The genetic landscape for amyloid beta fibril nucleation accurately discriminates familial Alzheimer’s disease mutations
Figures
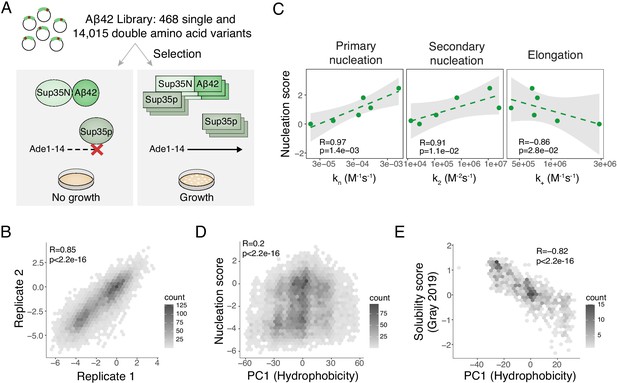
Deep mutagenesis of amyloid beta (Aß) nucleation.
(A) In vivo Aß selection assay. Aß fused to the Sup35N domain seeds aggregation of endogenous Sup35p causing a read-through of a premature UGA in the Ade1-14 reporter, allowing the cells to grow in medium lacking adenine. (B) Correlation of nucleation scores for biological replicates 1 and 2 for single and double amino acid (aa) mutants. Pearson correlation coefficient and p-value are indicated (Figure 1—figure supplement 1B) n = 10,157 genotypes. (C) Correlation of nucleation scores with in vitro primary and secondary nucleation and elongation rate constants (Yang et al., 2018). Weighted Pearson correlation coefficient and p-value are indicated. (D) Nucleation scores as a function of principal component 1 (PC1) aa property changes (Bolognesi et al., 2019) for single and double aa mutants (n = 14,483 genotypes). Weighted Pearson correlation coefficient and p-value are indicated. (E) Solubility scores (Gray et al., 2019) as a function of PC1 changes (Bolognesi et al., 2019) for n = 895 single and double mutants. Pearson correlation coefficient and p-value are indicated.
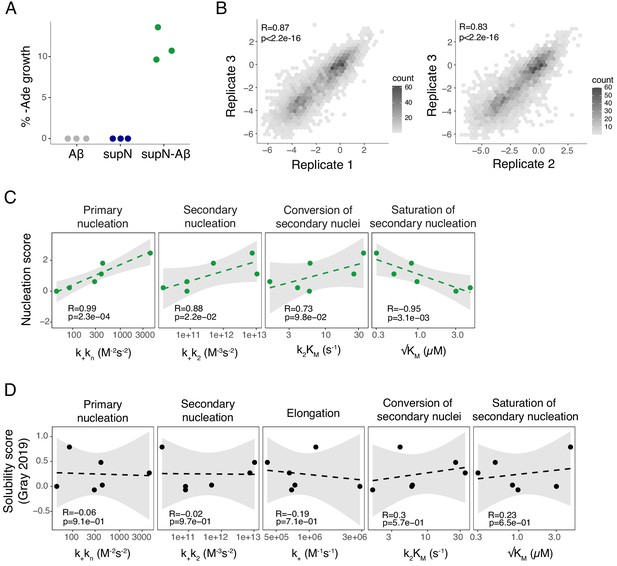
Reproducibility of the assay and correlation with in vitro fibril nucleation.
(A) Percentage of yeast growth in medium lacking adenine for sup35N, amyloid beta (Aß) and sup35N-Aß constructions. (B) Correlation of nucleation scores for biological replicates 1 and 3 (n = 4,124 genotypes) and 2 and 3 (n = 4093) for single and double amino acid (aa) mutants. Pearson correlation coefficient and p-value are indicated. (C) Correlation of nucleation scores with in vitro combined rate constants for primary nucleation, secondary nucleation, conversion of secondary nuclei and saturation of secondary nucleation (Yang et al., 2018). Weighted Pearson correlation coefficient and p-value are indicated. (D) Correlation of solubility scores (Yang et al., 2018) with in vitro combined rate constants for primary nucleation, secondary nucleation, elongation, conversion of secondary nuclei, and saturation of secondary nucleation (Yang et al., 2018). Weighted Pearson correlation coefficient and p-value are indicated.
-
Figure 1—figure supplement 1—source data 1
Raw colony counts from independent testing of the strains expressing the variants reported in Figure 1—figure supplement 1A.
- https://cdn.elifesciences.org/articles/63364/elife-63364-fig1-figsupp1-data1-v2.xlsx
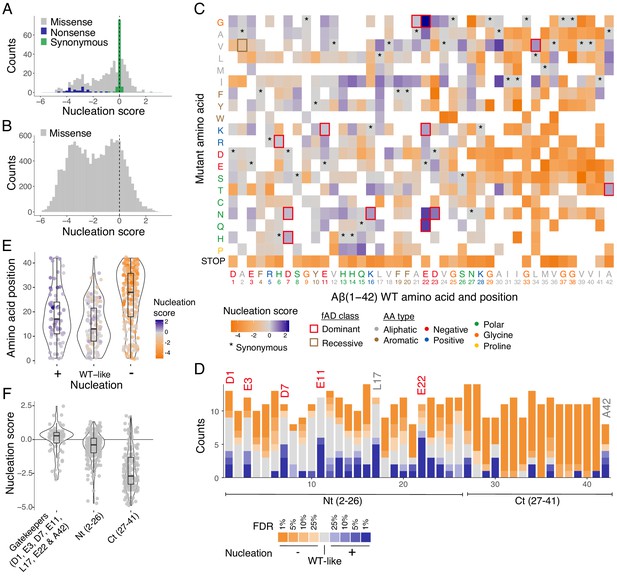
Modular organization of mutational effects in amyloid beta (Aß).
(A and B) Nucleation scores distribution for single (A) and double (B) amino acid (aa) mutants. n = 468 (missense), n = 31 (nonsense), n = 90 (synonymous) for singles, and n = 14,015 (missense) for doubles. Vertical dashed line indicates wild-type (WT) score (0). (C) Heatmap of nucleation scores for single aa mutants. The WT aa and position are indicated in the x-axis and the mutant aa is indicated on the y-axis, both coloured by aa class. Variants not present in the library are represented in white. Synonymous mutants are indicated with ‘*’ and familial Alzheimer’s disease (fAD) mutants with a box, coloured by fAD class. (D) Number of variants significantly increasing (blue) and decreasing (orange) nucleation at different false discovery rates (FDRs). Gatekeeper positions (D1, E3, D7, E11, L17, E22, and A42) are indicated on top of the corresponding bar and coloured on the basis of aa type. The N-terminal and C-terminal definitions are indicated on the x-axis. Gatekeeper positions are excluded from the N-terminal and C-terminal classes. (E) Aa position distributions for variants that increase (+), decrease (−), or have no effect on nucleation (WT-like) (FDR < 0.1). (F) Nucleation score distributions for the three clusters of positions defined on the basis of nucleation: Nt (2-26), Ct (27-41), and gatekeeper positions (clusters are mutually exclusive). Horizontal line indicates WT nucleation score (0). Nonsense (stop) mutants were only included in A and C. Boxplots represent median values and the lower and upper hinges correspond to the 25th and 75th percentiles, respectively. Whiskers extend from the hinge to the largest value no further than 1.5*IQR (interquartile range). Outliers are plotted individually or omitted when the boxplot is plotted together with individual data points or a violin plot.
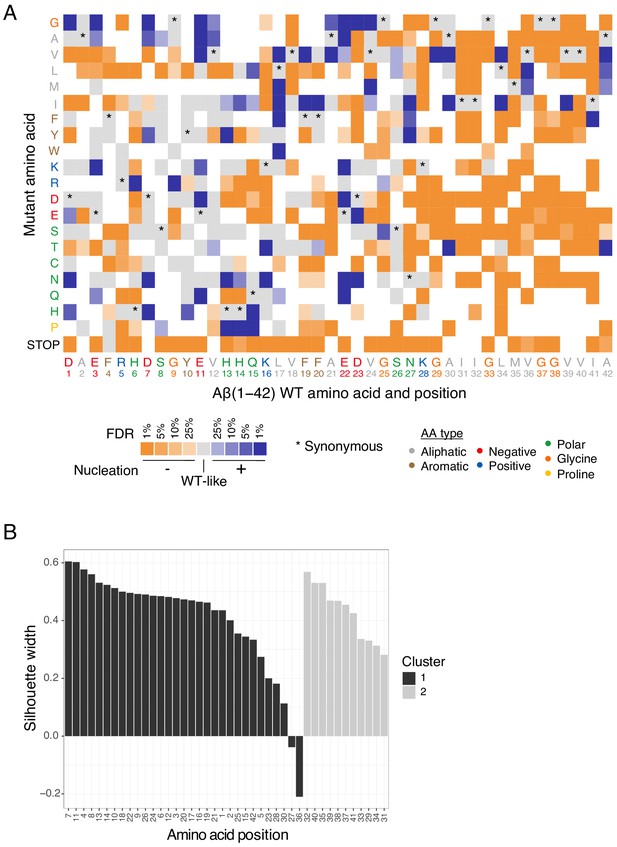
Mutational effects in amyloid beta (Aß).
(A) Heatmap of nucleation score categories for single amino acid (aa) variants at different false discovery rates (FDR) (blue: increased nucleation, orange: decreased nucleation, grey: not different from wild-type [WT]). The WT aa and position are indicated on the x-axis and the mutant aa is indicated on the y-axis, both coloured on the basis of aa type class (see legend). Variants not present in the library are represented in white. Synonymous mutants are indicated with ‘*’. Nonsense mutations (stop) were included. (B) K-medoids clustering of single aa variant nucleation scores estimates by residue position. Silhouette widths for all positions with K = 2.
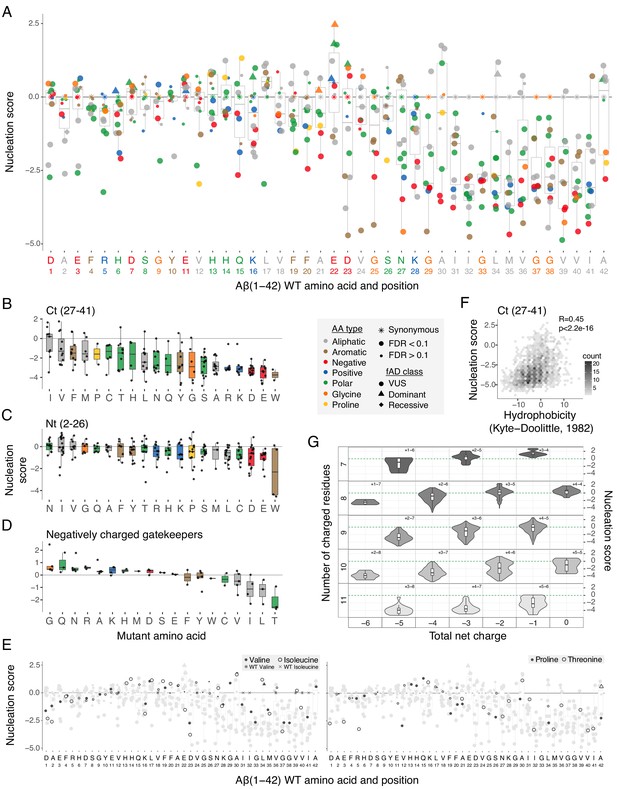
Determinants of amyloid beta (Aß) nucleation.
(A) Effect of single aa mutants on nucleation for each Aß position. The wild-type (WT) aa and position are indicated on the x-axis and coloured on the basis of aa type. The horizontal line indicates the WT nucleation score (0). (B to D) Effect of each mutant aa on nucleation for the Ct (27-41) (B), the Nt (2-26) (C), and the negatively charged gatekeeper positions (D1, E3, D7, E11, and E22) (D). The three position clusters are mutually exclusive. Colour code indicates aa type. The horizontal line is set at the WT nucleation score (0). (E) Effect on nucleation for single aa mutations to proline, threonine, valine, and isoleucine. Mutations to other aa are indicated in grey. The horizontal line indicates WT nucleation score (0). Point size and shape indicate false discovery rate (FDR) and familial Alzheimer’s disease (fAD) class, respectively (see legend). (F) Nucleation scores as a function of hydrophobicity changes (Kyte and Doolittle, 1982) for single and double aa mutants in the Ct (27-41) cluster. Only double mutants with both mutations in the indicated position-range were used. Weighted Pearson correlation coefficient and p-value are indicated. (G) Nucleation score distributions arranged by the number of charged residues (y-axis) and the total net charge (x-axis) for single and double aa mutants in the full peptide (1-42). Only polar, charged, and glycine aa types were taken into account, for both WT and mutant residues. Colour gradient indicates the total number of charged residues. Numbers inside each cell indicate the number of positive and negative residues. The horizontal line indicates WT nucleation score (0). Boxplots represent median values and the lower and upper hinges correspond to the 25th and 75th percentiles, respectively. Whiskers extend from the hinge to the largest value no further than 1.5*IQR (interquartile range). Outliers are plotted individually or omitted when the boxplot is plotted together with individual data points or a violin plot.
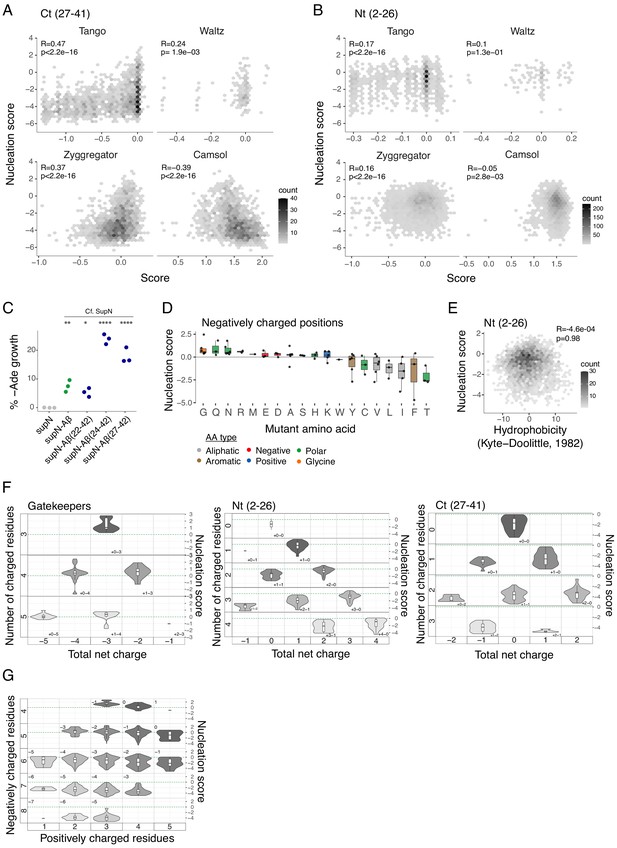
Determinants of amyloid beta (Aß) nucleation.
(A and B) Nucleation scores as a function of aggregation predictors (Tango, Waltz, Zyggregator, and Camsol; Tartaglia and Vendruscolo, 2008; Fernandez-Escamilla et al., 2004; Oliveberg, 2010; Sormanni et al., 2015) for single and double aa mutants in the Ct (27-41) (A) and Nt (2-26) (B) clusters (gatekeeper positions are excluded from the N-terminal and C-terminal classes). Only double mutants with both mutations in the indicated position-range were used. Weighted Pearson correlation coefficient and p-value are indicated. (C) Percentage of yeast growth in medium lacking adenine for sup35N, supN-Aß, and supN-Aß C-terminal fragments 22-42, 24-42, and 27-42. One-way ANOVA with Dunnett’s multiple comparisons test. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. (D) Effect of each mutant aa on nucleation for the six negatively charged positions (D1, E3, D7, E11, E22, and D23). Colour code indicates aa type (see legend). The horizontal line is set at the wild-type (WT) nucleation score (0). (E) Nucleation scores as a function of hydrophobicity changes (Kyte and Doolittle, 1982) for single and double aa mutants in the Nt (2-26) cluster. Only double mutants with both mutations in the indicated position-range were used. Weighted Pearson correlation coefficient and p-value are indicated. (F) Nucleation scores distributions arranged by the number of charged residues (y-axis) and the total net charge (x-axis) for single and double aa mutants in the Nt (2-26), Ct (27-41), and gatekeepers (D1, E3, D7, E11, L17, E22, and A42) clusters. Only double mutants with both mutations in the indicated position-range were used. Only polar, charged, and glycine aa types were taken into account, for both WT and mutant residues. Colour gradient indicates the total number of charged residues. Numbers inside each cell indicate the number of positive and negative residues. The horizontal line indicates WT nucleation score (0). (F) Nucleation score distributions arranged by the number of negatively charged residues (y-axis) and the number of positively charged (x-axis) for single and double aa mutants in the full peptide (1-42). Only polar, charged, and glycine aa types were taken into account, for both WT and mutant residues. Colour gradient indicates the total number of charged residues. Numbers inside each cell indicate the total net charge. The horizontal line indicates WT nucleation score (0). Boxplots represent median values and the lower and upper hinges correspond to the 25th and 75th percentiles, respectively. Whiskers extend from the hinge to the largest value no further than 1.5*IQR (interquartile range). Outliers are plotted individually or omitted when the boxplot is plotted together with individual data points or a violin plot.
-
Figure 3—figure supplement 1—source data 1
Raw colony counts from indepednet testing of the strains expressing the N-terminal truncated varaints reported in Figure 3—figure supplement 1C.
- https://cdn.elifesciences.org/articles/63364/elife-63364-fig3-figsupp1-data1-v2.xlsx
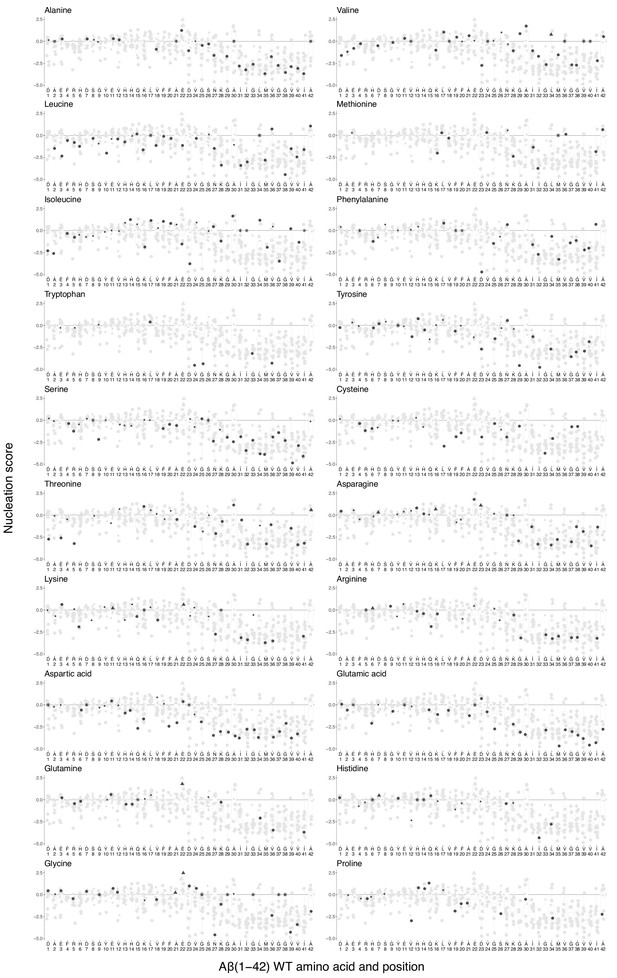
Effect of mutations to each specific amino acid (aa) on amyloid beta (Aß) nucleation.
Mutations to other aa are indicated in grey. The wild-type (WT) AA is indicated with ‘*’. The horizontal line indicates WT nucleation score (0). Point size indicates false discovery rate (FDR) (big, FDR < 0.1; small, FDR > 0.1) and shape indicates familial Alzheimer’s disease (fAD) class (round, variants of uncertain significance [VUS]; triangle, dominant; rhombus, recessive). Boxplots represent median values and the lower and upper hinges correspond to the 25th and 75th percentiles, respectively. Whiskers extend from the hinge to the largest value no further than 1.5*IQR (interquartile range). Outliers are plotted individually or omitted when the boxplot is plotted together with individual data points or a violin plot.
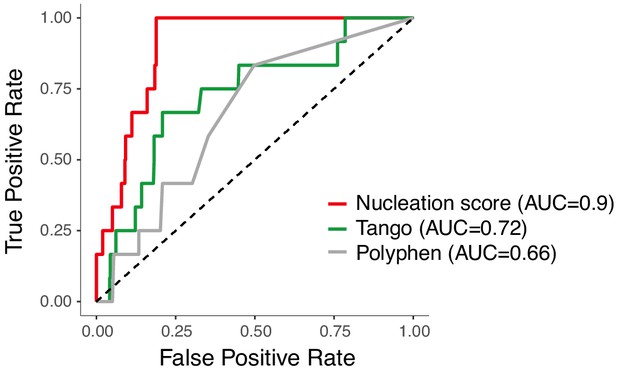
Amyloid beta (Aß) nucleation accurately discriminates dominant familial Alzheimer’s disease (fAD) variants.
Receiver operating characteristic (ROC) curves for 12 fAD mutants versus all other single aa mutants in the dataset. Area under the curve (AUC) values are indicated in the legend. Diagonal dashed line indicates the performance of a random classifier. The nucleation scores and categories for all fAD variants are reported in Supplementary file 1.
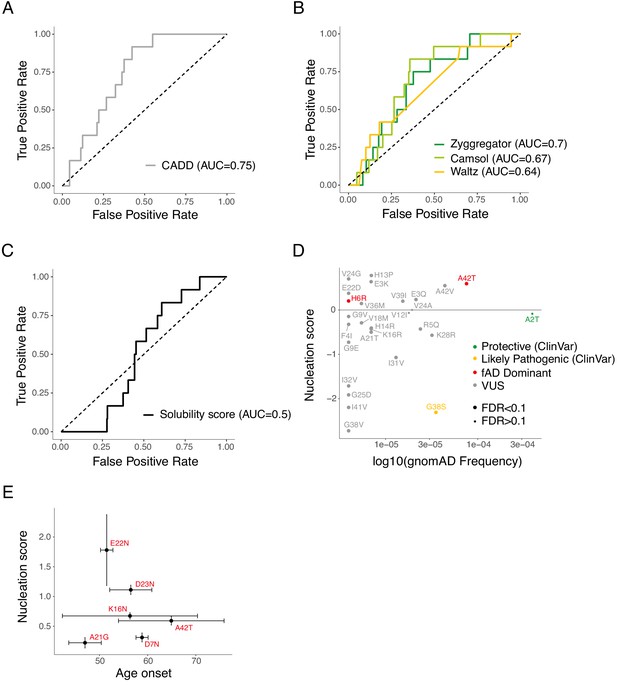
Discrimination of familial Alzheimer’s disease (fAD) variants by aggregation and variant effect predictors.
(A and B) Receiver operating characteristic (ROC) curves built using 12 fAD mutants versus all other single amino acid (aa) mutants in the dataset for variant effect predictors (A) and aggregation predictors (B). Area under the curve (AUC) values are indicated in the legend. Diagonal dashed lines indicate the performance of a random classifier. (C) ROC curve built using 12 fAD mutants versus all other single aa mutants available in the referenced study (Gray et al., 2019). AUC value is indicated in the legend. Diagonal dashed line indicates the performance of a random classifier. (D) Comparison between nucleation scores (NS) and gnomAD (Karczewski, 2020) allele frequencies (https://gnomad.broadinstitute.org/). The horizontal line indicates wild-type (WT) NS (0). The classification of variants is based on Clinvar annotations (https://www.ncbi.nlm.nih.gov/clinvar/). (E) Relation between NS and clinical age-of-onset (Ryman et al., 2014). Vertical and horizontal error lines indicate estimated error associated to NS and standard deviation for age-of-onset, respectively.
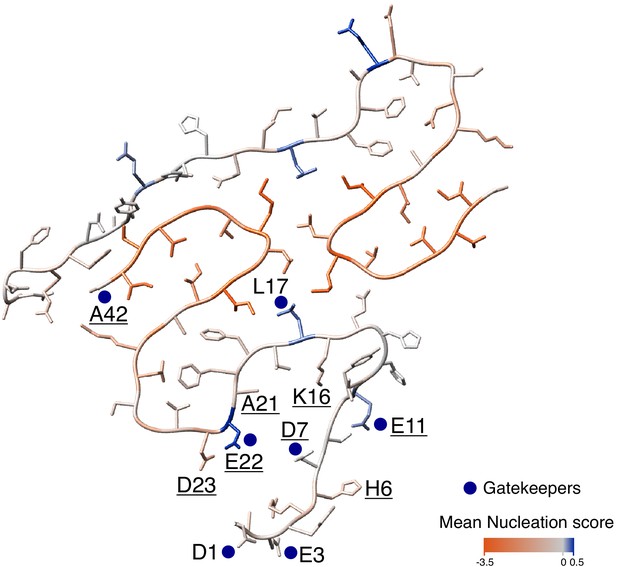
Mutational landscape of the amyloid beta (Aß) amyloid fibril.
Average effect of mutations visualized on the cross-section of an Aß amyloid fibril (PDB accession 5KK3; Colvin et al., 2016). Nucleation gatekeeper residues and known familial Alzheimer’s disease (fAD) mutations positions are indicated by the wild-type (WT) aa identity on one of the two monomers; gatekeepers are indicated with blue dots and fAD are underlined. A single layer of the fibril is shown and the unstructured N-termini (aa 1-14) are shown with different random coil conformations for the two Aß monomers. See Figure 5—figure supplement 2 for alternative Aß42 amyloid polymorphs.
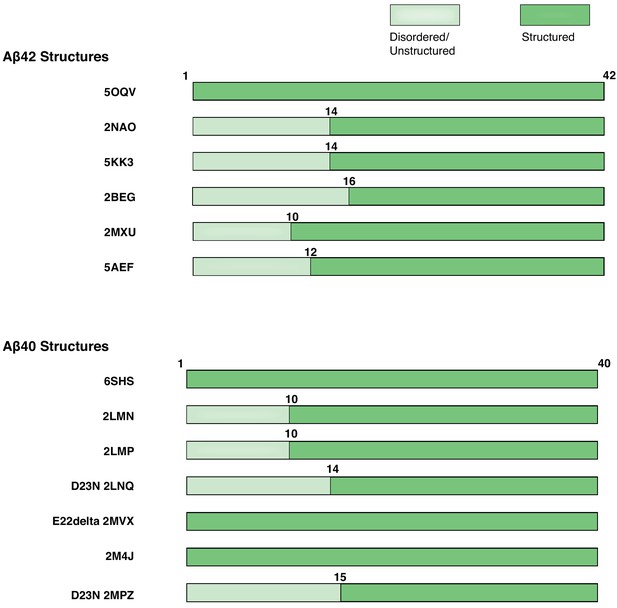
Modular organization of Aß42 and Aß40 polymorphs.
Linear organization of the Aß42 and Aß40 fibrils. Disordered/unstructured and structured residues are indicated.
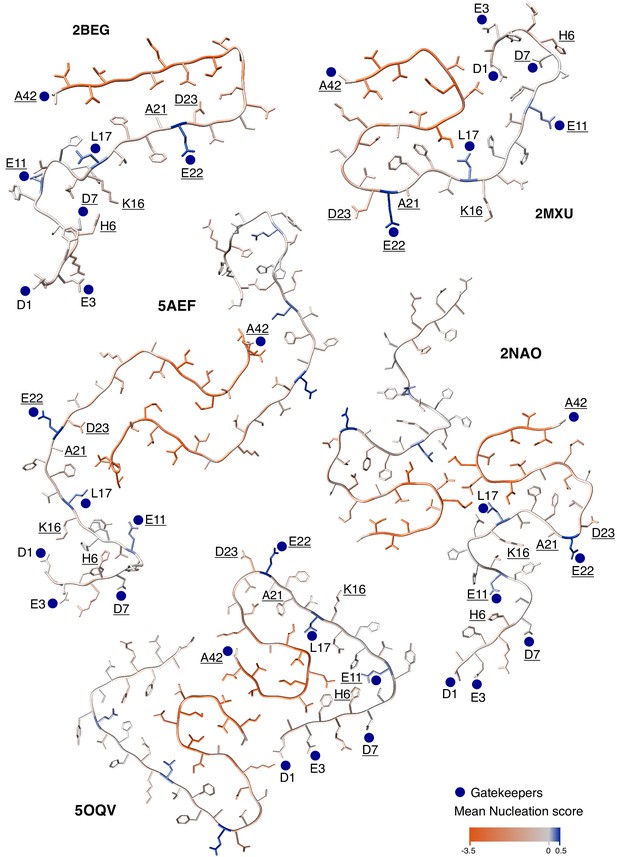
Modular organization of mutational effects and gatekeepers visualized on Aß42 polymorphs.
Average effect of mutations visualized on the cross-section of various amyloid beta (Aß) amyloid polymorphs: PDB accession 2BEG (Lührs et al., 2005), 2MXU (Xiao et al., 2015), 5AEF (Schmidt et al., 2015), 2NAO (Wälti et al., 2016), and 5OQV (Gremer et al., 2017). Nucleation gatekeeper residues and known familial Alzheimer’s disease (fAD) mutations positions are indicated by the wild-type (WT) aa identity on one of the two monomers; gatekeepers are indicated with blue dots and fAD are underlined. A single layer of the fibril is shown and the unstructured N-termini are shown with different random coil conformations for the two Aß monomers.
Additional files
-
Supplementary file 1
Table listing the impact on aggregation rates for 16 Aß42 variants for which these measurements could be retrieved from the literature.
For the same variants, the table also reports nucleation scores, as quantified in this study, and the qualitative agreement or disagreement with the previously published data.
- https://cdn.elifesciences.org/articles/63364/elife-63364-supp1-v2.xlsx
-
Supplementary file 2
Table listing the mutations in Aß42 that significantly increase nucleation score and that are therefore proposed as novel familial Alzheimer’s disease (fAD) candidates.
For each mutation, the corresponding nucleation score (NS) is reported.
- https://cdn.elifesciences.org/articles/63364/elife-63364-supp2-v2.xlsx
-
Supplementary file 3
List of oligonucleotides used in this study.
- https://cdn.elifesciences.org/articles/63364/elife-63364-supp3-v2.xlsx
-
Supplementary file 4
Processed data required to make all analyses and figures in this paper.
Read counts, nucleation scores, and associated error terms are reported for each Aß42 variant in each replicate. See sheet one for a deeper explanation of headers.
- https://cdn.elifesciences.org/articles/63364/elife-63364-supp4-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/63364/elife-63364-transrepform-v2.docx





