Mutations in SKI in Shprintzen–Goldberg syndrome lead to attenuated TGF-β responses through SKI stabilization
Figures
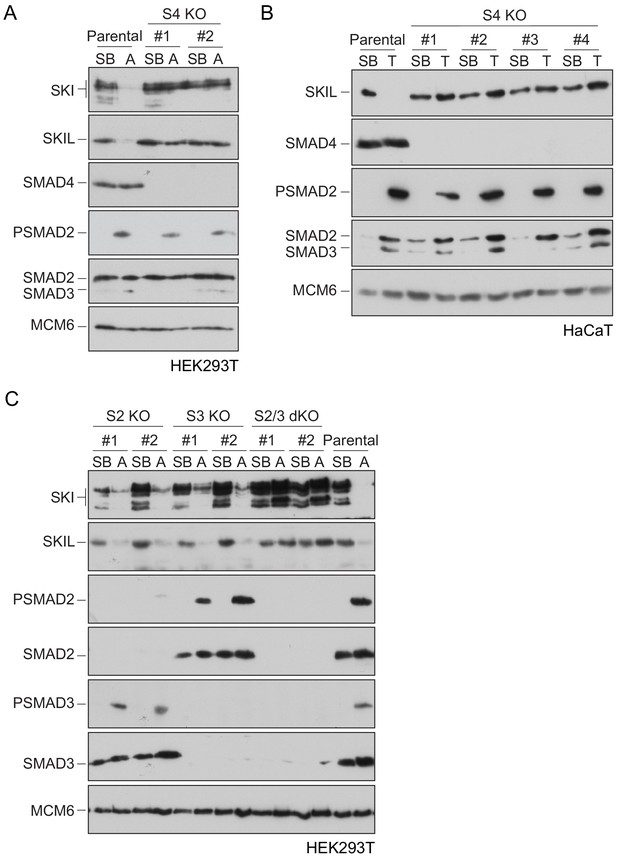
Requirement of SMAD2 or SMAD3 and SMAD4 for SKI and SKIL degradation.
(A and C) The parental HEK293T cell line and two individual SMAD4 knockout clones (A) or two individual SMAD2, SMAD3 knockout clones, or two SMAD2 and SMAD3 double knockout clones (C) were incubated overnight with 10 μM SB-431542, washed out, then incubated with full media containing either SB-431542 or 20 ng/ml Activin A for 1 hr, as indicated. Whole-cell extracts were immunoblotted with the antibodies indicated. (B) Parental HaCaT and four individual SMAD4 knockout clones were treated as above, except that they were treated with 2 ng/ml TGF-β for 1 hr instead of Activin A. Nuclear lysates were immunoblotted using the antibodies indicated. SB, SB-431542; A, Activin A; T, TGF-β; S2, SMAD2; S3, SMAD3; S2/3, SMAD2 and SMAD3; S4, SMAD4; KO, knockout; dKO, double knockout.
-
Figure 1—source data 1
Sequences of knockout alleles made in HEK293T cells.
- https://cdn.elifesciences.org/articles/63545/elife-63545-fig1-data1-v2.docx
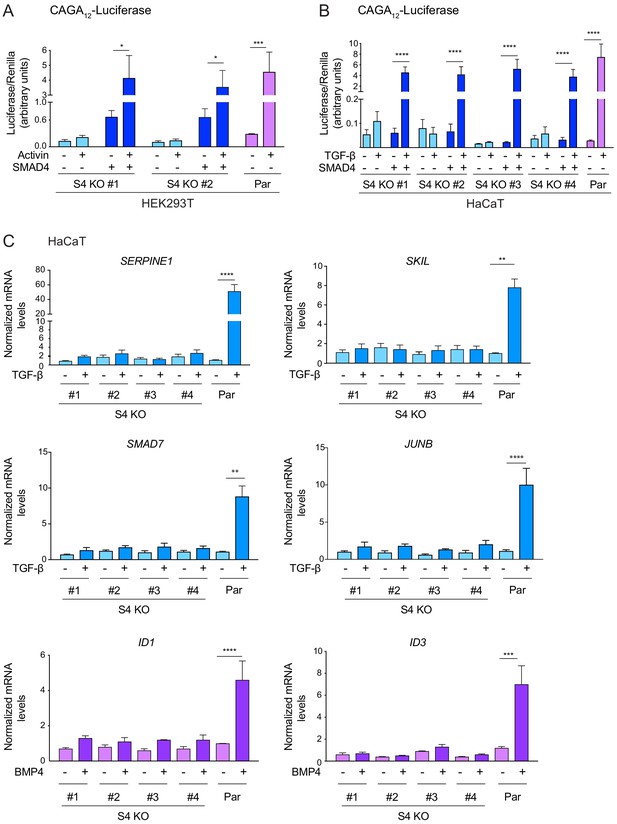
SMAD4 is essential for TGF-β/Activin-induced transcriptional responses.
(A and B) HEK293T (A) or HaCaT (B) parental cells and individual clones of SMAD4 knockout cells were transiently transfected with CAGA12-Luciferase together with TK-Renilla as an internal control and a plasmid expressing human SMAD4 as indicated (A and B). Cells were incubated with 0.5% fetal bovine serum-containing media overnight and then treated with 20 ng/ml Activin (A) or 2 ng/ml TGF-β (B) for 8 hr. Cell lysates were prepared and Luciferase/Renilla activity was measured. Plotted are the means ± SEM of three independent experiments. The p-values are from one-way ANOVA with Tukey’s post hoc correction. *p<0.05; ***p<0.001; ****p<0.0001. (C) HaCaT parental and four independent clones of SMAD4 knockout cells were either untreated or incubated with either 2 ng/ml TGF-β or 20 ng/ml BMP4 for 1 hr (SMAD7, JUNB, ID1, and ID3) or 6 hr (SERPINE1 and SKIL). Total RNA was extracted and qPCR was used to assess the levels of mRNA for the genes shown. The data are the average of three or four experiments ± SEM. The p-values are from one-way ANOVA with Tukey’s post hoc correction. **p<0.01; ***p<0.001; ****p<0.0001. Par, parental; S4 KO, SMAD4 knockout.
-
Figure 1—figure supplement 1—source data 1
Luciferase assay data for HEK293T S4 KO clones, as presented in Figure 1—figure supplement 1A.
- https://cdn.elifesciences.org/articles/63545/elife-63545-fig1-figsupp1-data1-v2.xlsx
-
Figure 1—figure supplement 1—source data 2
Luciferase assay data for HaCaT S4 KO clones, as presented in Figure 1—figure supplement 1B.
- https://cdn.elifesciences.org/articles/63545/elife-63545-fig1-figsupp1-data2-v2.xlsx
-
Figure 1—figure supplement 1—source data 3
qPCR data for HaCaT S4 KO clones, as presented in Figure 1—figure supplement 1C.
- https://cdn.elifesciences.org/articles/63545/elife-63545-fig1-figsupp1-data3-v2.xlsx
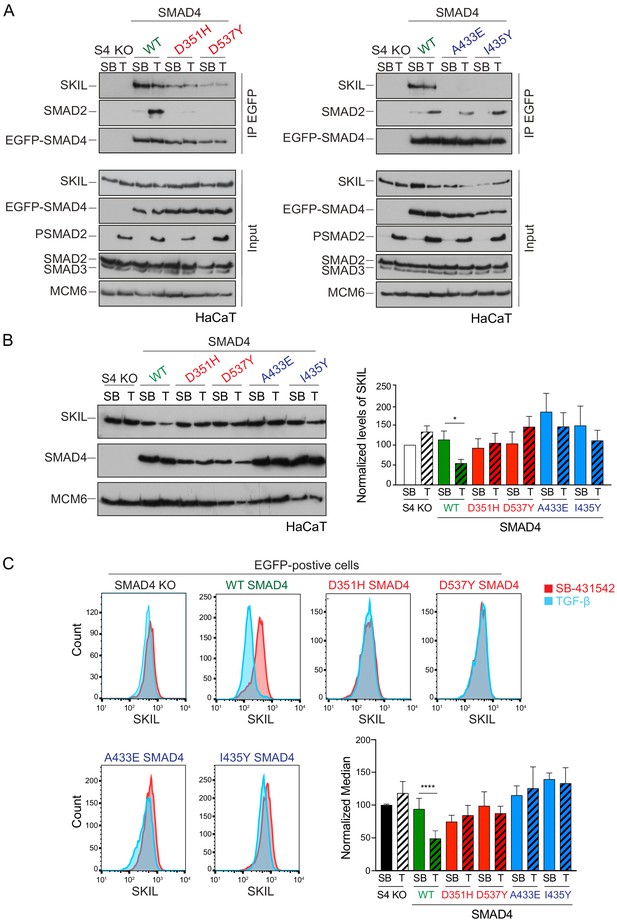
Characterization of the role of SMAD4 in TGF-β-induced SKIL degradation.
(A–C) HaCaT SMAD4 knockout (S4 KO) cells were stably transfected with EGFP alone, or EGFP SMAD4 (WT) or with four different EGFP-SMAD4 mutants (D351H, D537Y, which abolish interaction with the R-SMADs, and A433E and I435Y, which do not interact with SKIL). (A) Cells were incubated overnight with 10 µM SB-431542, washed out and pre-incubated with 25 μM MG-132 for 3 hr, and then treated either with 10 μM SB-431542 or 2 ng/ml TGF-β for 1 hr. Whole-cell extracts were immunoprecipitated (IP) with GFP-trap agarose beads. The IPs were immunoblotted using the antibodies shown. Inputs are shown below. (B) Nuclear lysates were prepared from the HaCaT S4 KO cells stably transfected with EGFP alone or with EGFP-SMAD4 constructs as indicated, treated as in (A), but without the MG-132 step and immunoblotted using the antibodies shown. On the right the quantifications are the normalized average ± SEM of five independent experiments. The quantifications are expressed as fold changes relative to SB-431542-treated S4 KO cells. (C) Levels of SKIL in the EGFP-positive S4 KO rescue cell lines treated as in (B), assayed by flow cytometry. Each panel shows an overlay of the indicated treatment conditions. The red line indicates the SB-431542-treated sample, whereas the cyan line indicates the TGF-β-treated sample. Quantifications are shown bottom right. For each group, the percentage of the median fluorescence intensity normalized to the SB-431542-treated sample is quantified. Data are the mean ± SEM of five independent experiments. The p-values are from one-way ANOVA with Sidak’s post hoc correction *p<0.05; ****p<0.0001. SB, SB-431542; T, TGF-β.
-
Figure 2—source data 1
Quantification of Western blot for HaCaT S4 KO rescue cell lines, as presented in Figure 2B.
- https://cdn.elifesciences.org/articles/63545/elife-63545-fig2-data1-v2.xlsx
-
Figure 2—source data 2
Flow cytometry data for HaCaT S4 KO rescue cell lines, as presented in Figure 2C.
- https://cdn.elifesciences.org/articles/63545/elife-63545-fig2-data2-v2.xlsx
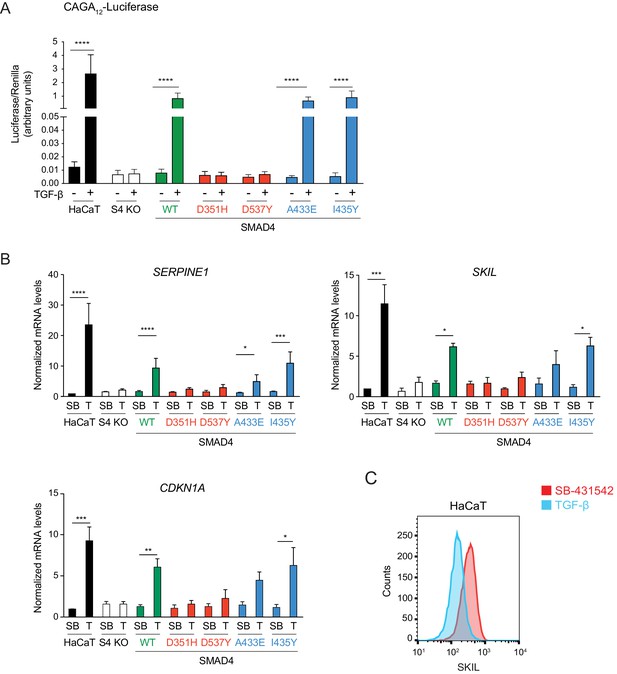
Transcriptional activity of the SMAD4 mutants compared to WT SMAD4.
(A) Parental HaCaT, SMAD4 knockout clone 2 stably transfected with EGFP alone (S4 KO), or with EGFP fusions of WT SMAD4 or the four indicated SMAD4 mutants were transiently transfected with CAGA12-Luciferase together with TK-Renilla as an internal control. Cells were untreated or treated with 2 ng/ml TGF-β for 8 hr. Luciferase/Renilla activity was measured on whole-cell lysates. Plotted are the means ± SEM of four independent experiments. The p-values are from one-way ANOVA with Tukey’s post hoc correction. ****p<0.0001. (B) Parental HaCaT and SMAD4 knockout clone 2 cells stably transfected as in (A) were incubated overnight with 10 μM SB-431542, washed out, and then retreated with SB-431542 (SB) or with 2 ng/ml TGF-β (T) for 6 hr. Total RNA was extracted and qPCR was performed for the genes shown. Plotted are the means ± SEM of four independent experiments. The p-values are from two-way ANOVA with Sidak’s post hoc test. *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001. (C) Levels of SKIL in parental HaCaT cells were measured by flow cytometry 1 hr after incubation with 10 μM SB-431542 or after treatment for 1 hr with 2 ng/ml TGF-β. The panel shows an overlay of the indicated treatment conditions. The red line indicates the SB-431542-treated sample, while the cyan line represents the TGF-β-treated sample.
-
Figure 2—figure supplement 1—source data 1
Luciferase assay data for HaCaT S4 KO rescue cell lines, as presented in Figure 2—figure supplement 1A.
- https://cdn.elifesciences.org/articles/63545/elife-63545-fig2-figsupp1-data1-v2.xlsx
-
Figure 2—figure supplement 1—source data 2
qPCR data for HaCaT S4 KO rescue cell lines, as presented in Figure 2—figure supplement 1B.
- https://cdn.elifesciences.org/articles/63545/elife-63545-fig2-figsupp1-data2-v2.xlsx
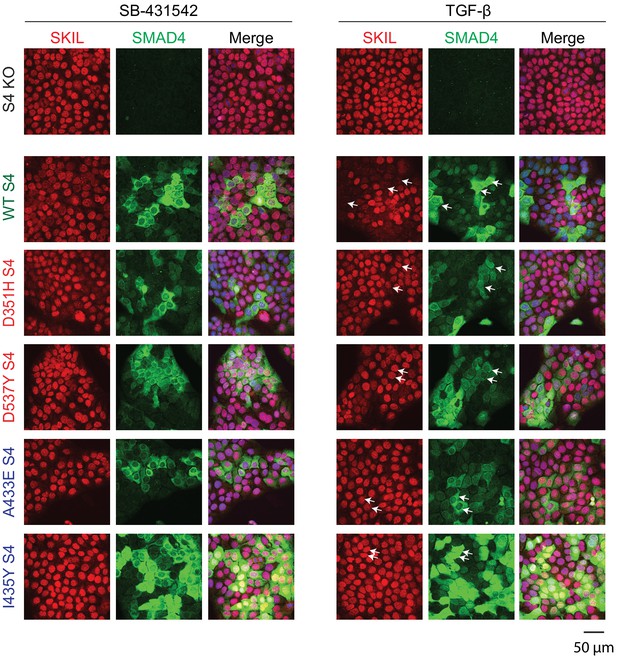
Visualization of TGF-β-induced SKIL degradation.
HaCaT SMAD4 knockout (S4 KO) cells or those stably expressing EGFP SMAD4 WT or EGFP SMAD4 mutants were incubated overnight with 10 μM SB-431542, washed out, and incubated for 1 hr with 10 µM SB-431542 or with 2 ng/ml TGF-β. Cells were fixed and stained for EGFP (for SMAD4), SKIL, and with DAPI (blue) to mark nuclei and imaged by confocal microscopy. The merge combines SKIL, SMAD4, and DAPI staining. Arrows indicate examples of EGFP-expressing cells and corresponding levels of nuclear SKIL. Scale bar corresponds to 50 µm.
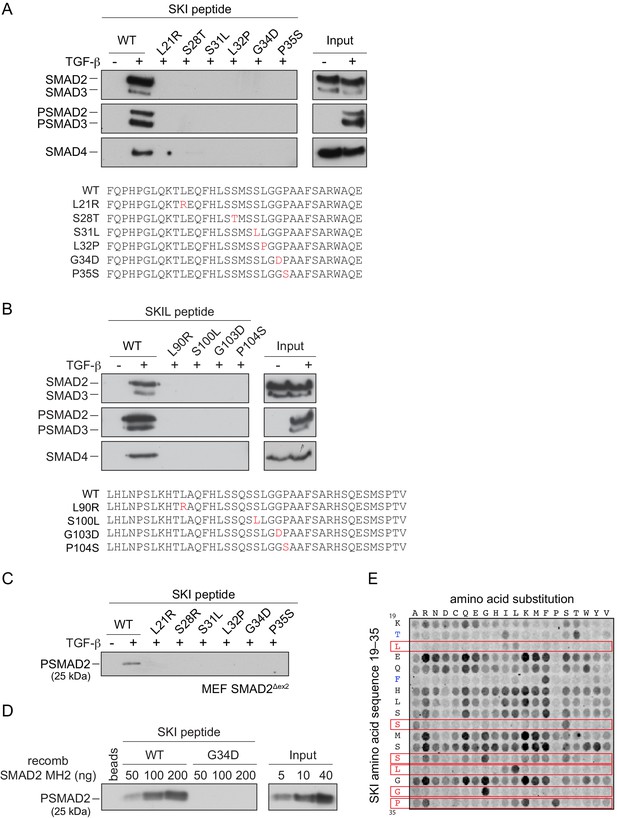
SGS mutations inhibit binding of SKI to SMAD2/3.
(A and B) HaCaT cells were treated or not with 2 ng/ml TGF-β. A peptide pulldown assay was performed on whole cells extracts and pulldowns were immunoblotted with the antibodies indicated. Inputs are shown on the right. (A) Wild-type (WT) SKI peptides corresponding to amino acids 11–45 or containing SGS point mutations as shown in red were used. (B) WT SKIL peptides corresponding amino acids 80–120 or containing mutations (in red) corresponding to SGS mutations in SKI were used. (C) WT SKI peptides or those containing SGS point mutations were used in pulldown assays with whole-cell extracts of SMAD2-null mouse embryonic fibroblasts that express just the MH2 domain of SMAD2 (MEF SMAD2Δex2) (Das et al., 2009), treated with 2 ng/ml TGF-β. The untreated sample is only shown for the WT SKI peptide. A PSMAD2 immunoblot is shown. (D) A recombinant trimer of phosphorylated SMAD2 MH2 domain was used in a peptide pulldown assay with WT and G34D SKI peptides. A PSMAD2 immunoblot is shown, with inputs on the right. (E) Mutational peptide array of SKI peptides (amino acids 11–45), mutated at all residues between amino acids 19 and 35, was probed with a recombinant PSMAD3–SMAD4 complex, which was visualized using a SMAD2/3 antibody conjugated to Alexa 488. On each row, the indicated amino acid is substituted for every other amino acid. A representative example is shown. See Figure 4—figure supplement 1C and Figure 4—source data 2 for quantification of the peptide arrays.
-
Figure 4—source data 1
Peptide sequences for peptide array.
- https://cdn.elifesciences.org/articles/63545/elife-63545-fig4-data1-v2.pdf
-
Figure 4—source data 2
Quantification of peptide arrays.
- https://cdn.elifesciences.org/articles/63545/elife-63545-fig4-data2-v2.xlsx
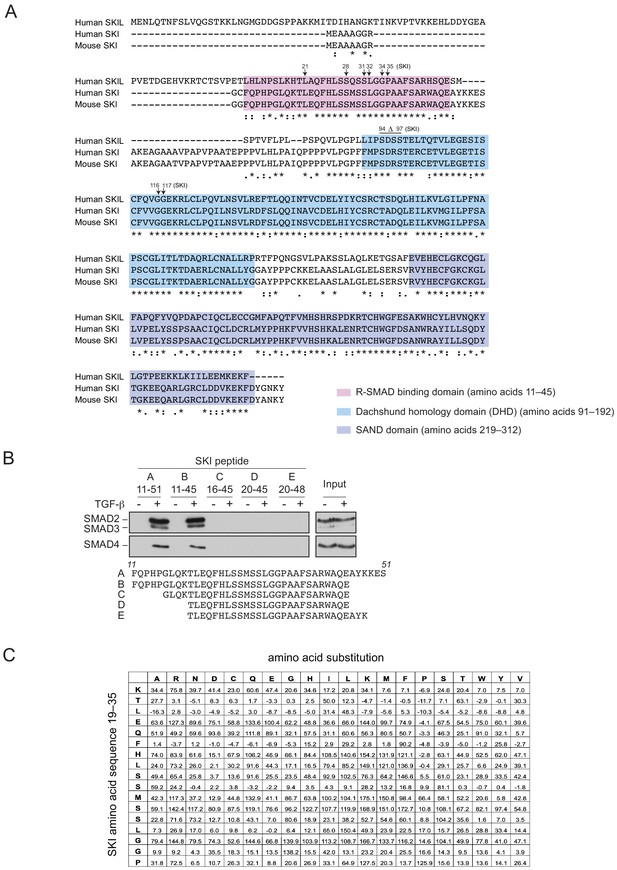
SGS mutations in SKI.
(A) Alignment of the first 317 amino acids of human SKI with the corresponding regions of mouse SKI and human SKIL is shown. Key domains are shown: pink corresponds to the R-SMAD-binding domain; blue, DHD domain; purple, SAND domain. SGS mutations are indicated by arrows. (B) SKI peptides corresponding to amino acids 11–51 and truncated versions as shown were analyzed in peptide pulldown assays with whole cell extracts from HaCaT cells treated with or without 2 ng/ml TGF-β. The pulldowns were immunoblotted using the antibodies shown. Inputs are shown on the right. (C) Quantification of the mutational peptide array of SKI peptides (amino acids 11–45), a representative of which is shown in Figure 4E. Each intensity measure is normalized to the average intensity of 60 positive controls of the WT peptide after subtracting the background, measured from the average intensity of 60 negative controls (truncated SKI peptide C as indicated in B). The values shown are the mean normalized intensities for each mutated peptide. See also Figure 4—source data 2.
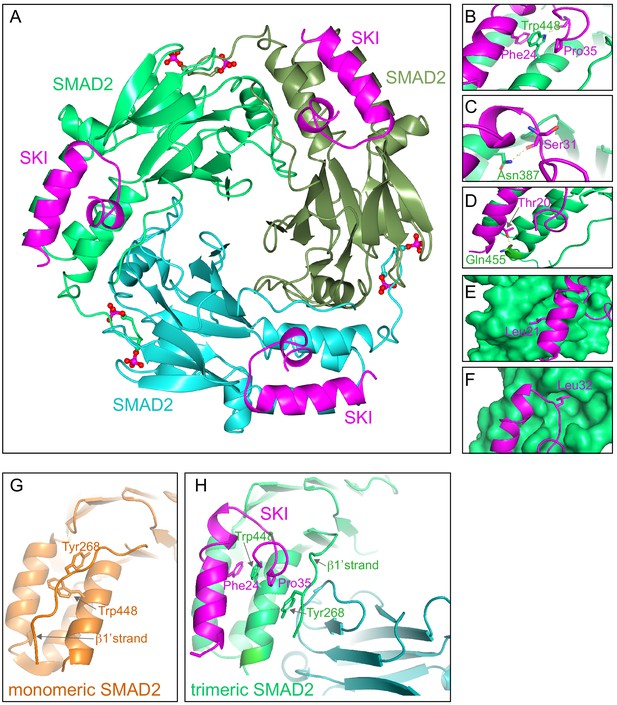
Crystal structure of PSMAD2 MH2 domain and N-terminal SKI peptide.
(A) Crystal structure of the phosphorylated SMAD2 MH2 domain trimer (the three monomers are shown in bright green, cyan, and olive) with the N-terminal SKI peptide amino acids 11–45 (magenta). A ribbon representation is shown. The C-terminal phosphates are indicated with a ball and stick representation (red and magenta). (B–F) Close ups on key residues for SKI binding. SKI residues are shown in magenta, and SMAD2 residues are in green. In (B–D), a ribbon representation is shown. In (E and F), SMAD2 is shown as a surface representation and SKI as a ribbon. (G) A detail from the structure of monomeric SMAD2 MH2 domain with a peptide from ZFYVE9 (formerly called SARA) (Wu et al., 2000). Note that the β1’ strand that contains Tyr268 is locked in a hydrophobic pocket, forcing Trp448 into flattened orientation, incompatible with SKI binding. (H) A detail from the structure in (A) indicating how SMAD2 complex formation shifts the position of the β1’ strand and more particularly, Tyr268, allowing Trp448 to flip 90°, enabling it to stack with SKI residues Phe24 and Pro35.
-
Figure 5—source data 1
Structure validation report for crystal structure (ID: 6ZVQ).
- https://cdn.elifesciences.org/articles/63545/elife-63545-fig5-data1-v2.pdf
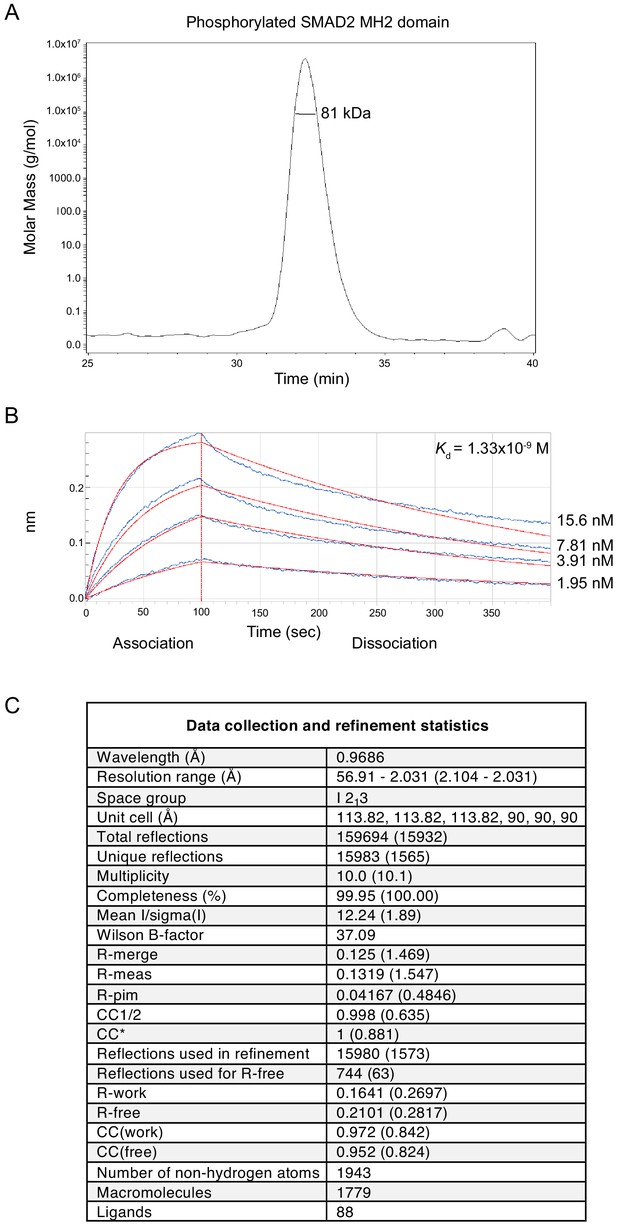
Analysis of the phosphorylated SMAD2 MH2 domain complex used for structural studies.
(A) The trimeric arrangement of the phosphorylated SMAD2 MH2 domain was confirmed by SEC-MALLS. The SEC-MALLS chromatogram is shown. The calculated molecular weight was 81 kDa, which was very close to the expected molecular weight of 78.5 kDa. (B) The interaction between the phosphorylated SMAD2 MH2 domain and the SKI peptide (amino acids 11–45) was measured by biolayer interferometry. The biosensors were loaded with biotinylated SKI peptide and incubated with different concentrations of phosphorylated SMAD2 MH2 domain as shown. The calculated Kd was 1.33 × 10−9 ± 2.12 x10−11 M. (C) Data collection and refinement statistics for the crystal structure of the phosphorylated SMAD2 MH2 domain complex with the N-terminal region of SKI shown in Figure 5A.
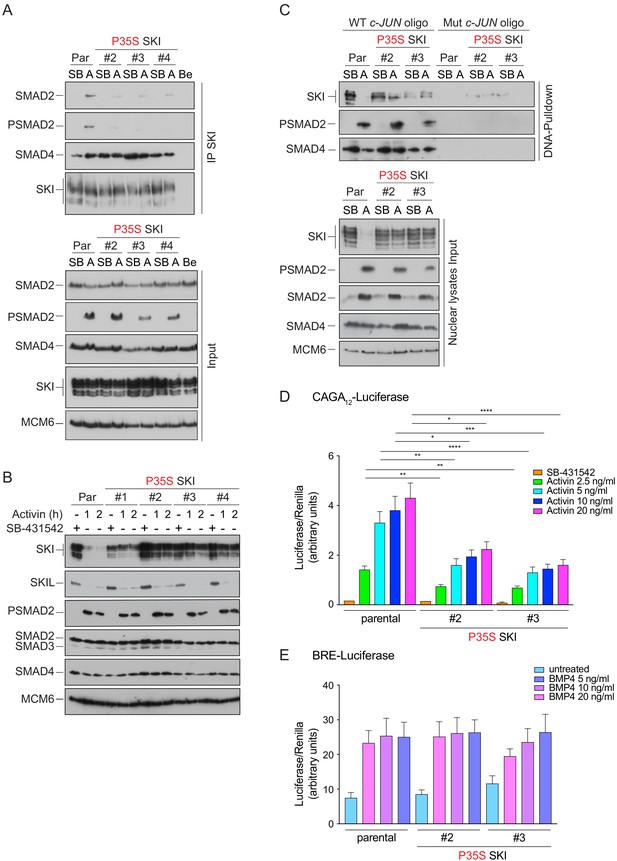
Knockin of an SGS mutation into SKI in HEK293T cells inhibits SKI degradation and inhibits Activin-induced transcription.
(A) Parental HEK293T and three independent P35S SKI knockin clones were incubated overnight with 10 μM SB-431542, washed out, and treated for 3 hr with 25 μM MG-132 and then with either SB-431542 or 20 ng/ml Activin A for an additional 1 hr. Whole-cell lysates were immunoprecipitated (IP) with SKI antibody or beads alone (Be). The IPs were immunoblotted using the antibodies shown. Inputs are shown below. (B) Parental HEK293T and four independent P35S SKI knockin clones were incubated with 10 μM SB-431542 overnight, washed out, and incubated with either SB-431542 or 20 ng/ml Activin for the times indicated. Whole-cell lysates were immunoblotted using the antibodies indicated. (C) Cells were treated as in (B), and nuclear lysates were prepared and analyzed by DNA pulldown assay using the wild-type c-Jun SBE oligonucleotide or a version mutated at the SMAD3–SMAD4 binding sites (top panel). Inputs are shown in the bottom panel. HEK293T parental and two independent P35S SKI knockin clones were stably transfected with the CAGA12-Luciferase reporters (D) or the BRE-Luciferase reporter (E) with TK-Renilla as an internal control. Cells were serum starved with media containing 0.5% fetal bovine serum and 10 μM SB-431542 overnight. Subsequently, cells were washed and treated with Activin A (D) or BMP4 (E) at the concentrations indicated for 8 hr. Cell lysates were prepared and assayed for Luciferase and Renilla activity. Plotted are the means and SEM of seven (D) or four (E) independent experiments, with the ratio of Luciferase:Renilla shown. *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001. The p-values are from two-way ANOVA with Tukey’s post hoc test. A, Activin; SB, SB-431542; Par, parental.
-
Figure 6—source data 1
Luciferase assays for Activin A-induced HEK293T P35S SKI clones, as presented in Figure 6D.
- https://cdn.elifesciences.org/articles/63545/elife-63545-fig6-data1-v2.xlsx
-
Figure 6—source data 2
Luciferase assays for BMP4-induced HEK293T P35S SKI clones, as presented in Figure 6E.
- https://cdn.elifesciences.org/articles/63545/elife-63545-fig6-data2-v2.xlsx
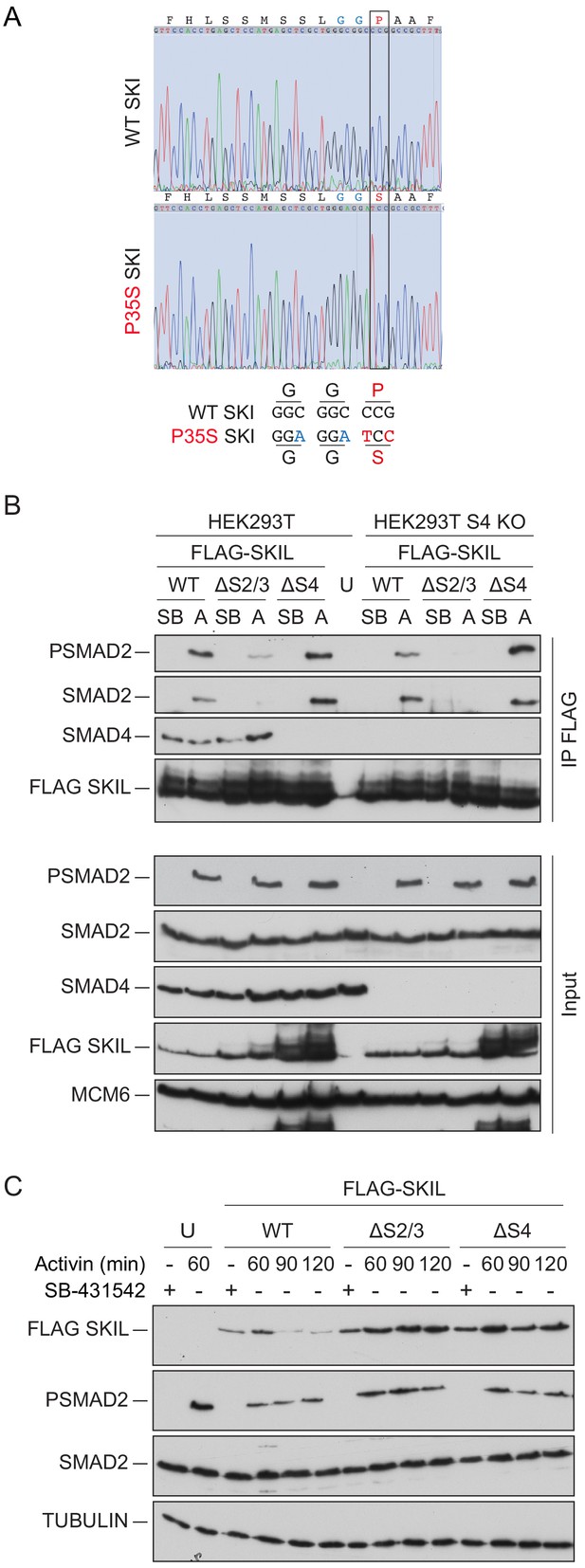
Mutation of the R-SMAD binding domain or SAND domain in SKI/SKIL prevents ligand-induced SKI/SKIL degradation.
(A) Characterization of P35S SKI mutation in HEK293T cells compared to parental cells by PCR followed by Sanger sequencing analysis. The black box indicates the change in nucleotides that give rise to the desired mutation. The details of the knocked in changes are given below. (B) HEK293T parental and SMAD4 knockout (S4 KO) cells were transiently transfected with FLAG-SKIL WT or FLAG-SKIL G103V (ΔS2/3) or FLAG-SKIL R314A, T315A, H317A, and W318E (ΔS4) as indicated, or left untransfected (U). Cells were incubated overnight with 10 μM SB-431542, then washed out, and pre-treated with 25 μM MG-132 for 3 hr, followed by incubation with SB-431542 or with 20 ng/ml Activin A for 1 hr. Whole cell extracts were immunoprecipitated (IP) with FLAG beads. The IPs were immunoblotted using the antibodies shown. Inputs are shown below. (C) HEK293T cells were untransfected (U) or transfected with the plasmids indicated as in (B). Cells were incubated overnight with 10 μM SB-431542, then washed out, and subsequently treated with SB-431542 or with 20 ng/ml Activin A for the times shown. Whole-cell extracts were immunoblotted using the antibodies shown. SB, SB-431542; A, Activin A.
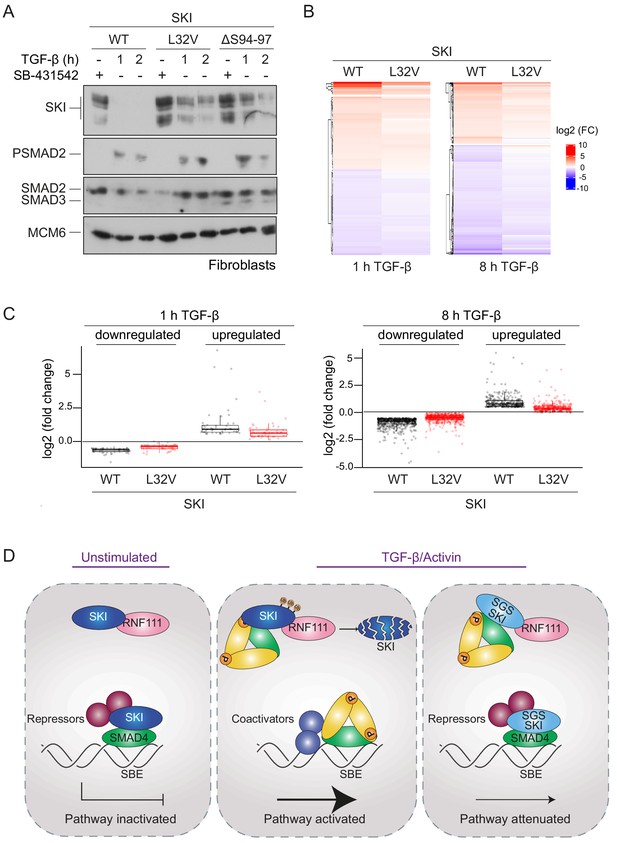
SGS mutations in SKI inhibit TGF-β-induced transcriptional responses in fibroblasts derived from SGS patients.
(A) Fibroblasts derived from a healthy subject carrying WT SKI and from two SGS patients carrying the L32V or the ΔS94-97 heterozygous mutations in SKI were incubated overnight with 10 μM SB-431542, washed out, and either re-incubated with SB-431542 or 2 ng/ml TGF-β for the times indicated. Whole-cell lysates were immunoblotted using the antibodies indicated. (B) Hierarchically clustered heatmaps of log2FC values (relative to the SB-431542-treated samples) showing the expression of TGF-β-responsive genes in the healthy fibroblasts and the L32V SKI fibroblasts after 1 hr and 8 hr of TGF-β treatment, analyzed by RNA-seq. Four biological replicates per condition were analyzed. The genes shown are those for which the TGF-β inductions were statistically significant in the healthy fibroblasts, but non-significant in the L32V fibroblasts. (C) The same data as in (B) are presented as box plots. (D) Model for the mechanism of action of WT SKI and mutated SKI. The left panel shows the unstimulated condition. In the nuclei, SKI (blue) is complexed with RNF111 (pink) and is also bound to DNA at SBEs with SMAD4 (green) forming a transcriptionally repressive complex with other transcriptional repressors (maroon). In the middle panel, TGF-β/Activin stimulation induces the formation of phosphorylated R-SMAD–SMAD4 complexes (yellow and green), which induce WT SKI degradation by RNF111. This allows an active PSMAD3–SMAD4 complex to bind SBEs and activate transcription. In the right panel, SGS-mutated SKI (light blue) is not degraded upon TGF-β/Activin stimulation, due to its inability to interact with PSMAD2 or PSMAD3. It therefore remains bound to SMAD4 on DNA, leading to attenuated transcriptional responses.
-
Figure 7—source data 1
RNA-seq raw data.
- https://cdn.elifesciences.org/articles/63545/elife-63545-fig7-data1-v2.xlsx
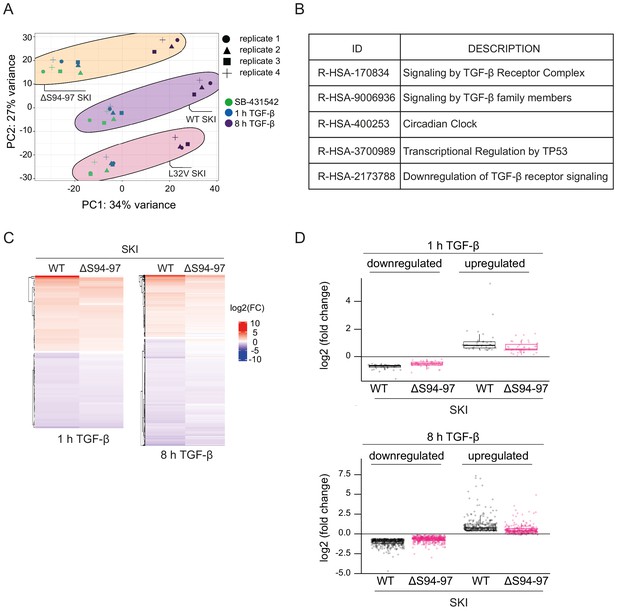
Dermal fibroblasts from SGS patients exhibit an attenuated TGF-β transcriptional response.
(A) Principal component analysis (PCA) plot is shown for RNA-seq on normal fibroblasts containing WT SKI, and fibroblasts from SGS patients containing either the L32V SKI mutation or the ΔS94-97 SKI mutation, treated as indicated. PCA measures sample to sample variation using rlog transformed read count of all genes expressed above one read in at least one sample. Four replicates are shown for each condition. (B) Enriched Reactome pathways common between the pairwise comparisons of time points (SB-431542-treated versus 1 hr TGF-β or 8 hr TGF-β) of fibroblasts derived from a healthy subject containing WT SKI. (C) Hierarchically clustered heatmaps of log2FC values (relative to SB-431542 condition) showing the expression of TGF-β-responsive genes in the healthy (WT SKI) and the ΔS94-97 SKI-containing fibroblasts after 1 hr and 8 hr treatment with TGF-β, analyzed by RNA-seq. Four biological replicates per condition were analyzed. The genes shown are those for which the TGF-β inductions were statistically significant in the healthy fibroblasts, but non-significant in the ΔS94-97 fibroblasts. (D) The same data as in (C) are presented as box plots.
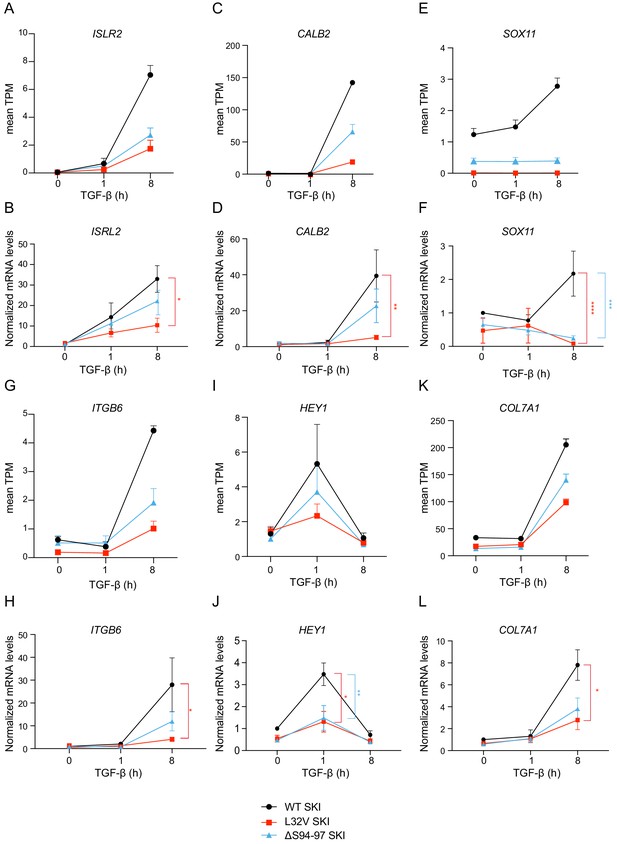
Validation of RNA-seq data by qPCR.
(A–L) Healthy dermal fibroblasts (WT SKI) and dermal fibroblasts from SGS patients containing either the L32V SKI mutation or the ΔS94-97 SKI mutation were treated overnight with 10 µM SB-431542, washed out, and incubated with either SB-431542 or 2 ng/ml TGF-β for 1 hr and 8 hr. Total RNA was extracted, and either RNA-seq or qPCR analysis was performed. Transcript levels for a subset of target genes are displayed as plots of the mean transcripts per million (TPM) from the RNA-seq data (A, C, E, G, I, K) or were measured by qPCR and plotted normalized to SB-431542-treated healthy fibroblasts (B, D, F, H, J, L). Plotted are the means and SEM of at least of four independent experiments. The p-values are from two-way ANOVA with Tukey’s post hoc test. *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001.
-
Figure 7—figure supplement 2—source data 1
qPCR validations of RNA-seq data for SGS and control dermal fibroblasts.
- https://cdn.elifesciences.org/articles/63545/elife-63545-fig7-figsupp2-data1-v2.xlsx
Videos
Mechanism of SKI binding to phosphorylated SMAD2 MH2 domain.
Animation of SMAD2 MH2 domain monomer (orange) forming a complex with two other SMAD2 MH2 domain monomers (cyan and olive). Note the movement of Trp448 on helix five and Tyr268 on the β1’ strand in the orange monomer upon trimerization. The flipped Trp448 is then in the correct orientation for binding to the SKI peptide (magenta).
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Homo sapiens) | HaCaT | Francis Crick Institute Cell Services | RRID:CVCL_0038 | Keratinocytes (immortalized adult) |
| Cell line (Homo sapiens) | HEK293T | Francis Crick Institute Cell Services | RRID:CVCL_0063 | Embryonic kidney cells (normal) |
| Cell line (Homo sapiens) | Dermal fibroblasts (normal, adult) | David Abraham, UCL, UK | Primary cells, male | |
| Cell line (Homo sapiens) | Dermal fibroblasts (carrying heterozygous mutation L32V SKI) | Carmignac et al., 2012 PMID:23103230 | Primary cells, female | |
| Cell line (Homo sapiens) | Dermal fibroblasts (carrying heterozygous mutation ΔS94-97 SKI) | Carmignac et al., 2012 PMID:23103230 | Primary cells, male | |
| Cell line (Homo sapiens) | HaCaT S4 KO #1; HaCaT S4 KO #2; HaCaT S4 KO #3; HaCaT S4 KO #4 | This paper | Keratinocytes in which SMAD4 is deleted by CRISPR/Cas9 technology | |
| Cell line (Homo sapiens) | HEK293T S2 KO #1; HEK293T S2 KO #2 | This paper | Embryonic kidney cells in which SMAD2 is deleted by CRISPR/Cas9 technology | |
| Cell line (Homo sapiens) | HEK293T S3 KO #1; HEK293T S3 KO #2 | This paper | Embryonic kidney cells in which SMAD3 is deleted by CRISPR/Cas9 technology | |
| Cell line (Homo sapiens) | HEK293T S2/S3 dKO #1; HEK293T S2/S3 dKO #2 | This paper | Embryonic kidney cells in which SMAD2 and SMAD3 are deleted simultaneously by CRISPR/Cas9 technology | |
| Cell line (Homo sapiens) | HEK293T S4 KO #1; HEK293T S4 KO #2 | This paper | Embryonic kidney cells in which SMAD4 is deleted by CRISPR/Cas9 technology | |
| Cell line (Homo sapiens) | HEK293T P35S SKI #1; HEK293T P35S SKI #2; HEK293T P35S SKI #3; HEK293T P35S SKI #4 | This paper | Embryonic kidney cells in which the P35S SKI mutation is introduced by CRISPR/Cas9 technology | |
| Cell line (Homo sapiens) | HEK293T CAGA12-Luciferase/Renilla | This paper | Embryonic kidney cells in which the CAGA12-Luciferase and TK-Renilla reporters are stably expressed | |
| Cell line (Homo sapiens) | HEK293T BRE-Luciferase/Renilla | This paper | Embryonic kidney cells in which the BRE-Luciferase and TK-Renilla reporters are stably expressed | |
| Cell line (Homo sapiens) | HEK293T P35S SKI CAGA12 Luciferase/Renilla #2; HEK293T P35S SKI CAGA12 Luciferase/Renilla #3 | This paper | HEK293T P35S SKI clones 2 and 3 in which the CAGA12-Luciferase and TK-Renilla reporters are stably expressed | |
| Cell line (Homo sapiens) | HEK293T P35S SKI BRE-Luciferase/Renilla #2; HEK293T P35S SKI BRE-Luciferase/Renilla #3 | This paper | HEK293T P35S SKI clones 2 and 3 in which the BRE-Luciferase and TK-Renilla reporters are stably expressed | |
| Cell line (Homo sapiens) | HaCaT S4 KO EGFP | This paper | HaCaT SMAD4 KO clone 2 cells stably expressing EGFP | |
| Cell line (Homo sapiens) | HaCaT SMAD4 KO rescue EGFP-SMAD4 WT | This paper | HaCaT SMAD4 KO clone 2 cells stably expressing EGFP-SMAD4 WT | |
| Cell line (Homo sapiens) | HaCaT SMAD4 KO rescue EGFP-SMAD4 D351H | This paper | HaCaT SMAD4 KO clone 2 cells stably expressing EGFP-SMAD4 D351H | |
| Cell line (Homo sapiens) | HaCaT SMAD4 KO rescue EGFP-SMAD4 D537Y | This paper | HaCaT SMAD4 KO clone 2 cells stably expressing EGFP-SMAD4 D537Y | |
| Cell line (Homo sapiens) | HaCaT SMAD4 KO rescue EGFP-SMAD4 A433E | This paper | HaCaT SMAD4 KO clone 2 cells stably expressing EGFP-SMAD4 A433E | |
| Cell line (Homo sapiens) | HaCaT SMAD4 KO rescue EGFP-SMAD4 I435Y | This paper | HaCaT SMAD4 KO clone 2 cells stably expressing EGFP-SMAD4 I435Y | |
| Cell line (M. musculus) | MEF SMAD2Δex2 | Piek et al., 2001 PMID:11262418 | Mouse embryo-derived fibroblasts carrying the homozygous null allele Smad2ex2 | |
| Cell line (Spodoptera frugiperda) | Sf21 | Fitzgerald et al., 2006 PMID:17117155 | Insect cells used to express recombinant proteins | |
| Transfected construct | pGFP-C1 | VectorBuilder | Clontech, Cat# 6084–1 | Construct used to make HaCaT SMAD4 KO stably expressing EGFP |
| Transfected construct (human) | pEGFP-SMAD4 WT | Nicolás et al., 2004 PMID:15280432 | Construct used to make HaCaT SMAD4 KO stably expressing EGFP SMAD4 WT | |
| Transfected construct (human) | pEGFP-SMAD4 D351H | This paper | Construct used to make HaCaT SMAD4 KO stably expressing EGFP-SMAD4 D351H | |
| Transfected construct (human) | pEGFP-SMAD4 D537Y | This paper | Construct used to make HaCaT SMAD4 KO stably expressing EGFP-SMAD4 D537Y | |
| Transfected construct (human) | pEGFP-SMAD4 A433E | This paper | Construct used to make HaCaT SMAD4 KO stably expressing EGFP-SMAD4 A433E | |
| Transfected construct (human) | pEGFP-SMAD4 I435Y | This paper | Construct used to make HaCaT SMAD4 KO stably expressing EGFP-SMAD4 I435Y | |
| Transfected construct | pRL-TK Vector | Promega | Cat# E2241 | Construct used to make HEK 293T cells stably expressing TK Renilla |
| Transfected construct | pGL3-BRE-Luc | Korchynskyi and ten Dijke, 2002 PMID:11729207 | Addgene Cat# 45126 | Construct used to make HEK 293T cells stably expressing Luciferase under control of the BRE |
| Transfected construct | pGL3 CAGA12-Luc | Dennler et al., 1998 PMID:9606191 | Construct used to make HEK 293T cells stably expressing Luciferase under control of the CAGA12 sequence | |
| Transfected construct | pSUPER.retro.puro | OligoEngine | Cat# VEC-pRT-0002 | Construct used to express resistance to puromycin in order to make stable cell lines |
| Transfected construct | pSpCas9(BB)−2A-GFP (PX458) | Ran et al., 2013 PMID:24157548 | Addgene Cat# 48138 | Different oligonucleotides corresponding to gRNAs have been cloned into this plasmid in order to make several different CRISPR/Cas9 knockout cell lines. |
| Antibody | Anti-phosphorylated SMAD2 (Rabbit monoclonal) | Cell Signaling Technology | Cat# 3108; RRID:AB_490941 | WB (1:1000) |
| Antibody | Anti-SMAD2/3 (mouse monoclonal) | BD Biosciences | Cat# 610843; RRID:AB_398162 | IF (1:500) WB (1:1000) |
| Antibody | Anti-phospho-SMAD3 (rabbit monoclonal) | Cell Signaling Technology | Cat# 9520; RRID:AB_2193207 | WB (1:500) |
| Antibody | Anti-SMAD4 (B-8) (mouse monoclonal) | Santa Cruz | Cat# sc-7966; RRID:AB_627905 | WB (1:1000) |
| Antibody | Anti-SMAD3 (Rabbit monoclonal) | Abcam | Cat# 40854; RRID:AB_777979 | WB (1:1000) |
| Antibody | Anti-FLAG DYKDDDDK Tag (L5) (Rat monoclonal) | Thermo Fisher | Cat# MA1-142, RRID:AB_2536846 | IP (5 µg) WB (1:1000) |
| Antibody | Anti-MCM6 (C-20) (Goat polyclonal) | Santa Cruz | Cat# sc-9843; RRID:AB_2142543 | WB (1:2000) |
| Antibody | Anti MCM6 (H-8) (Mouse monoclonal) | Santa Cruz | Cat# sc-393618 RRID:AB_2885187 | WB (1:2000) |
| Antibody | Anti-Tubulin (Rat monoclonal) | Abcam | Cat# ab6160; RRID:AB_305328 | WB (1:5000) |
| Antibody | Anti-SKI (Rabbit Polyclonal) | GeneTex | Cat# GTX133764 RRID:AB_2885186 | IP (5 µg) WB (1:2000) |
| Antibody | Anti-GFP (Goat polyclonal) | Abcam | Cat# ab6673, RRID:AB_305643 | IF (1:200) |
| Antibody | Anti SKIL (SnoN) (H-317) (Rabbit polyclonal) | Santa Cruz | Cat# sc-9141, RRID:AB_671124 | WB (1:1000) IF (1:1000) Flow cytometry (1:200) |
| Antibody | Donkey anti-Goat IgG (H+L) Cross-adsorbed secondary antibody, Alexa Fluor 546 | Thermo Fisher Scientific | Cat# A-11056, RRID:AB_2534103 | IF (1:1000) |
| antibody | Anti-Rabbit Alexa Fluor 594 | Thermo Fisher Scientific | Cat# A-21244; RRID:AB_10562581 | IF (1:1000) |
| Antibody | Anti-Rabbit Alexa Fluor 647 | Thermo Fisher Scientific | Cat# A-21244, RRID:AB_2535812 | Flow cytometry (1:1000) |
| Antibody | Goat anti-rabbit HRP | Dako | Cat# P0448; RRID:AB_2617138 | WB (1:5000) |
| Antibody | Goat anti-mouse HRP | Dako | Cat#P0447; RRID:AB_2617137 | WB (1:5000) |
| Antibody | Donkey anti-rat HRP | Jackson Lab | Cat#712-035-153; RRID:AB_2340639 | WB (1:5000) |
| Antibody | Rabbit anti-goat HRP | Dako | Cat# P0449, RRID:AB_2617143 | WB (1:5000) |
| Recombinant DNA reagent | pEF-FLAG-SKIL (plasmid) | This paper | Transient transfection in HEK293T cells to express FLAG SKIL WT | |
| Recombinant DNA reagent | FLAG SKIL G103V (SKIL ΔS2/S3) (plasmid) | This paper | Transient transfection in HEK293T cells to express FLAG SKIL G103V | |
| Recombinant DNA reagent | FLAG SKIL R314A, T315A, H317A, W318E (SKIL ΔS4) (plasmid) | This paper | Transient transfection in HEK293T cells to express FLAG SKIL R314A, T315A, H317A, W318E | |
| Recombinant DNA reagent | pFast Dual ALK5* GST-MH2-hSMAD2 (plasmid) | This paper | Used to express recombinant proteins in insect cells | |
| Recombinant DNA reagent | pFastDual ALK5*/GST-hSMAD3 (plasmid) | This paper | Used to express recombinant proteins in insect cells | |
| Recombinant DNA reagent | pBacPAK-SMAD4 (plasmid) | This paper | Used to express recombinant proteins in insect cells | |
| Sequence-based reagent | SMAD4_F1 | This paper | CACCGACAACTCGTTCGTAGTGATA CRISPR/Cas9-mediated knockout guide, forward oligo 1 | |
| Sequence-based reagent | SMAD4_R1 | This paper | AAACTATCACTACGAACGAGTTGTC CRISPR/Cas9 mediated knockout guide, reverse oligo 1 | |
| Sequence-based reagent | SMAD4_F2 | This paper | CACCGTGAGTATGCATAAGCGACGA CRISPR/Cas9 mediated knockout guide, forward oligo 2 | |
| Sequence-based reagent | SMAD4_R2 | This paper | AAACTCGTCGCTTATGCATACTCAC CRISPR/Cas9 mediated knockout guide, reverse oligo 2 | |
| Sequence-based reagent | SMAD2 _F1 | This paper | CACCGCTATCGAACACCAAAATGC CRISPR/Cas9 mediated knockout guide, forward oligo | |
| Sequence-based reagent | SMAD2 _R1 | This paper | AAACGCATTTTGGTGTTCGATAGC CRISPR/Cas9 mediated knockout guide, reverse oligo | |
| Sequence-based reagent | SMAD3_F1 | This paper | CACCGGAATGTCTCCCCGACGCGC CRISPR/Cas9 mediated knockout guide, forward oligo | |
| Sequence-based reagent | SMAD3_R1 | This paper | AAACGCGCGTCGGGGAGACATTCC CRISPR/Cas9 mediated knockout guide, reverse oligo | |
| Sequence-based reagent | SKI_F1 | This paper | CACCGCAGCGCGCCGAGAAAGCGGC CRISPR/Cas9 mediated knockin guide, forward oligo | |
| Sequence-based reagent | SKI_R1 | This paper | AAACGCCGCTTTCTCGGCGCGCTGC CRISPR/Cas9 mediated knockin guide, reverse oligo | |
| Sequence-based reagent | SKI_ssODN | This paper | G*C*AGTTCCACCTGAGCTCCATGAGC TCGCTGGGAGGATCCGCCGCTTTCTC GGCGCGCTGGGCGCAGGAGGCCTA CAAGAAGGAGAGCGCCAAGGAGGCG GGCGCGGCCGCGGTGCCG*G*C Repair template | |
| Sequence-based reagent | SKI_R2 | This paper | GCCCATGACTTTGAGGATCTCC Universal reverse primer for clone screening | |
| Sequence-based reagent | SKI_F2 | This paper | ATGAGCTCGCTGGGCGGCCCG WT SKI forward primer for clone screening | |
| Sequence-based reagent | SKI_F3 | This paper | ATGAGCTCGCTGGGAGGATCC P35S SKI forward primer for clone screening | |
| Sequence-based reagent | Biotinylated WT cJUN SBE oligonucleotide (top) | Levy et al., 2007 PMID:17591695 | 5′-Biotin- GGAGGTGCGCGGAGTCAGG CAGACAGACAGACACAGCCA GCCAGCCAGGTCGGCA DNAP oligo - Top | |
| Sequence-based reagent | WT cJUN SBE oligonucleotide (bottom) | Levy et al., 2007 PMID:17591695 | TGCCGACCTGGCTGGCTGGC TGTGTCTGTCTGTCTGCCTG ACTCCGCGCACCTCC DNAP oligo - Bottom | |
| Sequence-based reagent | Biotinylated MUT cJUN SBE oligonucleotide (top) | This paper | 5′-biotin- GGATTTGCTAATGATATAGT AATATATATATATACATATAT ATATATTGATCTTCA Mutated DNAP oligo -Top | |
| Sequence-based reagent | MUT cJUN SBE oligonucleotide (bottom) | This paper | TGAAGATCAATATATATATAT GTATATATATATATTACTAT ATCATTAGCAAATCC Mutated DNAP oligo -Bottom | |
| Sequence-based reagent | MUT cJUN SBE oligo-nucleotide (top) | This paper | GGATTTGCTAATGATATAGTA ATATATATATATACATATAT ATATATTGATCTTCA Competitor mutant oligo - Top | |
| Sequence-based reagent | MUT cJUN SBE oligo-nucleotide (bottom) | This paper | TGAAGATCAATATATATATATG TATATATATATATTACTATA TCATTAGCAAATCC Competitor mutant oligo – bottom | |
| Sequence-based reagent | GAPDH_F1 | Grönroos et al., 2012 PMID:22615489 | CTTCAACAGCGACACCCACT PCR primer | |
| Sequence-based reagent | GAPDH_R1 | Grönroos et al., 2012 PMID:22615489 | GTGGTCCAGGGGTCTTACTC PCR primer | |
| Sequence-based reagent | SMAD7_F1 | Grönroos et al., 2012 PMID:22615489 | CTTAGCCGACTCTGCGAACT PCR primer | |
| Sequence-based reagent | SMAD7_R1 | Grönroos et al., 2012 PMID:22615489 | CCAGGCTCCAGAAGAAGTTG PCR primer | |
| Sequence-based reagent | SKIL_F1 | This paper | CTGGGGCTTTGAATCAGCTA PCR primer | |
| Sequence-based reagent | SKIL_R1 | This paper | CATGGTCACCTTCCTGCTTT PCR primer | |
| Sequence-based reagent | SERPINE1_F1 | Grönroos et al., 2012 PMID:22615489 | TGATGGCTCAGACCAACAAG PCR primer | |
| Sequence-based reagent | SERPINE1_R1 | Grönroos et al., 2012 PMID:22615489 | GTTGGTGAGGGCAGAGAGAG PCR primer | |
| Sequence-based reagent | JUNB_F1 | Ramachandran et al., 2018 PMID:29376829 | ATACACAGCTACGGGATACGG PCR primer | |
| Sequence-based reagent | JUNB_R1 | Ramachandran et al., 2018 PMID:29376829 | GCTCGGTTTCAGGAGTTTGT PCR primer | |
| Sequence-based reagent | ID1_F1 | Ramachandran et al., 2018 PMID:29376829 | GCCGAGGCGGCATGCGTTC PCR primer | |
| Sequence-based reagent | ID1_R1 | Ramachandran et al., 2018 PMID:29376829 | CTTGCCCCCTGGATGGCTGG PCR primer | |
| Sequence-based reagent | ID3_F1 | Grönroos et al., 2012 PMID:22615489 | GGCCCCCACCTTCCCATCC PCR primer | |
| Sequence-based reagent | ID3_R1 | Grönroos et al., 2012 PMID:22615489 | GCCAGCACCTGCGTTCTGGAG PCR primer | |
| Sequence-based reagent | CDKN1A_F1 | Miller et al., 2018 PMID:30428352 | ACTCTCAGGGTCGAAAACGG PCR primer | |
| Sequence-based reagent | CDKN1A_R1 | Miller et al., 2018 PMID:30428352 | ATGTAGAGCGGGCCTTTGAG PCR primer | |
| Sequence-based reagent | ISLR2_F1 | This paper | AGTCGGCGAATATTGGGAGC PCR primer | |
| Sequence-based reagent | ISLR2_R1 | This paper | ATGATCCGGCCACTCCTAGA PCR primer | |
| Sequence-based reagent | CALB2_F1 | This paper | ATGGCAAATTGGGCCTCTCA PCR primer | |
| Sequence-based reagent | CALB2_R1 | This paper | GGTCAGCTTCATGCCCTGAAAT PCR primer | |
| Sequence-based reagent | SOX11_F1 | This paper | AGCGGAGGAGGTTTTCAGTG PCR primer | |
| Sequence-based reagent | SOX11_R1 | This paper | TTCCATTCGGTCTCGCCAAA PCR primer | |
| Sequence-based reagent | ITGB6_F1 | This paper | TGCGACCATCAGTGAAGAAG PCR primer | |
| Sequence-based reagent | ITGB6_R1 | This paper | GACAACCCCGATGAGAAGAA PCR primer | |
| Sequence-based reagent | HEY1_F1 | This paper | GCTTTTGAGAAGCAGGGATCT PCR primer | |
| Sequence-based reagent | HEY1_R1 | This paper | GATAACGCGCAACTTCTGCC PCR primer | |
| Sequence-based reagent | COL7A1_F1 | This paper | CAAGGGGGACATGGGTGAAC PCR primer | |
| Sequence-based reagent | COL7A1_R1 | This paper | CGGATACCAGGCACTCCATC PCR primer | |
| Sequence-based reagent | SMAD4_F3 | This paper | GTTCATAAGATCTACCCAAGTG AATATATAAAGGTCTTTGATTTG PCR primer for cloning, forward primer carrying the mutation A433E | |
| Sequence-based reagent | SMAD4_R3 | This paper | ACGCAAATCAAAGACCTTTATA TATTCACTTGGGTAGATCTTATG PCR primer for cloning, reverse primer carrying the mutation A433E | |
| Sequence-based reagent | SMAD4_F4 | This paper | AAGATCTACCCAAGTGCATATT ACAAGGTCTTTGATTTGCGTCAG PCR primer for cloning, forward primer carrying the mutation I435Y | |
| Sequence-based reagent | SMAD4_R4 | This paper | CTGACGCAAATCAAAGACCTTGT AATATGCACTTGGGTAGATCTT PCR primer for cloning, reverse primer carrying the mutation I435Y | |
| Sequence-based reagent | SKIL_F2 | This paper | CAGAGCTCGCTGGGTGTA CCAGCAGCATTTTC PCR primer for cloning, forward primer carrying the mutation G103V | |
| Sequence-based reagent | SKIL_R2 | This paper | GAAAATGCTGCTGGTAC ACCCAGCGAGCTCTG PCR primer for cloning, reverse primer carrying the mutation G103V | |
| Peptide, recombinant protein | SKI peptide A | This paper | Biotin-aminohexanoic acid- FQPHPGLQKTLEQFHLSSMSS LGGPAAFSARWAQEAYKKES | |
| Peptide, recombinant protein | SKI peptide B | This paper | Biotin-aminohexanoic acid- FQPHPGLQKTLEQFHLS SMSSLGGPAAFSARWAQE | |
| Peptide, recombinant protein | SKI peptide C | This paper | Biotin-aminohexanoic acid- GLQKTLEQFHLSSMSSLG GPAAFSARWAQE | |
| Peptide, recombinant protein | SKI peptide D | This paper | Biotin-aminohexanoic acid- TLEQFHLSSMSSLGG PAAFSARWAQE | |
| Peptide, recombinant protein | SKI peptide E | This paper | Biotin-aminohexanoic acid- TLEQFHLSSMSSLGGPA AFSARWAQEAYK | |
| Peptide, recombinant protein | SKI L21R | This paper | Biotin-aminohexanoic acid- FQPHPGLQKTREQFHLSS MSSLGGPAAFSARWAQE | |
| Peptide, recombinant protein | SKI S28T | This paper | Biotin-aminohexanoic acid- FQPHPGLQKTLEQFHLS TMSSLGGPAAFSARWAQE | |
| Peptide, recombinant protein | SKI S31L | This paper | Biotin-aminohexanoic acid- FQPHPGLQKTLEQFHLS SMSLLGGPAAFSARWAQE | |
| Peptide, recombinant protein | SKI L32P | This paper | Biotin-aminohexanoic acid- FQPHPGLQKTLEQFHLS SMSSPGGPAAFSARWAQE | |
| Peptide, recombinant protein | SKI G34D | This paper | Biotin-aminohexanoic acid – FQPHPGLQKTLEQFHLSS MSSLGDPAAFSARWAQE | |
| Peptide, recombinant protein | SKI P35S | This paper | Biotin-aminohexanoic acid – FQPHPGLQKTLEQFHLSS MSSLGGSAAFSARWAQE | |
| Peptide, recombinant protein | SKIL WT | This paper | Biotin-aminohexanoic acid – LHLNPSLKHTLAQFHLSSQSS LGGPAAFSARHSQESMSPTV | |
| Peptide, recombinant protein | SKIL L90R | This paper | Biotin-aminohexanoic acid – LHLNPSLKHTRAQFHLSSQS SLGGPAAFSARHSQESMSPTV | |
| Peptide, recombinant protein | SKIL S100L | This paper | Biotin-aminohexanoic acid - LHLNPSLKHTLAQFHLSSQS LLGGPAAFSARHSQESMSPTV | |
| Peptide, recombinant protein | SKIL G103D | This paper | Biotin-aminohexanoic acid - LHLNPSLKHTLAQFHLSSQSS LGDPAAFSARHSQESMSPTV | |
| Peptide, recombinant protein | SKIL P104S | This paper | Biotin-aminohexanoic acid - LHLNPSLKHTLAQFHLSSQSS LGGSAAFSARHSQESMSPTV | |
| Peptide, recombinant protein | SKI WT | This paper | FQPHPGLQKTLEQFHLSSMSS LGGPAAFSARWAQE | |
| Peptide, recombinant protein | Human recombinant TGF-β1 | Peprotech | Cat# 100–21 | |
| Peptide, recombinant protein | Human recombinant BMP4 | Peprotech | Cat# 120-05ET | |
| Peptide, recombinant protein | Human recombinant Activin A | Peprotech | Cat# 120–14 | |
| Commercial assay or kit | PowerUp SYBR Green Master Mix | Thermo Fisher Scientific | Cat# A25742 | |
| Commercial assay or kit | Quickextract DNA extraction solution | Lucigen | Cat# QE09050 | |
| Commercial assay or kit | Dual-Glo Luciferase Assay System | Promega | Cat# E2920 | |
| Commercial assay or kit | Fugene 6 transfection reagent | Promega | Cat# E2691 | |
| Commercial assay or kit | Superdex 200 10/300 size exclusion column | Cytiva | Cat# 28990944 | |
| Commercial assay or kit | Streptavidin (SA) Biosensors | ForteBio | Cat# 18–5019 | |
| Commercial assay or kit | Glutathione 4B Sepharose | Cytiva | Cat# 17075601 | |
| Commercial assay or kit | Pierce NeutrAvidin Agarose | Thermo Fisher Scientific | Cat# 29200 | |
| Commercial assay or kit | GFP-Trap Agarose (IP) | Cromotek | Cat# gta-20 | |
| Commercial assay or kit | Protein G Sepharose, Fast Flow | Sigma–Aldrich | Cat# P3296 | |
| Chemical compound, drug | cOmplete, EDTA-free Protease Inhibitor Cocktail | Sigma–Aldrich | Cat# 000000011873580001 | |
| Chemical compound, drug | TRIzol | Thermo Fisher Scientific | Cat# 15596026 | |
| Chemical compound, drug | DAPI | Sigma-Aldrich | Cat# 10236276001 | |
| Chemical compound, drug | SB-431542 | Tocris; Inman et al., 2002: PMID:12065756 | Cat# 1614 | |
| Chemical compound, drug | MG-132 | Tocris | Cat# 1748 | |
| Software, algorithm | FIJI (ImageJ) | https://imagej.net/Fiji/Downloads | RRID:SCR_002285 | |
| Software, algorithm | FlowJo 10 | FlowJo | RRID:SCR_008520 | |
| Software, algorithm | GraphPad Prism 8 | GraphPad | RRID:SCR_002798 | |
| Software, algorithm | ASTRA6.1 | Wyatt | RRID:SCR_001625 | |
| Software, algorithm | Octet CFR software | ForteBio | ||
| Software, algorithm | DLS Xia2/XDS pipeline | Winter, 2010; Kabsch, 2010 PMID:20124692 | ||
| Software, algorithm | CCP4 suite | Winn et al., 2011 PMID:21460441 | RRID:SCR_007255 | |
| Software, algorithm | Phaser | McCoy et al., 2007 PMID:19461840 | RRID:SCR_014219 | |
| Software, algorithm | Refmac | Murshudov et al., 2011 PMID:21460454 | RRID:SCR_014225 | |
| Software, algorithm | Coot | Emsley et al., 2010 PMID:20383002 | RRID:SCR_014222 | |
| Software, algorithm | PHENIX Suite | Liebschner et al., 2019 PMID:31588918 | RRID:SCR_014224 | |
| Software, algorithm | Phenix.Refine | Afonine et al., 2012 PMID:22505256 | RRID:SCR_016736 | |
| Software, algorithm | RSEM-1.3.0/STAR-2.5.2 | Li and Dewey, 2011; PMID:21816040 Dobin et al., 2013 PMID:23104886 | RRID:SCR_013027 | |
| Software, algorithm | DESeq2 package (version 1.24.0) | Love et al., 2014 PMID:25516281 | RRID:SCR_015687 |





