Glycine acylation and trafficking of a new class of bacterial lipoprotein by a composite secretion system
Figures
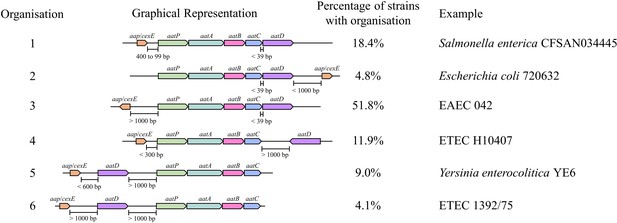
Organisation of the Aat operon.
Unique Aat amino acid sequences were detected using PSI-BLAST. These sequences were used to identify the strains that encoded them in the NCBI identical protein groups. As the aat genes were present on contigs of whole genome sequencing projects, it was not possible to assess if a strain encoded a gene on the chromosome or plasmid. Instead, contigs were used to identify the complete Aat system. This analysis does not include strains that might encode the aat genes or aap/cexE on a separate genomic element. However, a total of 827 complete Aat systems were identified in the same nucleotide accession. The positions of these genes were used to assess the organisation of the aat operon. From this assessment five different classes of aat operon organisation were defined.
-
Figure 1—source data 1
aatA gene positions.
- https://cdn.elifesciences.org/articles/63762/elife-63762-fig1-data1-v2.txt
-
Figure 1—source data 2
aatB gene positions.
- https://cdn.elifesciences.org/articles/63762/elife-63762-fig1-data2-v2.txt
-
Figure 1—source data 3
aatC gene positions.
- https://cdn.elifesciences.org/articles/63762/elife-63762-fig1-data3-v2.txt
-
Figure 1—source data 4
aatD gene positions.
- https://cdn.elifesciences.org/articles/63762/elife-63762-fig1-data4-v2.txt
-
Figure 1—source data 5
aatP gene positions.
- https://cdn.elifesciences.org/articles/63762/elife-63762-fig1-data5-v2.txt
-
Figure 1—source data 6
aap or cexE gene positions.
- https://cdn.elifesciences.org/articles/63762/elife-63762-fig1-data6-v2.txt
-
Figure 1—source data 7
Calculated distance between genes.
- https://cdn.elifesciences.org/articles/63762/elife-63762-fig1-data7-v2.txt
-
Figure 1—source data 8
Distances and designations.
- https://cdn.elifesciences.org/articles/63762/elife-63762-fig1-data8-v2.txt
AatD post-translational modification of Aap and CexE.
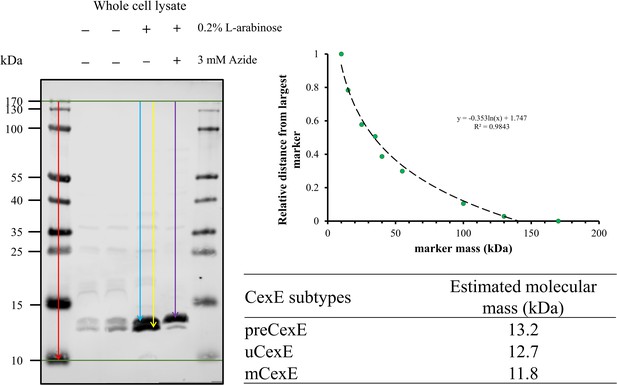
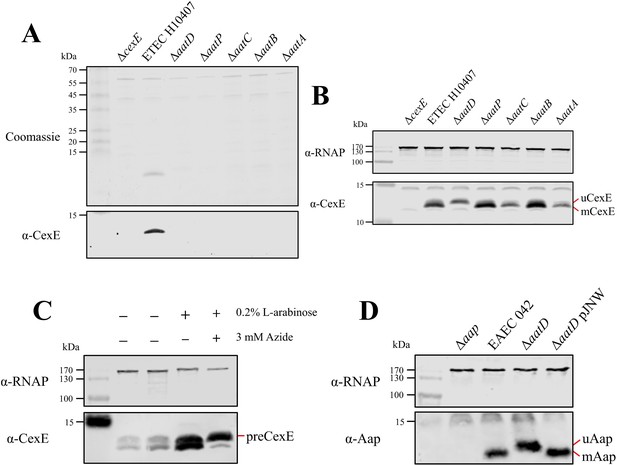
(A) Culture supernatant of ETEC H10407 and aat mutants harbouring pCfaD grown in lysogeny broth (LB) supplemented with L-arabinose. Cells were removed from the culture supernatant and remaining protein precipitated with trichloroacetic acid. Protein samples were separated by Tris-tricine SDS-PAGE and stained with Coomassie or transferred to nitrocellulose for western blotting with polyclonal antibodies against CexE. (B) Whole-cell lysates of the aat mutants and parental strain separated by Tris-tricine SDS-PAGE. (C) Whole-cell lysates of ETEC H10407 grown in LB with or without azide. (D) Whole-cell lysates of EAEC 042, aap, aatD, and aatD complemented strains grown in DMEM (Dulbecco's Modified Eagle Medium) high glucose. The positions of uCexE, mCexE, uAap, mAap, and preCexE are indicated as appropriate. CexE and Aap were detected by western blotting (α-CexE and α-Aap, respectively) and RNA polymerase (α-RNAP) was included as a loading control where appropriate.
-
Figure 2—source data 1
Western blot and Coomassie stained gels of culture supernatant fractions.
- https://cdn.elifesciences.org/articles/63762/elife-63762-fig2-data1-v2.pptx
-
Figure 2—source data 2
Western blot of whole-cell lysates derived from wild-type and mutant strains.
- https://cdn.elifesciences.org/articles/63762/elife-63762-fig2-data2-v2.pptx
-
Figure 2—source data 3
Western blot of whole-cell lysates after growth in sodium azide.
- https://cdn.elifesciences.org/articles/63762/elife-63762-fig2-data3-v2.pptx
-
Figure 2—source data 4
Western blot of whole-cell lysates showing Aap modification in the presence and absence of aatD.
- https://cdn.elifesciences.org/articles/63762/elife-63762-fig2-data4-v2.pptx
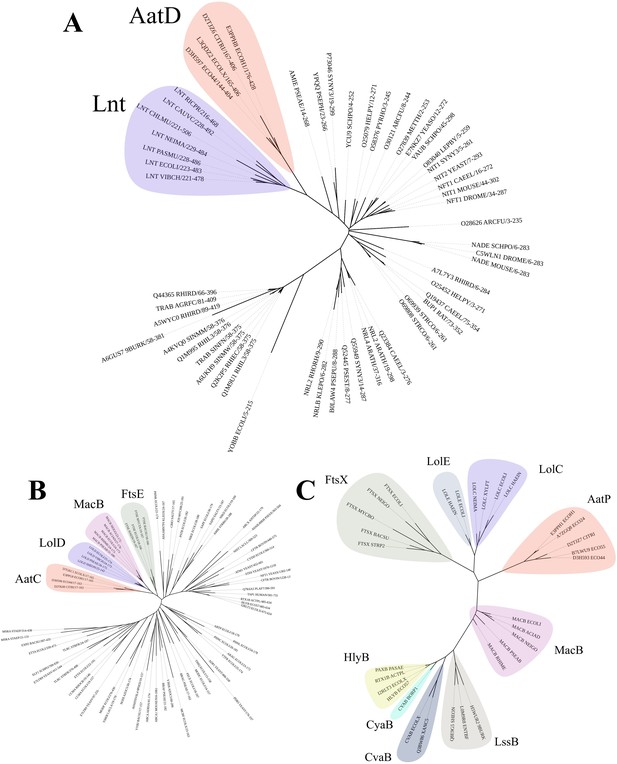
Phylogenetic analysis of AatD, AatP, and AatC.
Sequences were aligned using Clustal omega and the tree was calculated using RAxML. (A) The carbon-nitrogen(C-N) hydrolase domains of C-N hydrolase family seed sequences were aligned to the C-N hydrolase domains of AatD sequences from ETEC H10407, EAEC 042, Citrobacter rodentium ICC168, and Escherichia coli KTE75. (B) Phylogram of the ATPase domains of the pfam ABC transporters (PF00005) and AatC sequences. (C) Phylogram of type I secretion system (T1SS) ABC transporters, Lol ABC transporters, and AatP sequences.
-
Figure 3—source data 1
AatD and carbon-nitrogen (C-N) hydrolase Pfam family sequences used for tree.
- https://cdn.elifesciences.org/articles/63762/elife-63762-fig3-data1-v2.txt
-
Figure 3—source data 2
Sequences used to generate AatP tree.
- https://cdn.elifesciences.org/articles/63762/elife-63762-fig3-data2-v2.txt
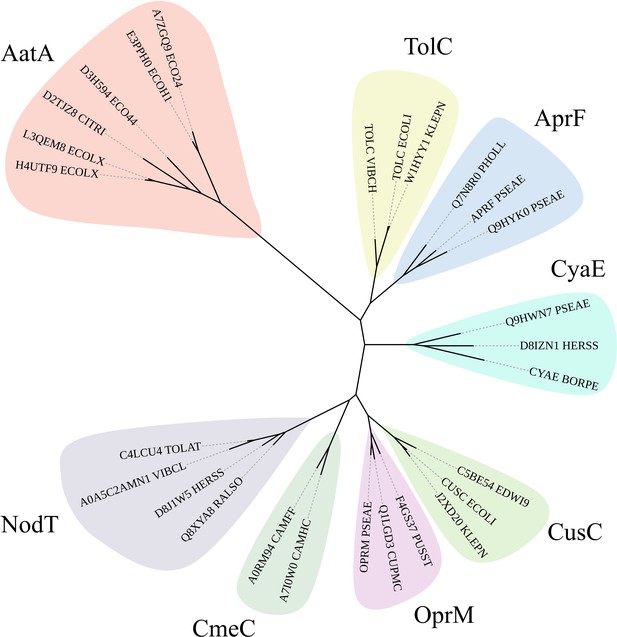
Phylogram of AatA, TolC, and other outer membrane protein (OMP) associated with T1SS and efflux pumps.
The hidden Markov model (HMM) of AatA proteins was used to confirm AatA as a homolog of TolC and OMP proteins. Representative sequences of secretion-associated OMPs (AprF, CyaE, and TolC) and efflux-associated OMPs (CusC, OprM, CmeC, and NodT) were used to construct a phylogram. AatA is more closely related to the secretion-associated OMPs AprF and CyaE than the efflux-associated OMPs. TolC is a promiscuous protein that functions as the OMP for secretion and efflux systems. All phylograms were constructed using RAxML and drawn using iTOL.
-
Figure 3—figure supplement 1—source data 1
AatA and outer membrane protein (OMP) sequences used for tree.
- https://cdn.elifesciences.org/articles/63762/elife-63762-fig3-figsupp1-data1-v2.txt
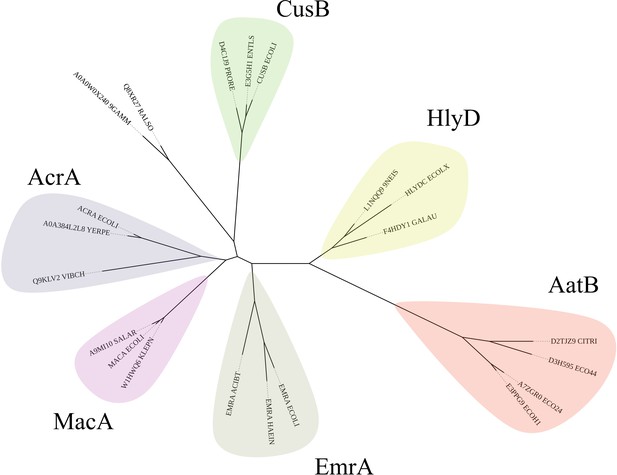
Phylogram of AatB and periplasmic adaptor proteins (PAPs) involved in type I secretion system (T1SS) and efflux pumps.
The AatB hidden Markov model (HMM) identified it as a homolog of the PAPs. Representative sequences of PAPs involved in T1SS and efflux were used to construct a phylogenetic tree using RAxML. The resulting tree was drawn in iTOL. AatB clades with the T1SS PAP HlyD which is involved in haemolysin secretion instead of the efflux-associated PAPs of EmrA, MacA, AcrA, and CusB.
-
Figure 3—figure supplement 2—source data 1
AatB and periplasmic adaptor protein (PAP) sequences used for tree.
- https://cdn.elifesciences.org/articles/63762/elife-63762-fig3-figsupp2-data1-v2.txt
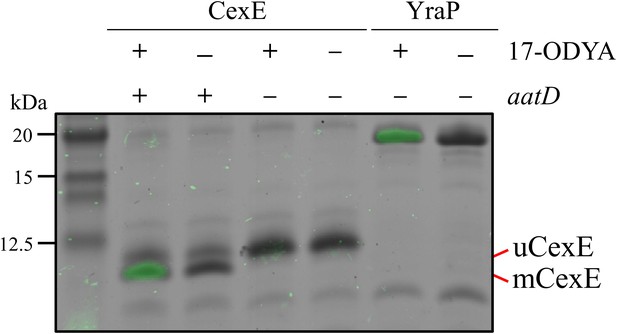
Incorporation of 17-ODYA into CexE in the presence of AatD.
His-tagged CexE was isolated from cexE or cexE aatD double mutants harbouring pACYC-cexE-6His grown in the presence or absence of 17-ODYA and separated by SDS-PAGE. An azide linked Alexa Fluor 488 was conjugated to the alkyne moiety present in 17-ODYA by CuAAC. The incorporation of 17-ODYA into the target protein was detected by fluorescence (green bands) and the image was overlaid on the image of the SDS-PAGE gel. A known lipoprotein YraP was used as a positive control.
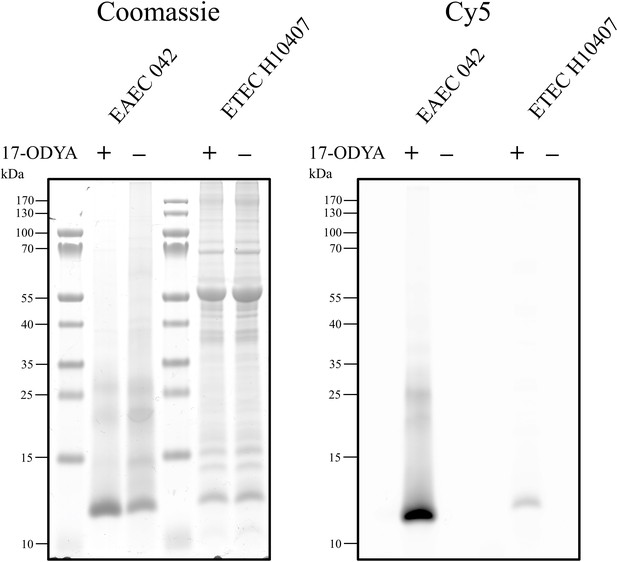
17-ODYA labelling of secreted Aap and CexE.
EAEC 042 and ETEC H10407 were grown in DMEM-high glucose (HG) or IMDM, respectively. After 2 hr of growth, 17-ODYA or an equal volume of DMSO was added. Cultures were grown for a further 3 hr. Triton X-100 was added to each culture to a final concentration of 0.1% for a final hour of growth. The culture supernatant was separated by centrifugation and filtered prior to TCA precipitation. Precipitated proteins were resuspended in 2% SDS. The incorporation of 17-ODYA into Aap and CexE was detected by CuAAC.
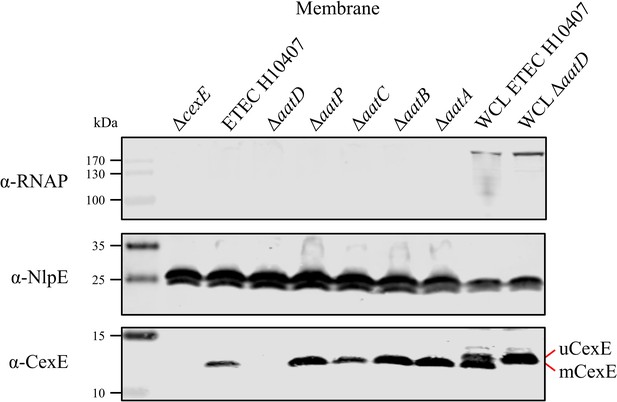
Membrane localisation of CexE.
The membrane fraction of ETEC H10407 and aat mutant harbouring pCfaD were separated by SDS-PAGE. Transferred to nitrocellulose and probed for the cytoplasmic protein RNAP, lipoprotein NlpE, and CexE using protein-specific antibodies.
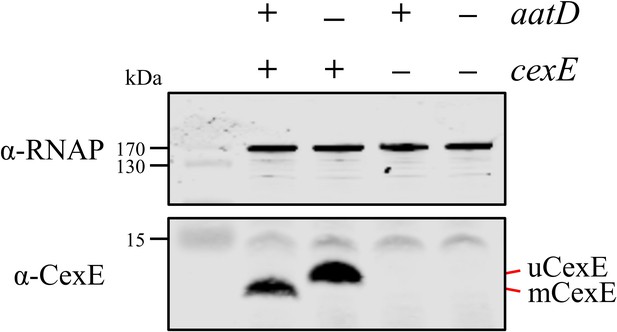
Recapitulation of CexE acylation in E. coli BL21(DE3).
E. coli BL21(DE3) was transformed with pET26b, or pET26b-cexE and pACYCDuet-1 or pACYC-aatD. Cultures were grown in lysogeny broth (LB) and protein production was stimulated with isopropyl β-D-1-thiogalactopyranoside (IPTG). Whole-cell lysate samples were taken and separated by Tris-tricine SDS-PAGE. CexE was detected using a polyclonal antibody. RNAP was used as a loading control. The position of mCexE and uCexE is indicated.
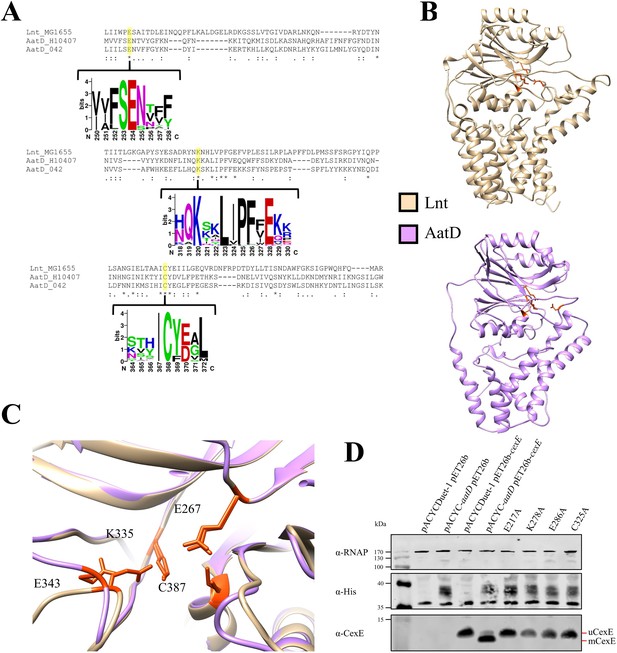
Catalytic residues of AatD.
(A) AatD from EAEC 042 and ETEC H10407 were aligned with Lnt from Escherichia coli MG1655. AatD sequences identified by PSI-BLAST were aligned and used to create a WebLogo. The catalytic residues of the carbon-nitrogen (C-N) hydrolase family are highlighted. (B) The structure of Lnt compared to the predicted structure of ETEC H10407 AatD. The catalytic residues of Lnt and AatD are highlighted in orange. (C) Magnified view of the catalytic site of Lnt with the predicted structure of AatD superimposed. Residues are numbered as they appear in Lnt. (D) WCL of E. coli BL21(DE3) expressing CexE in the presence of AatD and each of the four single site substitution derivatives of AatD. CexE was detected using an anti-CexE polyclonal antibody, AatD was detected with an anti-His tag antibody, and RNAP was used as a loading control.
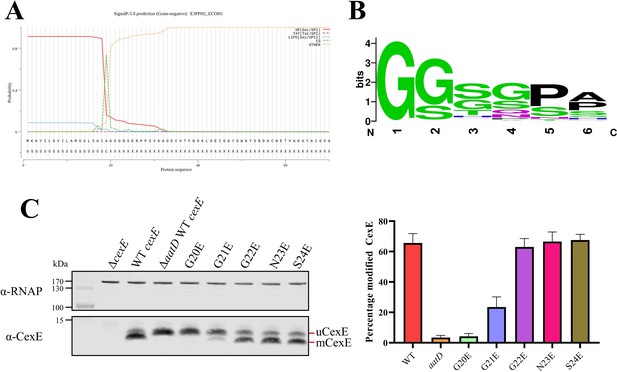
Single site substitution of the 5 N-terminal amino acids of CexE.
(A) SignalP result of CexE sequence from ETEC H10407 (E3PPH2_ECOH1) (B) WebLogo of the 5 N-terminal residues of Aap and CexE sequences post Sec signal sequence cleavage. (C) ETEC H10407 cexE mutants transformed with pACYC184 (ΔcexE) or pACYC-cexE-6His with either the wild-type sequence (WT cexE) or one of the first five amino acids mutated to glutamic acid. CexE was detected by polyclonal antibodies and RNAP was used as a loading control. The percentage of mCexE was determined from three biological replicates.
-
Figure 7—source data 1
ETEC H10407 CexE sequence.
- https://cdn.elifesciences.org/articles/63762/elife-63762-fig7-data1-v2.txt
-
Figure 7—source data 2
Aap and CexE sequences.
- https://cdn.elifesciences.org/articles/63762/elife-63762-fig7-data2-v2.txt
-
Figure 7—source data 3
Aap and CexE sequences with signal sequences removed by SignalP.
- https://cdn.elifesciences.org/articles/63762/elife-63762-fig7-data3-v2.txt
-
Figure 7—source data 4
T-coffee alignment of sequences.
- https://cdn.elifesciences.org/articles/63762/elife-63762-fig7-data4-v2.txt
-
Figure 7—source data 5
Western blots of CexE production.
- https://cdn.elifesciences.org/articles/63762/elife-63762-fig7-data5-v2.pptx
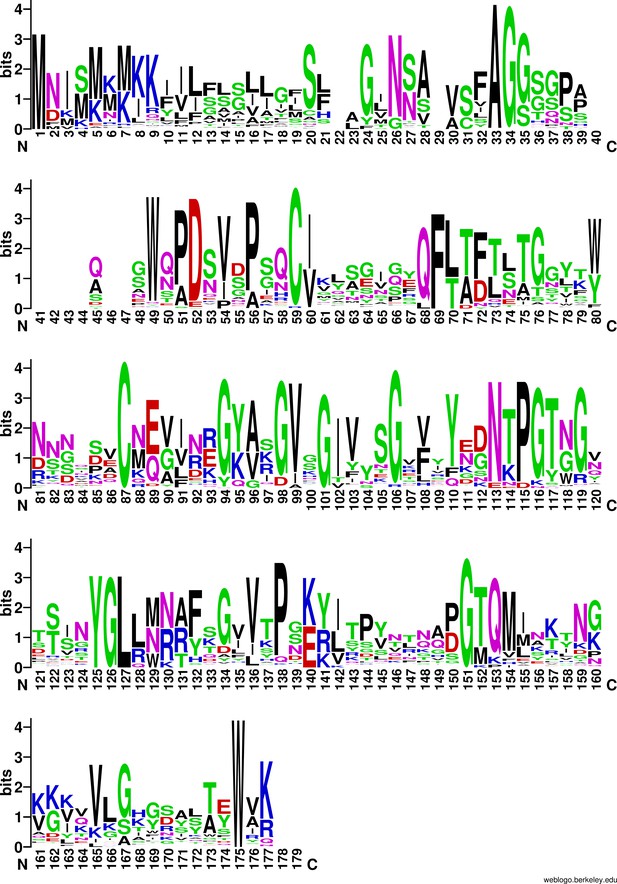
WebLogo of Aap and CexE sequences.
Aap and CexE sequences were aligned using ClustalO. The alignment was then placed into WebLogo.
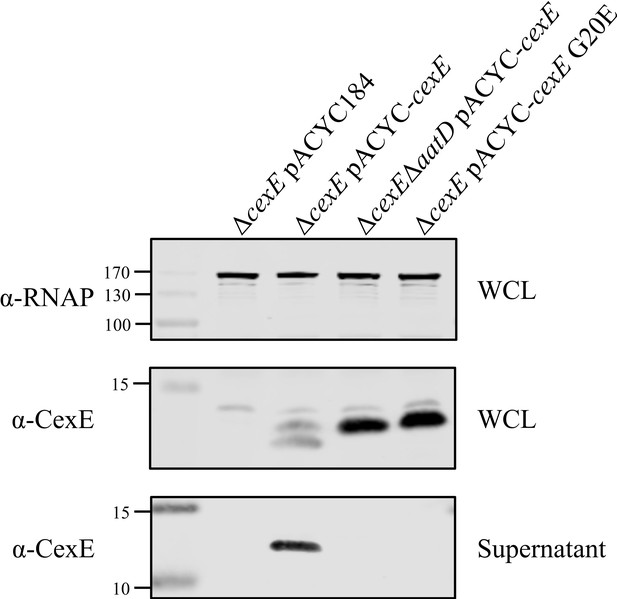
Culture supernatant of CexE G20E mutant.
Enterotoxigenic Escherichia coli (ETEC) H10407ΔcexE pCfaD was transformed with pACYC184, pACYC-cexE, or pACYC-cexE G20E. A double mutant of cexE and aatD harbouring pCfaD was transformed with pACYC-cexE. Both WCL and culture supernatant samples of these strains grown in the presence of arabinose were taken. Supernatant proteins were isolated by TCA precipitation after removal of cells by centrifugation and filtration. Protein samples were separated by SDS-PAGE and transferred to nitrocellulose for western blotting for RNAP and CexE.
-
Figure 7—figure supplement 2—source data 1
Coomassie stained gel and western blots of secreted CexE.
- https://cdn.elifesciences.org/articles/63762/elife-63762-fig7-figsupp2-data1-v2.pptx
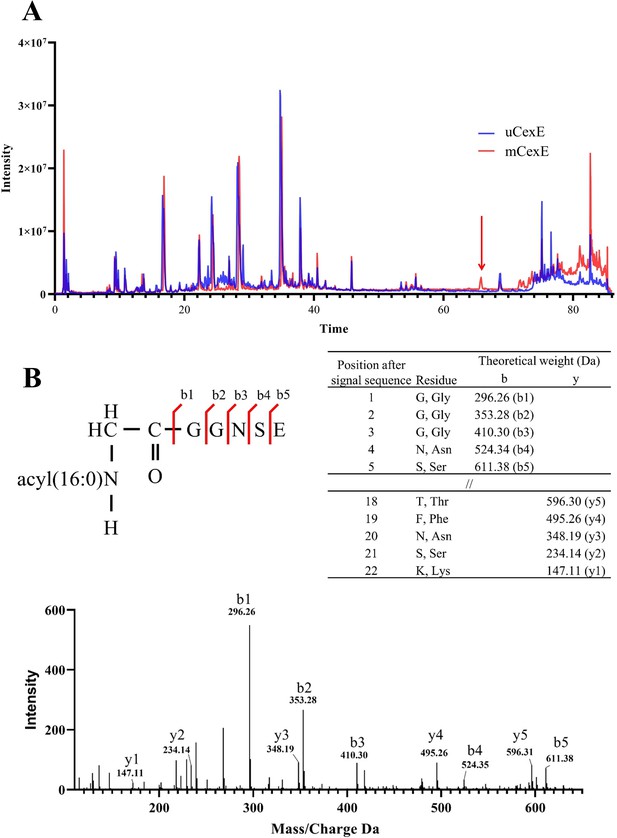
Mass spectrometric analysis of modified CexE.
(A) CexE-6His was isolated from a cexE mutant and a cexE aatD double mutant and subjected to LC-MS/MS. CexE and pro-CexE were trypsinated and separated by HPLC. (B) The indicated peak was subjected to MS/MS to identify the amino acid sequence.
-
Figure 8—source data 1
HLPC of mCexE.
- https://cdn.elifesciences.org/articles/63762/elife-63762-fig8-data1-v2.txt
-
Figure 8—source data 2
HLPC of uCexE.
- https://cdn.elifesciences.org/articles/63762/elife-63762-fig8-data2-v2.txt
-
Figure 8—source data 3
Peptide results.
- https://cdn.elifesciences.org/articles/63762/elife-63762-fig8-data3-v2.txt
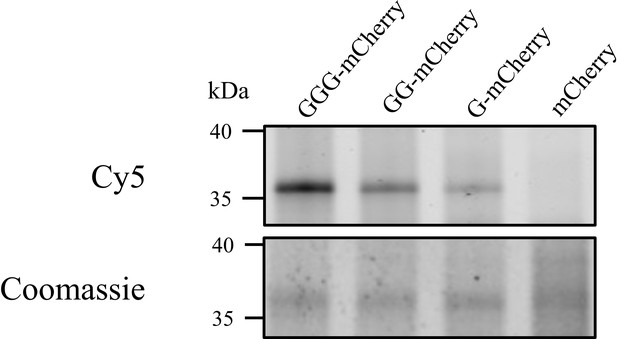
Heterologous acylation by AatD.
Plasmid encoding mCherry with the CexE signal sequence followed by none, one (G), two (GG), or three (GGG) glycine residues post signal sequence was produced in an ETEC H10407 cexE mutant grown in the presence of 17-ODYA. The mCherry proteins were isolated using a C-terminal 8 His tag and azide linked Cy5 was incorporated into mCherry proteins using CuAAC. The acylation of mCherry was detected using fluorescence.
-
Figure 9—source data 1
ODYA labelling of chimeric proteins.
- https://cdn.elifesciences.org/articles/63762/elife-63762-fig9-data1-v2.pptx
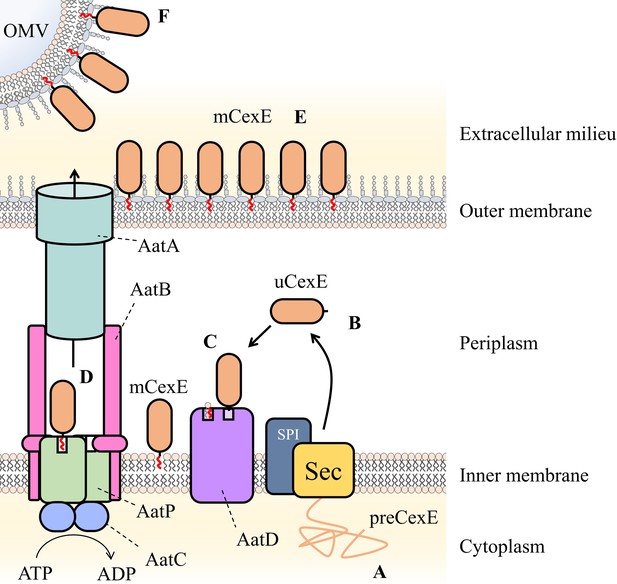
Schematic of proposed Aat system mechanism.
(A) PreCexE produced in the cytoplasm is secreted into the periplasm by the Sec pathway (Sec). (B) The signal sequence is cut by signal peptidase I (SPI) resulting in soluble uCexE. (C) An acyl chain is added to the N-terminal glycine of uCexE by AatD. Accordingly, mCexE associates with the membrane. (D) mCexE is extracted out of the inner membrane by the AatP and AatC complex. A single channel is formed comprised of AatP, AatA, AatB, and AatC that allows the secretion of mCexE. (E) mCexE is inserted into the outer leaflet of the outer membrane. mCexE remains associated with the outer membrane by the single acyl chain incorporated onto the N-terminal glycine. (F) mCexE is secreted into the extracellular milieu attached to outer membrane vesicles (OMVs).
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Escherichia coli) | H10407 | Evans and Evans (1973) | Prototypical ETEC strain | |
| Strain, strain background (Escherichia coli) | 042 | Evans and Evans (1973) | Prototypical EAEC strain | |
| Strain, strain background (Escherichia coli) | DH5α | New England Bioscience | C2987I | Chemically competent cells |
| Strain, strain background (Escherichia coli) | BL21(DE3) | Invitrogen | EC0114 | Protein production strain |
| Recombinant DNA reagent | pKD46 | Datsenko and Wanner, 2000 | Plasmid encoding λ Red recombinase genes | |
| Recombinant DNA reagent | pCP20 | Datsenko and Wanner, 2000 | Plasmid encoding Flp recombinase gene | |
| Recombinant DNA reagent | pET26b | Novagen | T7 expression vector with C-terminal 6 His tag | |
| Recombinant DNA reagent | pACYC184 | Chang and Cohen, 1978 | Plasmid with p15A origin of replication | |
| Recombinant DNA reagent | pACYCDuet-1 | Novagen | Plasmid with p15A origin of replication and two T7 promoters | |
| Antibody | Anti-CexE (rabbit polyclonal) | This paper | WB (1:2000) | |
| Antibody | Anti-His Tag (mouse monoclonal) | GenScript | Cat# A00186, RRID:AB_914704 | WB (1:5000) |
| Antibody | Anti E. coli RNA polymerase beta antibody (mouse monoclonal) | BioLegend | Cat# 663903, RRID:AB_2564524 | WB (1:10000) |
Additional files
-
Supplementary file 1
Nucleotide accession numbers for aat-containing bacteria.
- https://cdn.elifesciences.org/articles/63762/elife-63762-supp1-v2.xlsx
-
Supplementary file 2
SignalP predictions for CexE proteins.
- https://cdn.elifesciences.org/articles/63762/elife-63762-supp2-v2.xlsx
-
Supplementary file 3
Bacterial strains used in this study.
- https://cdn.elifesciences.org/articles/63762/elife-63762-supp3-v2.xlsx
-
Supplementary file 4
Primers used in this study.
- https://cdn.elifesciences.org/articles/63762/elife-63762-supp4-v2.xlsx
-
Supplementary file 5
Plasmids used in this study.
- https://cdn.elifesciences.org/articles/63762/elife-63762-supp5-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/63762/elife-63762-transrepform-v2.docx