Action detection using a neural network elucidates the genetics of mouse grooming behavior
Figures
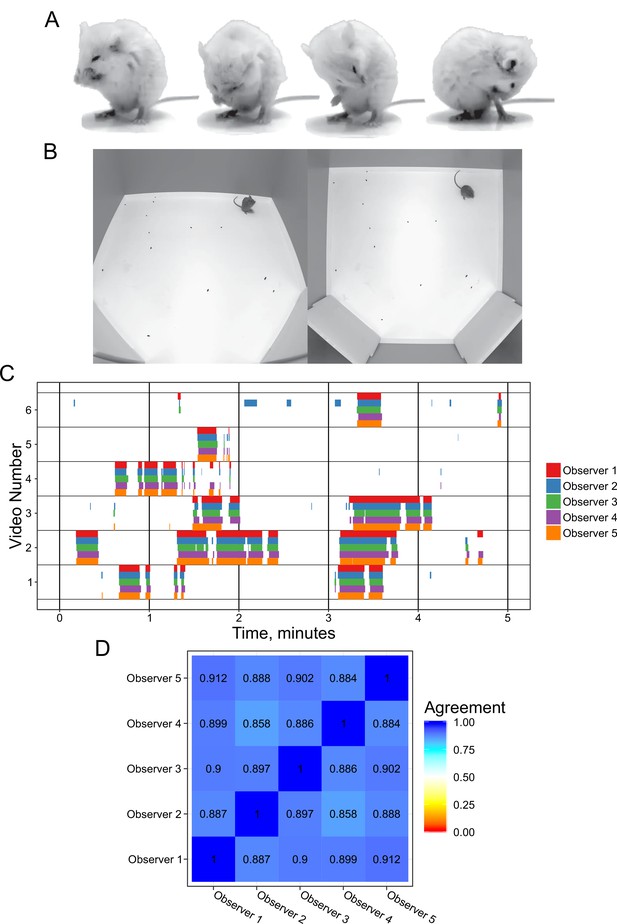
Annotation of mouse grooming behavior.
(A) Mouse grooming contains a wide variety of postures. Paw licking, face-washing, flank linking, as well as other syntaxes all contribute to this visually diverse behavior. (B) We provided synchronized two-view data for observers to annotate. (C) Grooming ethograms for six videos by five different trained annotators. Overall, there is very high agreement between human annotators. (D) Quantification of the agreement overlap between individual annotators. Average agreement between all annotators is 89.13%.
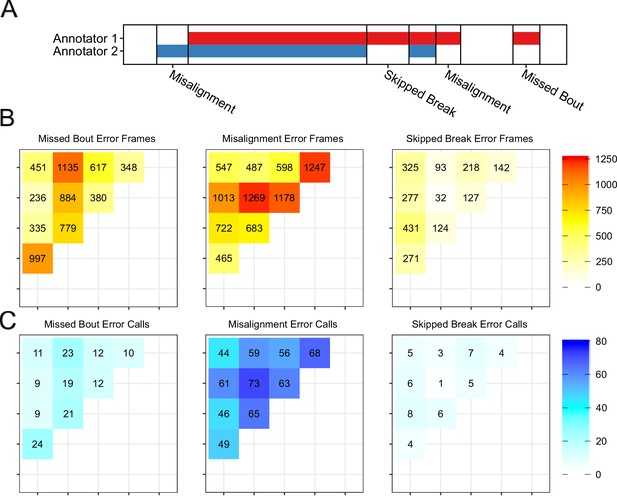
Additional details of annotator disagreements.
(A) We classify the types of annotation disagreement into three classes of errors: missed bouts, misalignment, and skipped breaks. (B–C) We quantify the types of errors in Figure 1. (B) The sum of frames that fall into each error category. 37.5% of frames are missed bouts, 50% are misalignment, and 12.4% are skipped breaks. The types of errors are not uniformly distributed across annotators as annotator two accounts for the most missed bout frame counts, annotator four accounts for the most misalignment frame counts, and annotator one accounts for the most skipped break frame counts. (C) Counting the number of multi-frame occurrences of errors, we observe a similar distribution. 19.2% of call are missed bouts, 74.6% are misalignment, and 6.3% are skipped breaks.
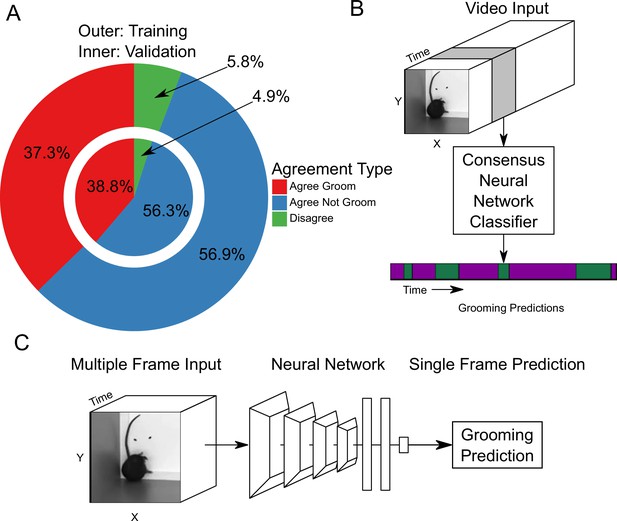
Neural network based action detection.
(A) A total of 2,637,363 frames were annotated across 1253 video clips by two different human annotators to create this data set for training and analyzing our neural network. The outer ring represents the training data set agreement between human annotators while the inner ring represents the validation data set agreement between human annotators. (B) A visual description of the classification approach that we implemented. To analyze an entire video, we pass a sliding window of frames into a neural network. (C) Our network takes video input and produces a grooming prediction for a single frame.
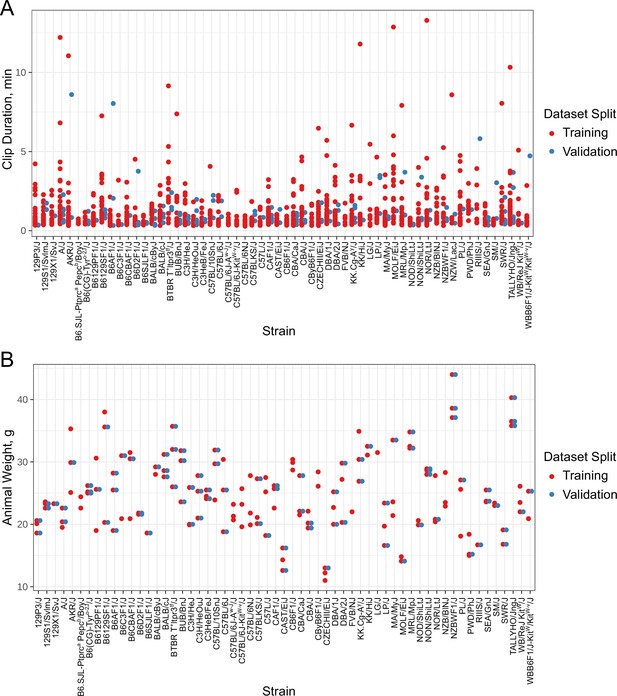
Additional details about the annotated dataset.
Our training dataset contains 1253 video clips that originate from 157 independent mice across 60 strains. (A) Distribution of clip durations by mouse strain. (B) Distribution of body weights present in our annotated dataset. Also see Supplementary file 1 for all associated annotated dataset metadata.
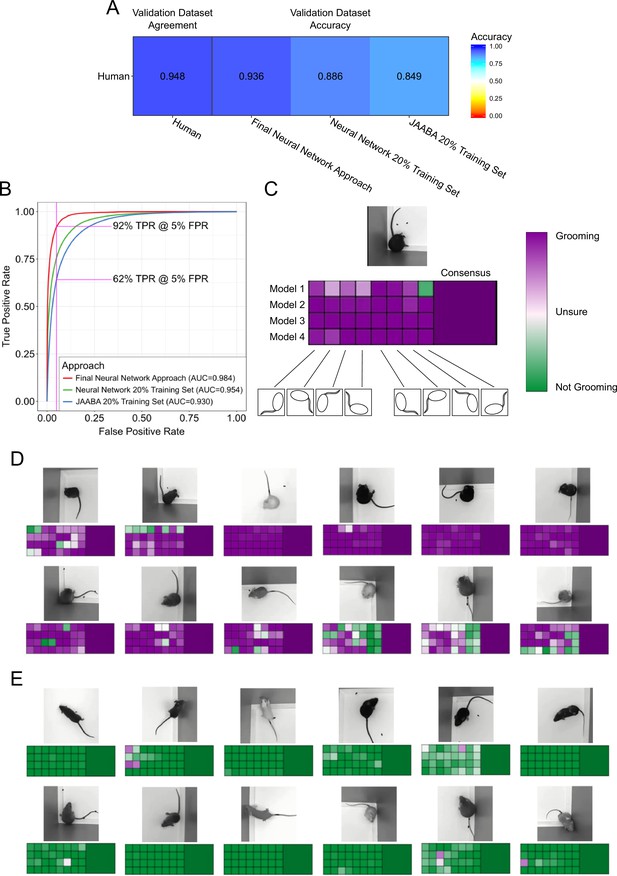
Validation of neural network model.
(A) Agreement between the annotators while creating the data set compared to the accuracy of the algorithms predicting on this data set. We compared the machine learning models against only annotations where the annotators agree. (B) Receiver operating characteristic (ROC) curve for three machine learning techniques trained on the training set and applied to the validation set. Our final neural network model approach achieves the highest area under curve (AUC) value of 0.9843402. True positive at 5% False positive is indicated with pink line. (C) A visual description of our proposed consensus solution. We use a 32x consensus approach where we trained four separate models and give eight frame viewpoints to each. To combine these predictions, we averaged all 32 predictions. While one viewpoint from one model can be wrong, the mean prediction using this consensus improves accuracy. (D–E) Example frames where the model is correctly predicting grooming and not-grooming behavior. Also see Figure 3—videos 1–9.
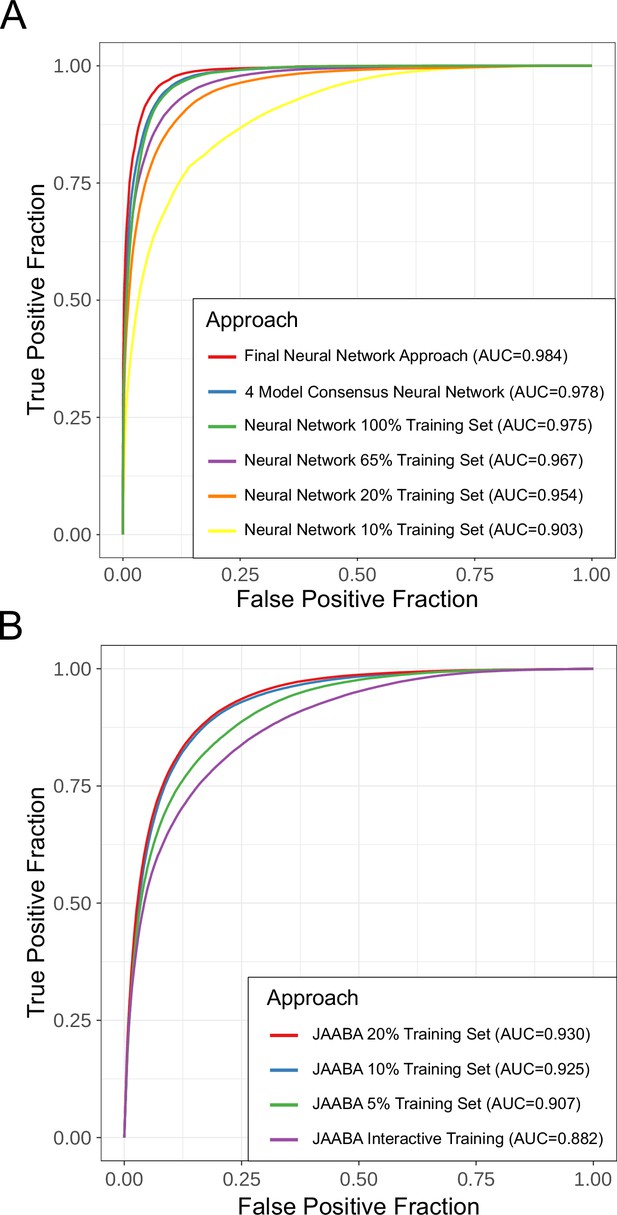
Additional ROC curve subsets.
(A) Performance of the network using different training data set sizes. As the number of training data samples increases, the ROC curve performance increases up to a point. (B) Performance of JAABA using different training data set sizes. Performance stops improving after using 10% of our annotations.
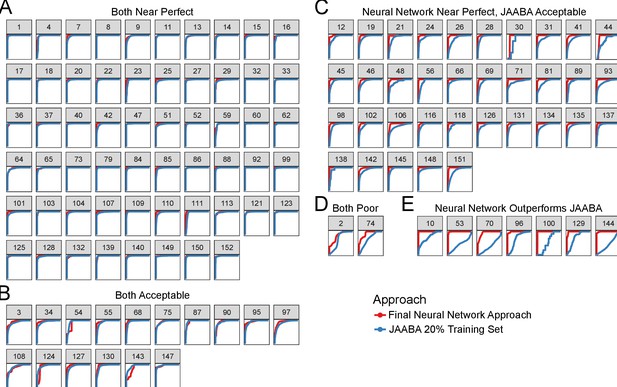
Validation performance of algorithm split by video.
Clusters of validation video ROC performance by machine learning approaches. (A) The majority of validation videos have good performance. (B) Some validation videos suffer from slightly degraded performance. (C) Validation videos where the JAABA approach performs slightly worse than our neural network approach. (D) Two validation videos showed poor performance using both machine learning approaches. Upon inspection of these videos, the annotated frames for grooming are visually difficult to classify. (E) seven validation videos showed good performance using the neural network but a clear drop in performance using JAABA. All videos that contain no positive grooming annotated frames do not have a ROC curve and were excluded from this figure.
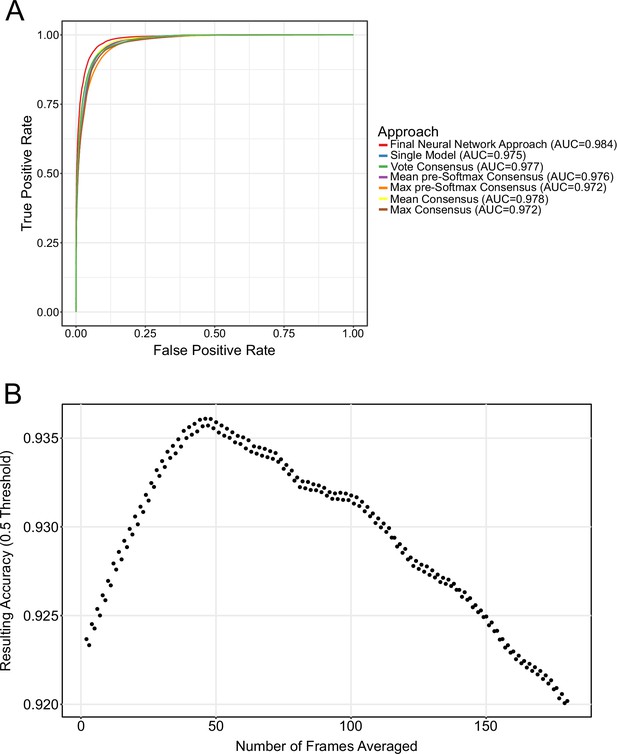
Comparison of different consensus modalities and temporal smoothing.
(A) ROC performance using different consensus modalities. All consensus modalities provide approximately the same result. (B) Temporal filter analysis.
Sample video of a black coat color mouse.
The ensemble networks are shown on the upper left (small squares) and consensus is shown in large square. See Figure 3C - green (negative for grooming) to purple (positive for grooming).
Sample video of an agouti coat color mouse.
The ensemble networks are shown on the upper left (small squares) and consensus is shown in large square. See Figure 3C - green (negative for grooming) to purple (positive for grooming).
Sample video of an agouti coat color mouse.
The ensemble networks are shown on the upper left (small squares) and consensus is shown in large square. See Figure 3C - green (negative for grooming) to purple (positive for grooming).
Sample video of an albino coat color mouse.
The ensemble networks are shown on the upper left (small squares) and consensus is shown in large square. See Figure 3C - green (negative for grooming) to purple (positive for grooming).
Sample video of an off-white coat color mouse.
The ensemble networks are shown on the upper left (small squares) and consensus is shown in large square. See Figure 3C - green (negative for grooming) to purple (positive for grooming).
Sample video of a gray coat color mouse.
The ensemble networks are shown on the upper left (small squares) and consensus is shown in large square. See Figure 3C - green (negative for grooming) to purple (positive for grooming).
Sample video of a chinchila coat color mouse.
The ensemble networks are shown on the upper left (small squares) and consensus is shown in large square. See Figure 3C - green (negative for grooming) to purple (positive for grooming).
Sample video of a nude coat color mouse.
The ensemble networks are shown on the upper left (small squares) and consensus is shown in large square. See Figure 3C - green (negative for grooming) to purple (positive for grooming).
Sample video of a kit mouse.
The ensemble networks are shown on the upper left (small squares) and consensus is shown in large square. See Figure 3C - green (negative for grooming) to purple (positive for grooming).
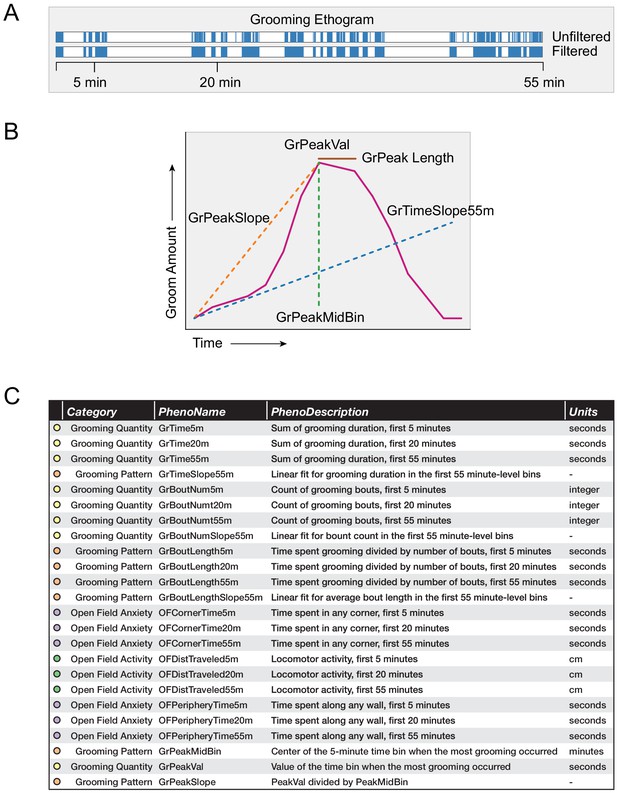
Grooming and open field behavioral metrics.
(A) Example grooming ethogram for a single animal. Time is on the x-axis and a blue colored bar signifies that the animal was performing grooming behavior during that time. We calculated summaries at 5, 20, and 55 min ranges. (B) A visual description of how we define our grooming pattern phenotypes. (C) A table summarizing the 24 behavioral metrics we analyzed. We grouped the phenotypes into four groups, including grooming quantity, grooming pattern, open field anxiety, and open field activity.
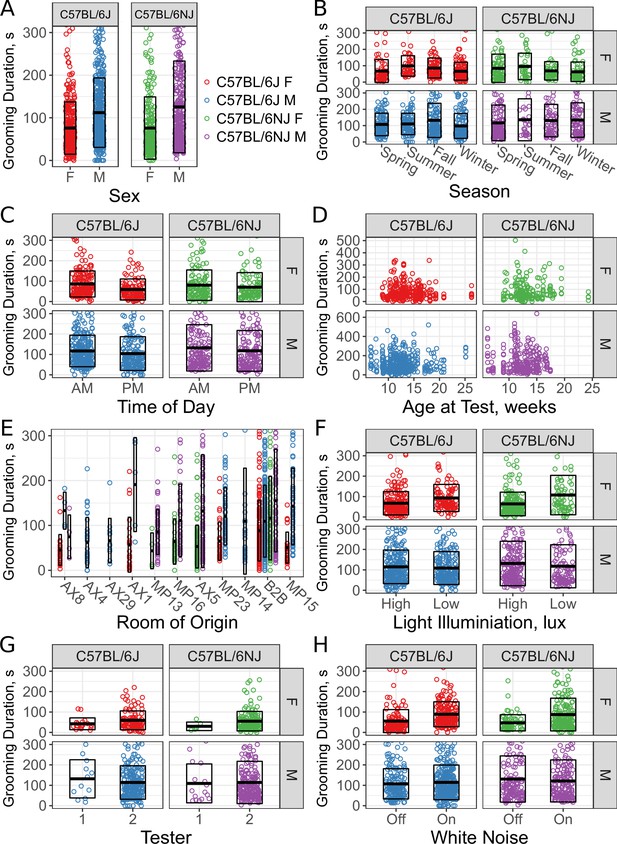
Sex and environmental covariate analysis of grooming behavior (total time grooming) in the open field for C57BL/6J and C57BL/6NJ strains.
Effect of sex (A), season (B), time of day (C), age (D), room of origin (E), light level (F), tester (G), and white noise (H).
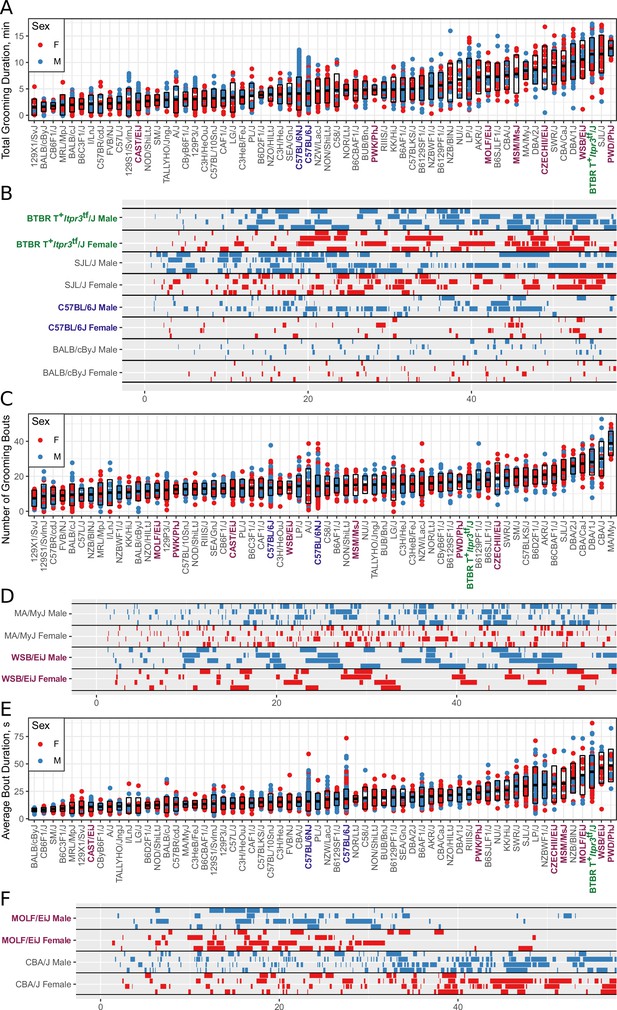
Strain survey of grooming phenotypes with representative ethograms (2457 animals and 2252 hr of data, each dot represents 55 min of a single animal).
(A) Strain survey results for total grooming time. Strains present a smooth gradient of time spent grooming, with wild derived strains (purple) showing enrichment on the high end. (B) Representative ethograms showing strains with high and low total grooming time. (C) Strain survey results for the number of grooming bouts. (D) Comparative ethograms for two strains with different number of bouts, but similar total time spent grooming. (E) Strain survey results for average grooming bout duration. (F) Comparative ethograms for two strains with different average bout length, but similar total time spent grooming.
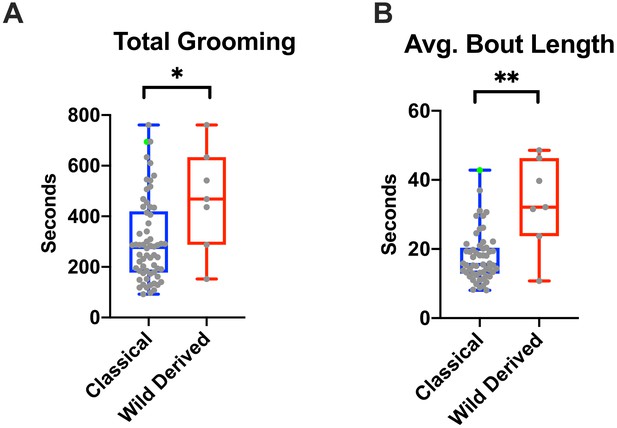
Grooming in wild-derived vs. classical inbred lines.
(A) Total grooming time of wild derived and classical strains are significantly different (*p<0.05, Mann-Whitney Test). (B) Wild derived lines have significantly longer grooming bouts (**p<0.01, Mann-Whitney Test). In both graphs, BtBR strain is indicated in green.
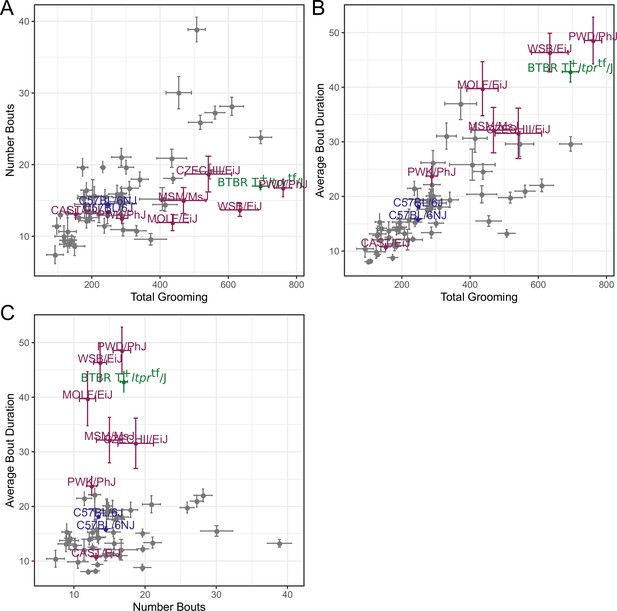
Relatedness of grooming phenotypes.
Points indicate strain-level means. Lines indicate 1SD. (A) Strain survey comparing total grooming time and number of bouts. Wild derived strains and BTBR show enrichment for having high grooming but low bout numbers. (B) Strain survey comparing total grooming time and average bout duration. Strains that groom more also tend to have a longer average bout length. (C) Strain survey comparing number of bouts to average bout duration.
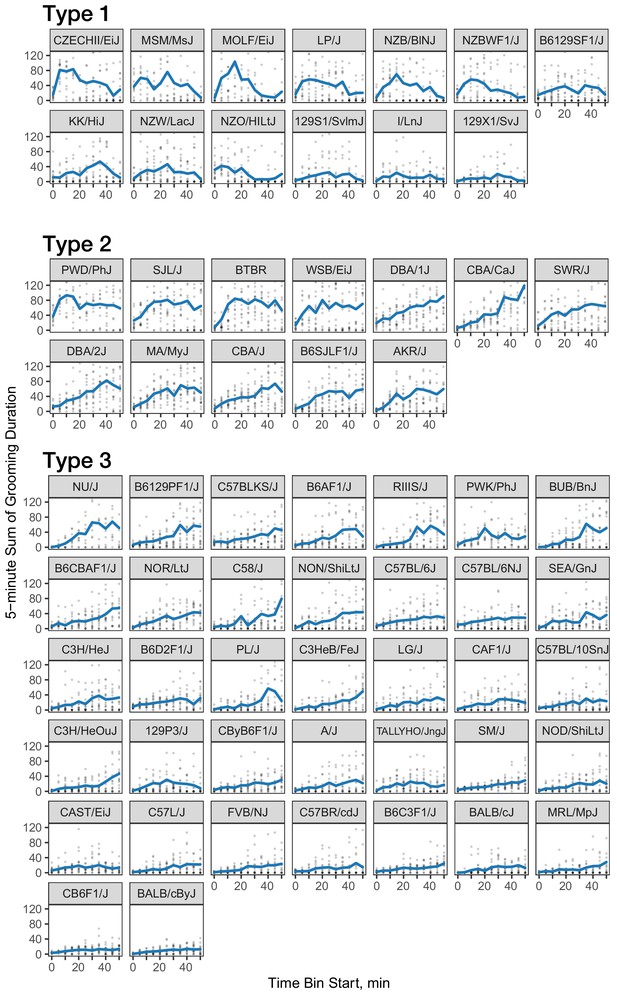
Three classes of grooming patterns in the open field revealed by clustering.
Grooming duration in 5 min bins is shown over the course of the open field experiment (blue line) and data from individual mice (gray points).
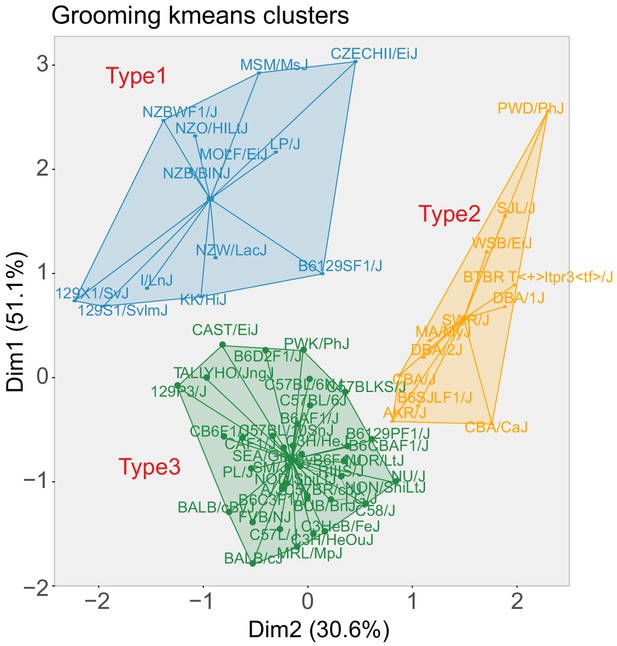
K-means clustering of grooming patterns Results from the k-means clustering.
The first two principal components from this clustering accounts for 81.7% of variance. three clusters were identified.
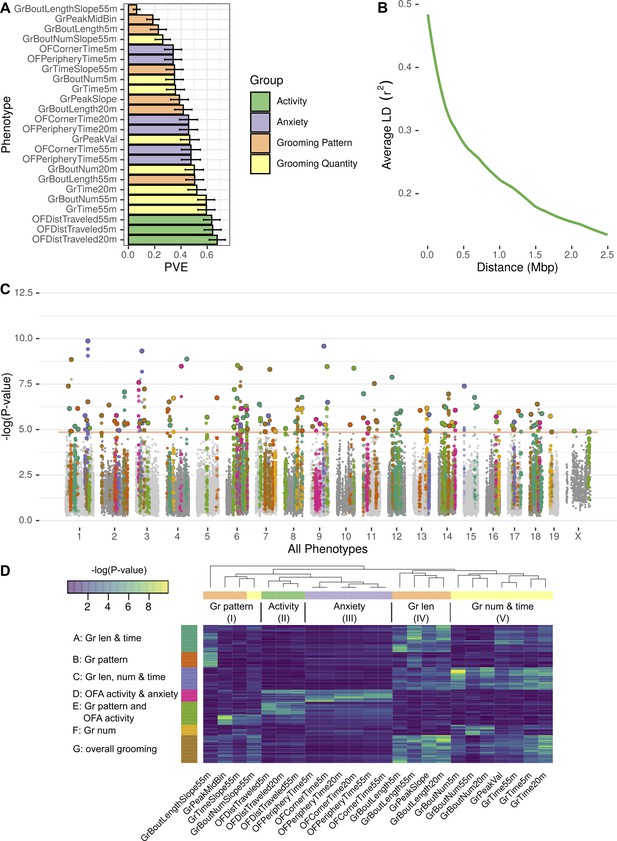
GWAS analysis of grooming and open field behaviors.
(A) Heritability (PVE) estimates of the computed phenotypes. (B) Linkage disequilibrium (LD) blocks size, average genotype correlations for SNPs in different genomic distances. (C) GWAS results shown as a Manhattan plot of all of the phenotypes combined, colors are according to peak SNPs clusters (from D), all the SNPs in the same LD block are colored according to the peak SNP. Minimal p-value over all of the phenotypes for each SNP (D) Heatmap of all the significant peak SNPs for each. Each row (SNP) is colored according to the assigned cluster in the k-means clustering. The color from k-means cluster is used in C.
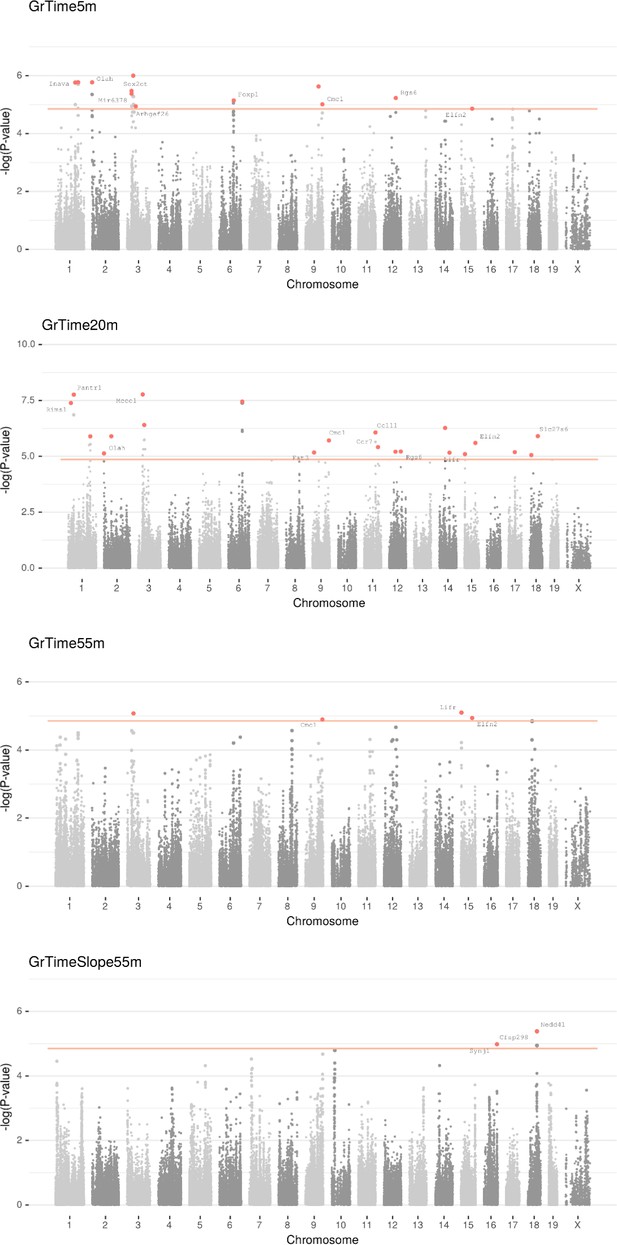
Manhattan plot for individual phenotypes.
Linear Mixed Model (LMM) results for each SNP genotype using Wald test, part 1 .
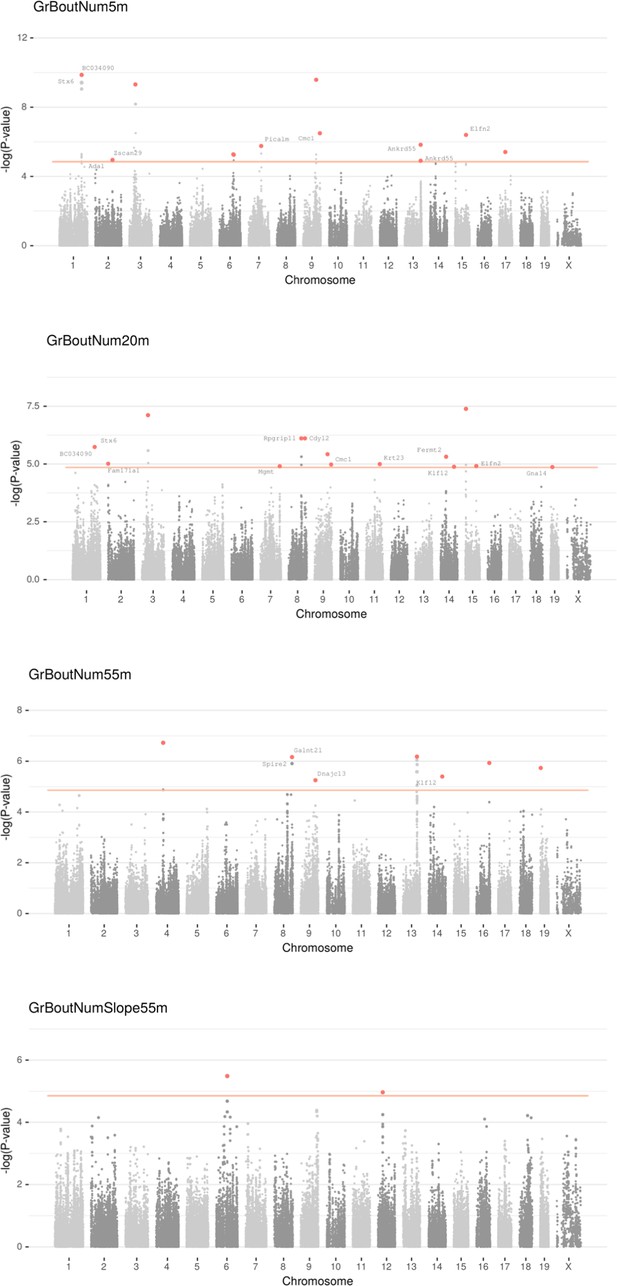
Manhattan plot for individual phenotypes.
Linear Mixed Model (LMM) results for each SNP genotype using Wald test, part 2 .
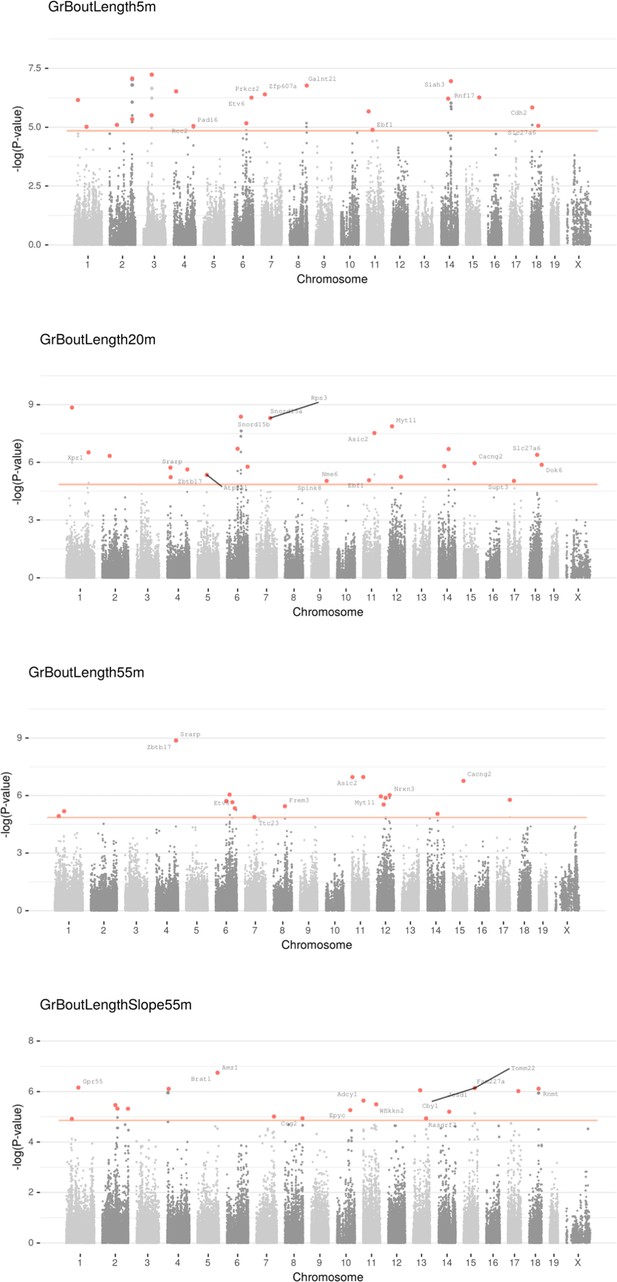
Manhattan plot for individual phenotypes.
Linear Mixed Model (LMM) results for each SNP genotype using Wald test, part 3 .
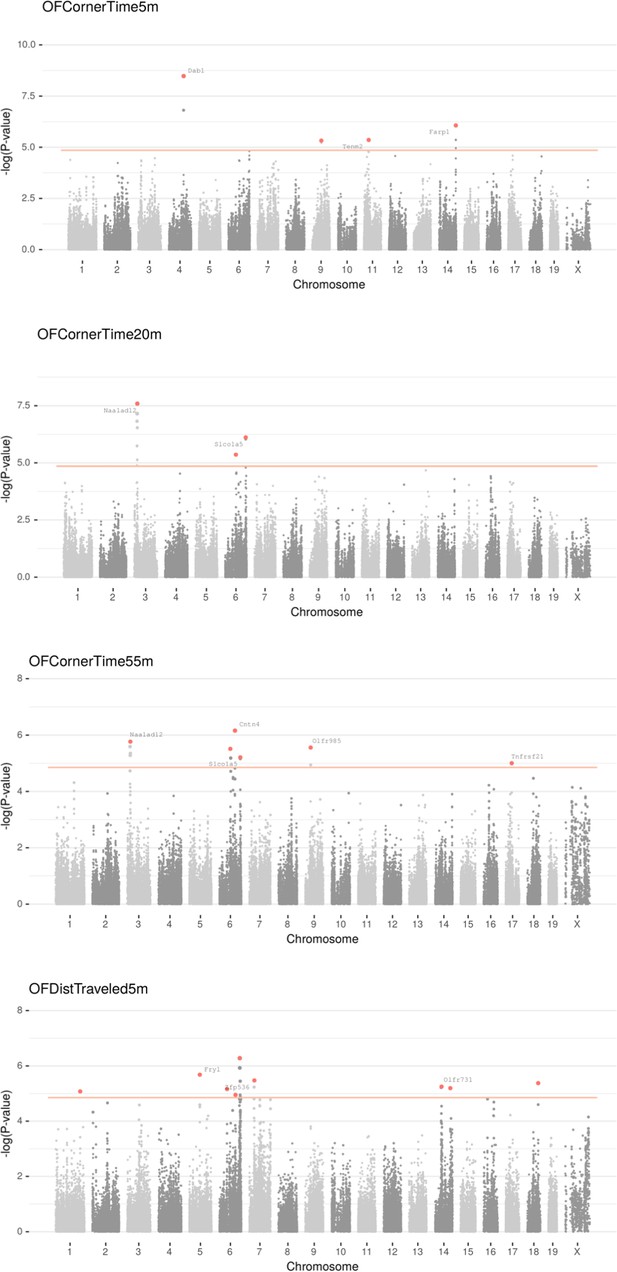
Manhattan plot for individual phenotypes.
Linear Mixed Model (LMM) results for each SNP genotype using Wald test, part 4 .
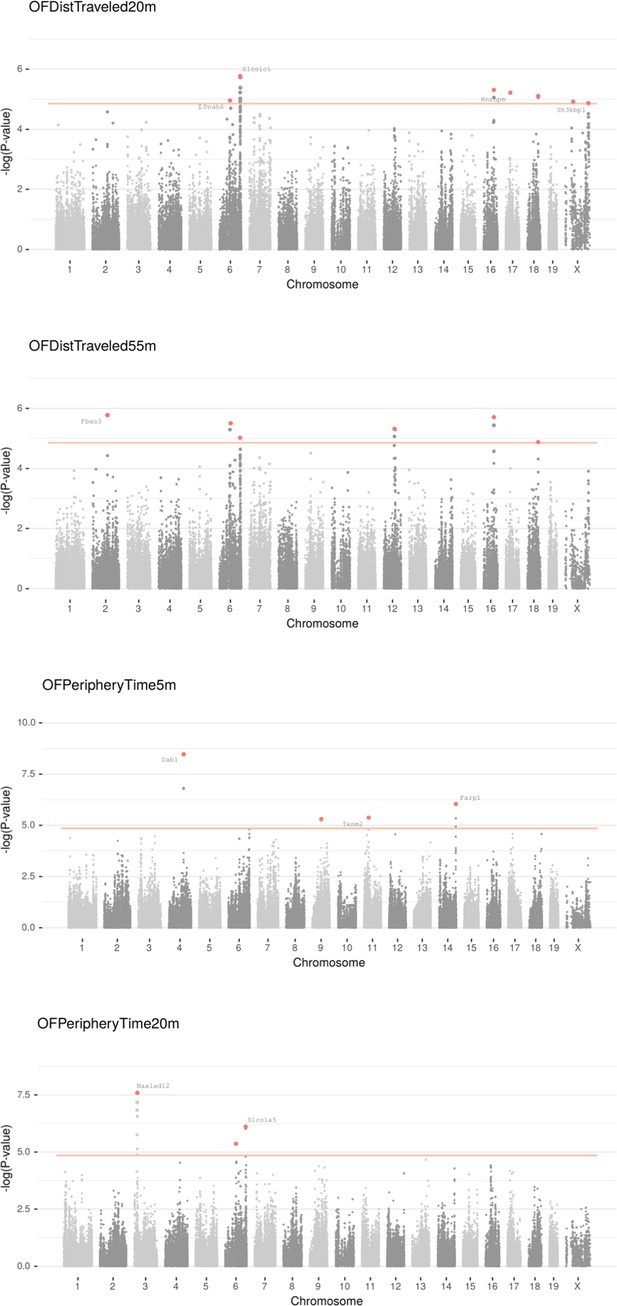
Manhattan plot for individual phenotypes.
Linear Mixed Model (LMM) results for each SNP genotype using Wald test, part 5 .
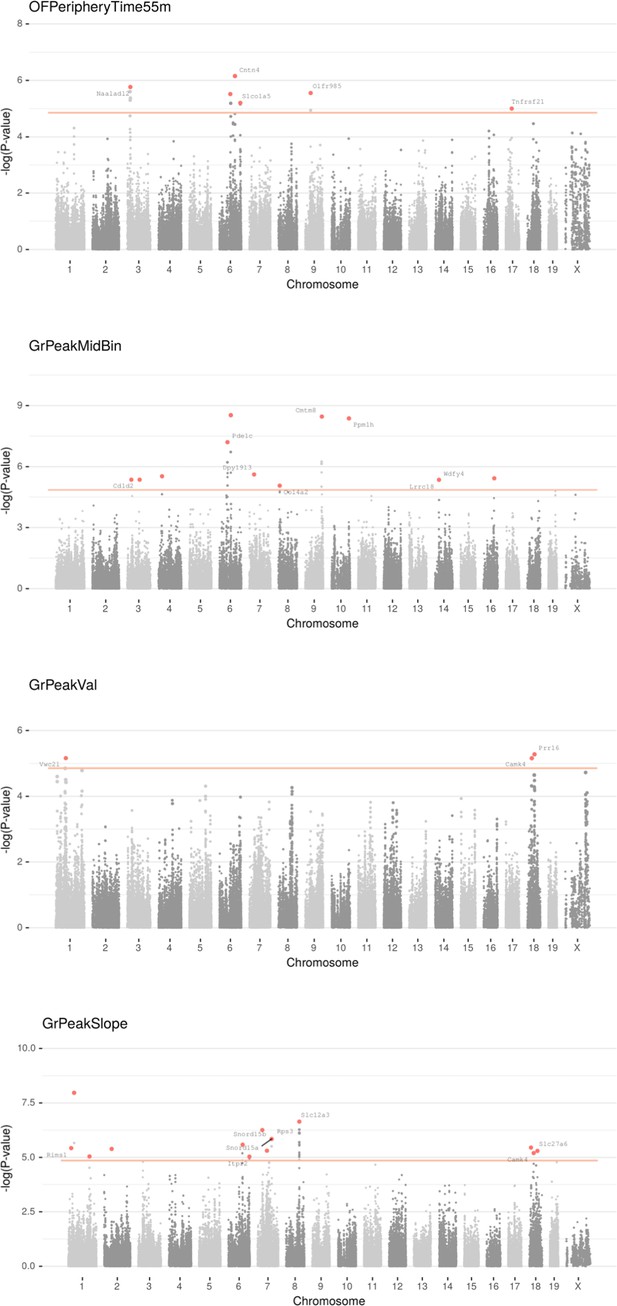
Manhattan plot for individual phenotypes.
Linear Mixed Model (LMM) results for each SNP genotype using Wald test, part 6.
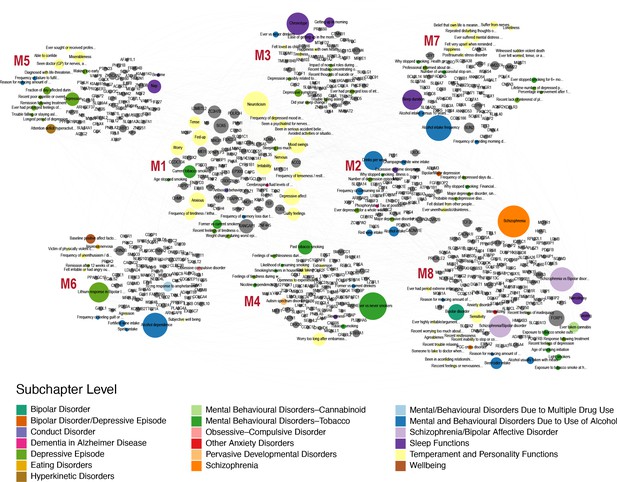
Human-Mouse trait relations through weighted bipartite network of PheWAS results.
We identified 509 human orthologs out of 860 mouse genes and focused on their association with traits in the Psychiatric domain of gwasATLAS with gene-level p value ≤ 0.001. We represented the relationships between these genes and Psychiatric traits by a weighted bipartite network. The width of an edge between a gene node (gray color) and a Psychiatric trait node is proportional to the association strength (-log10(p value)). The size of a node is proportional to the number of associated genes or traits and the color of a trait node corresponds to the subchapter level in the Psychiatric domain. Eight modules were identified and visualized using Gephi 0.9.2 software.
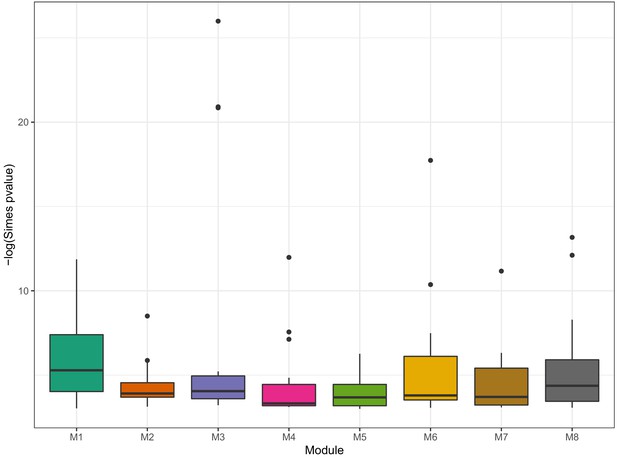
Box plot of Simes P values in eight modules.
Simes test was used to combine the p values of genes to obtain an overall p value for the association of each Psychiatric trait. Box plot showed the distribution of Simes p values in eight modules in Figure 10.
Additional files
-
Supplementary file 1
Full metadata related to the created annotated dataset.
Metadata columns includes strain, sex, arena, testing date, weight, training/validation split, animal number, starting frame for clip, duration of clip, clip identity in released dataset, and clip identity referenced in this paper.
- https://cdn.elifesciences.org/articles/63207/elife-63207-supp1-v2.xlsx
-
Supplementary file 2
Full results of the GWAS and PheWAS.
- https://cdn.elifesciences.org/articles/63207/elife-63207-supp2-v2.zip
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/63207/elife-63207-transrepform-v2.docx





