof. M. Mercola, Stanford University, USA | Kita-Matsuo et al., 2009 | |
| Cell line (Homo sapiens) | iPS cells | Professor D. Kaufman, UCSD, USA | Li et al., 2018 | |
| Strain, strain background (Escherichia coli) | BL21(DE3) | Invitrogen | C600003 | |
| Recombinant DNA reagent | Super TOPFlash (STF) | Addgene | Plasmid #12456 | RRID:Addgene_12456 |
| Recombinant DNA reagent | pFuse-hIgG1-Fc2 | Invivogen | pfuse-hg1fc2 | |
| Recombinant DNA reagent | pGEX-4T3 | Cytiva | 28-9545-52 | |
| Antibody | F7-Ab, chimeric human-mouse monoclonal | This paper | 1 μg/mL, available upon request from corresponding author | |
| Antibody | V5, mouse monoclonal | GeneTeX | Cat# GTX628529 | Immunoblot, 1:4000 Immunofluorescence, 1:500 |
| Antibody | Anti-β-catenin, mouse monoclonal | Sigma-Aldrich | Cat# C7207 | 1:2000 RRID:AB_476865 |
| Antibody | Anti-β-actin, mouse monoclonal | Sigma-Aldrich | Cat# A2228 | 1:5000 RRID:AB_476697 |
| Antibody | Goat anti-human IgG HRP-conjugated, goat polyclonal | ThermoFisher Scientific | Cat# 62–8420 | 1:20,000 RRID:AB_88136 |
| Antibody | Goat anti-mouse IgG HRP-conjugated, goat polyclonal | Southern Biotech | Cat#: 1030–05 | 1:20,000 RRID:AB_2619742 |
| Antibody | APC anti-human CD34, mouse monoclonal | Biolegend | Cat# 343608 | 1:100 RRID:AB_2228972 |
| Antibody | PE anti-human CD45, mouse monoclonal | Biolegend | Cat# 304008 | 1:100 RRID:AB_314396 |
| Antibody | APC mouse IgG2a k isotype control, mouse monoclonal | Biolegend | Cat# 400222 | 1:100 |
| Antibody | PE mouse IgG1, k isotype control, mouse monoclonal | Biolegend | Cat# 400112 | 1:100 RRID:AB_2847828 |
| Antibody | Alexa-Fluor 488 Goat anti-Mouse IgG, goat polyclonal | Invitrogen | Cat# A11001 | 1:1000 RRID:AB_2534069 |
| Recombinant protein | F7L6 | This paper | Purified from CHO_His-F7L6-Fc, available upon request from corresponding author | |
| Recombinant protein | F7L6-sc | This paper | Purified from CHO_His-F7L6, available upon request from corresponding author | |
| Recombinant protein | Wnt3a | Produced in Willert lab Willert et al., 2003 Willert, 2008 | Purified from CHO_Wnt3a cells | |
| Recombinant protein | RSPO1 | Prof. Xi He, Harvard Medical School | Purified from HEK293_Rspo1 cells | |
| Recombinant protein | FLAg FP+P-L61+3 | Prof. S. Angers, Toronto University, Canada Tao et al., 2019 | ||
| Recombinant protein | ActivinA | R and D Systems | Cat# 338-AC | 100 ng/mL |
| Recombinant protein | BMP4 | R and D Systems | Cat# 314 BP | 40 ng/mL |
| Recombinant protein | SCF | R and D Systems | Cat# 7466-SC | 40 ng/mL |
| Recombinant protein | VEGF | R and D Systems | Cat# 293-VE | 20 ng/mL |
| Recombinant protein | FGF2 | StemCell Technologies | Cat#78003 | 10 ng/mL |
| Commercial assay or kit | SuperSignal West Dura Western Blot Substrate | ThermoFisher Scientific | Cat# 34075 | |
| Commercial assay or kit | Pierce Coomassie (Bradford) Protein Assay Kit | ThermoFisher Scientific | Cat# 23200 | |
| Commercial assay or kit | TRIzol Reagent | ThermoFisher Scientific | Cat# 15596026 | |
| Commercial assay or kit | Direct-zol RNA MiniPrep Kit | Zymo Research | Cat# R2051 | |
| Commercial assay or kit | iScript Reverse Transcription Supermix | Bio-Rad | Cat# 1708840 | |
| Commercial assay or kit | iTaq Universal SYBR Green Supermix | Bio-Rad | Cat# 1725120 | |
| Chemical compound, drug | Zeocin | ThermoFisher Scientific | R25005 | 1 mg/mL |
| Chemical compound, drug | Puromycin | ThermoFisher Scientific | A1113802 | 4 µg/mL |
| Chemical compound, drug | Rock inhibitor Y-27631 | Tocris | Cat# 1254 | 5 μM |
| Chemical compound, drug | GSK3 inhibitor CHIR98014 | Sigma-Aldrich | Cat# SML1094 | 250 nM |
| Chemical compound, drug | ATP | Sigma-Aldrich | Cat# A2383 | |
| Chemical compound, drug | D-luciferin-Potassium Salt | ThermoFisher Scientific | Cat# 50227 | |
| Other | DAPI stain | Cell Signaling Technology | Cat# 4083S | (1 µg/mL) |
| Other | Protein G Sepharose | BioVision | Cat# 6511 | |
| Other | HiTrap IMAC HP, 1 mL | Cytiva | Cat# 17092003 | |
| Other | Superdex 200 10/300 GL | Cytiva | Cat# GE28-9909-44 | |
| Other | Matrigel | BD Biosciences | Cat# 356234 | |
| Other | mTeSR1 | StemCell Technologies | Cat# 85850 | |
| Other | APEL2 | StemCell Technologies | Cat# 05270 |
Additional files
-
Supplementary file 1
RNA-seq data set for all significantly differentially expressed genes in hPS cells (H1/WA01) treated with F7L6, Wnt3a, or CHIR.
(A) Reads per kilobase per million mapped reads (RPKM) values for 1814 genes with significant fold changes in expression in response to F7L6, Wnt3a, or CHIR. (B) 525 genes with significant fold changes in expression in response to F7L6. (C) 708 genes with significant fold changes in expression in response to Wnt3a. (D) 1646 genes with significant fold changes in expression in response to CHIR. (E) 428 genes overlapping in F7L6 and Wnt3a treatments, with significant fold changes in expression. (F) RPKM values for 805 genes with significant fold changes in expression in response to F7L6 or Wnt3a. (G) 1009 genes unique to CHIR treatment, with significant fold changes in expression. (H) 391 genes overlapping across F7L6, Wnt3a, and CHIR treatments, with significant fold changes in expression. (I) 411 genes overlapping in F7L6 and CHIR treatments, with significant fold changes in expression. (J) 617 genes overlapping in Wnt3a and CHIR treatments, with significant fold changes in expression. (K) 77 genes unique to F7L6 treatment with significant fold changes in expression. (L) 54 genes unique to Wnt3a treatment, with significant fold changes in expression.
- https://cdn.elifesciences.org/articles/63060/elife-63060-supp1-v2.xlsx
-
Supplementary file 2
Gene Set Enrichment Analysis (GSEA) data for genes significantly differentially expressed in response to F7L6, Wnt3a, and/or CHIR in hPS cells (H1/WA01).
(A) Top ten gene set hits from GSEA for overlapping genes significantly differentially expressed in response to F7L6, Wnt3a, and CHIR. The corresponding gene list is provided in Supplementary file 1H. (B) Top ten gene set hits from GSEA for genes significantly differentially expressed in response to F7L6. The corresponding gene list is provided in Supplementary file 1B. (C) Top ten gene set hits from GSEA for genes significantly differentially expressed in response to Wnt3a. The corresponding gene list is provided in Supplementary file 1C. (D) Top 10 gene set hits from GSEA for genes significantly differentially expressed in response to CHIR. The corresponding gene list is provided in Supplementary file 1D. (E) Top 10 gene set hits from GSEA for unique genes significantly differentially expressed in response to CHIR. The corresponding gene list is provided in Supplementary file 1G. (F) Top 10 gene set hits from GSEA for overlapping genes significantly differentially expressed in response to F7L6 and Wnt3a. The corresponding gene list is provided in Supplementary file 1E. (G) Top 10 gene set hits from GSEA for overlapping genes significantly differentially expressed in response to F7L6 and CHIR. The corresponding gene list is provided in Supplementary file 1I. (H) Top 10n gene set hits from GSEA for overlapping genes significantly differentially expressed in response to Wnt3a and CHIR. The corresponding gene list is provided in Supplementary file 1J.
- https://cdn.elifesciences.org/articles/63060/elife-63060-supp2-v2.xlsx
-
Supplementary file 3
Lists of genes activated by Wnt3a in H1/WA01 at 3 hr (A) and 6 hr (B).
Expression level changes for these gene sets are shown in Figure 5A and B.
- https://cdn.elifesciences.org/articles/63060/elife-63060-supp3-v2.xlsx
-
Supplementary file 4
Lists of plasmids available upon request from corresponding author.
- https://cdn.elifesciences.org/articles/63060/elife-63060-supp4-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/63060/elife-63060-transrepform-v2.pdf