Exocyst-mediated membrane trafficking of the lissencephaly-associated ECM receptor dystroglycan is required for proper brain compartmentalization
Figures
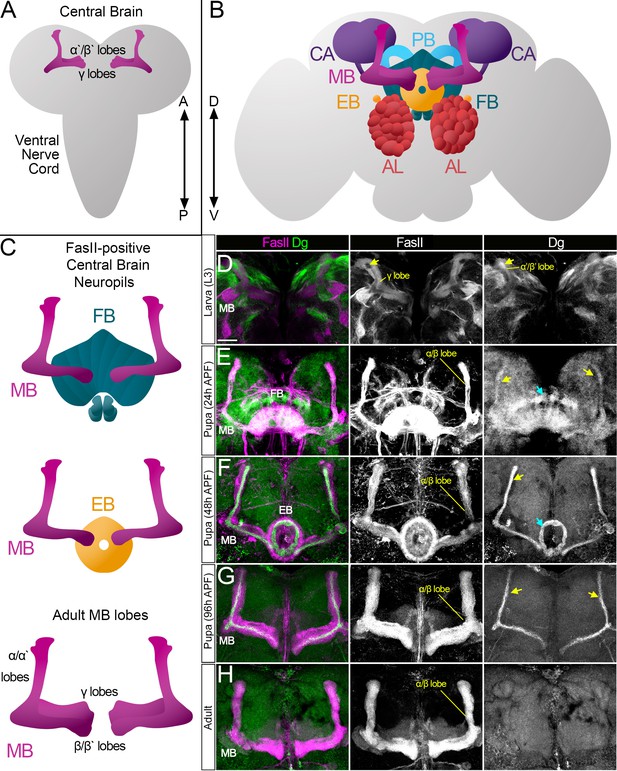
Survey of spatiotemporal dystroglycan (Dg) expression in the developing Drosophila brain.
(A–C) Schematic representation of larval (A) and adult (B) Drosophila brains. Antennal lobes (AL, tomato), ellipsoid body (EB, orange), fan-shaped body (FB, teal), mushroom body (MB, fuchsia), calyx (CA, violet), and protocerebral bridge (PB, cyan) are shown. Major neuropils of the central brain are also shown separately (C). Anterior ←→ posterior (A←→P). Dorsal ←→ ventral (D←→V). (D–H) Anterior view of the midbrain region of the larval brain (D) and frontal views of the pupal (E–G) and adult (H) brains stained with antibodies against Dg (green) and the homophilic cell adhesion molecule Fasciclin II (FasII, magenta). Expression patterns for FasII and Dg are also shown in separate channels. Scale bar 20 µm. (D) In the larval brain, FasII is expressed in the γ lobes of the MB neuropil, which are formed in early larval stages. At L3 stage, α′/β′ lobe formation takes place. These lobes can be seen by Dg expression (arrow) and the absence of FasII expression. (E) After the pupa is formed, MB neuroblasts give rise to α/β lobe neurons that are positive for FasII. Dg expression is observed in newly generated neurons of α/β lobes (yellow arrows). In addition, a distinct Dg pattern is seen in neurons forming the FB (blue arrow) neuropil. (F) At mid-pupal stage, Dg expression remains in inner MB α/β lobe neurons (freshly generated differentiating axons, yellow arrow) and disappears from outer α/β neurons, which were born at earlier pupal stages (FasII marker demonstrates their belonging to α/β MB lobe). In addition, the Dg pattern diminishes from the FB but appears in the developing EB (blue arrow). (G) In final pupal stages, when most neuropils, except for MB α/β lobe neurons, are established, Dg protein is enriched in a small subset of inner α/β lobe axons (yellow arrows) and significantly reduced in other neuropils. Note the diminished Dg staining in fully formed FB and EB neuropils. (H) In the adult brain, Dg expression is visibly reduced.
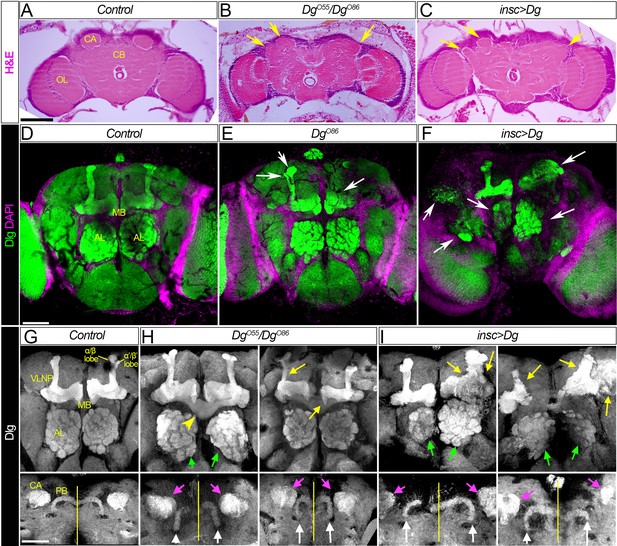
Dg is required for proper development of neuropils.
(A–C) Hematoxylin and eosin (H&E) stained histological sections of adult brains of control, Dg loss- and gain-of-function mutants (A: w1118/OregonR; B: DgO55/DgO86; and C: insc>Dg). Note the appearance of the cobblestone brain phenotype in Dg mutants and abnormal formation of brain neuropils (yellow arrows). OL: optic lobe; CA: calyx; CB: central brain. (D–F) Frontal-anterior view of adult brains of control, Dg loss- and gain-of-function mutants (D: w1118/OregonR; E: DgO86; and F: insc>Dg). Anti-discs large (Dlg, green) antibody marks septate junctions and is used to label membranes of neuronal cell bodies, neuronal fibers, and synapses, while DAPI (magenta) marks nuclei. Note that midbrain neuropils are abnormal in mutants with deregulated dystroglycan (Dg) expression (white arrows). MB: mushroom body; AL: antennal lobe. (G–I) Frontal-anterior view of the central brain in control, Dg loss- and gain-of-function mutants (G: w1118/OregonR; H: DgO55/DgO86; and I: insc>Dg). Anti-Dlg – grayscale. Upper panels show the α/β (bright) and γ (dim) MB lobes marked with anti-Dlg marker. Note that upon Dg deregulation ALs and MB neuropils (green and yellow arrows, respectively) are disorganized. Lower panels show the frontal-posterior views of adult brains showing abnormal shape of MB calyces (CA, magenta arrows) and the protocerebral bridge (PB, white arrows) in Dg loss- and gain-of-function mutants in comparison to the control. Yellow vertical line shows the midline. Scale bar 50 µm.
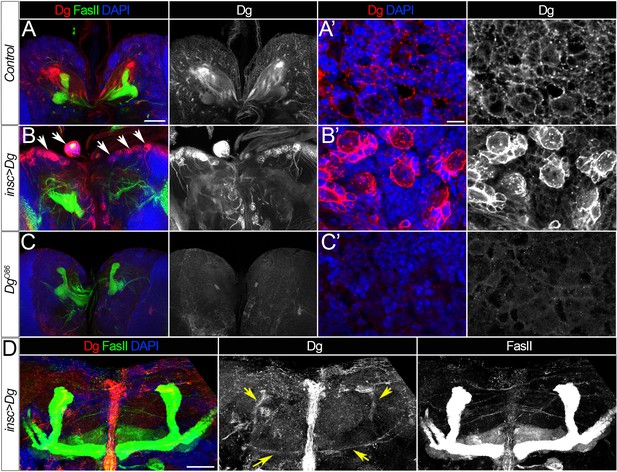
Dystroglycan (Dg) expression patterns.
(A–A’) Expression of Dg protein in the middle section of the brain (A) or at the brain surface in wild-type larvae. (B–B’) As a result of Dg overexpression using the insc-Gal4 driver, neuroblasts and their progeny have dramatically increased levels of Dg (B’). Note the cobblestone-like brain appearance in insc>Dg mutants (B’, arrows). (C–C’) In Dg loss-of-function mutant brains, Dg protein is undetectable. (D) In the adult brain of mutant flies expressing Dg under control of insc-Gal4, Dg protein persists in mushroom bodies (MBs) (yellow arrows), while in the adult brain of controls, Dg is no longer detected (Figure 1H). Dg (red), Fasciclin II (FasII) marks MBs (green), and DAPI marks nuclei (blue). (A–C) show middle sections of larval brain, scale bar 50 µm. (A’–C’) show the surface of the larval brain, scale bar 5 µm. (D) shows the central brain in adults, scale bar 25 µm.
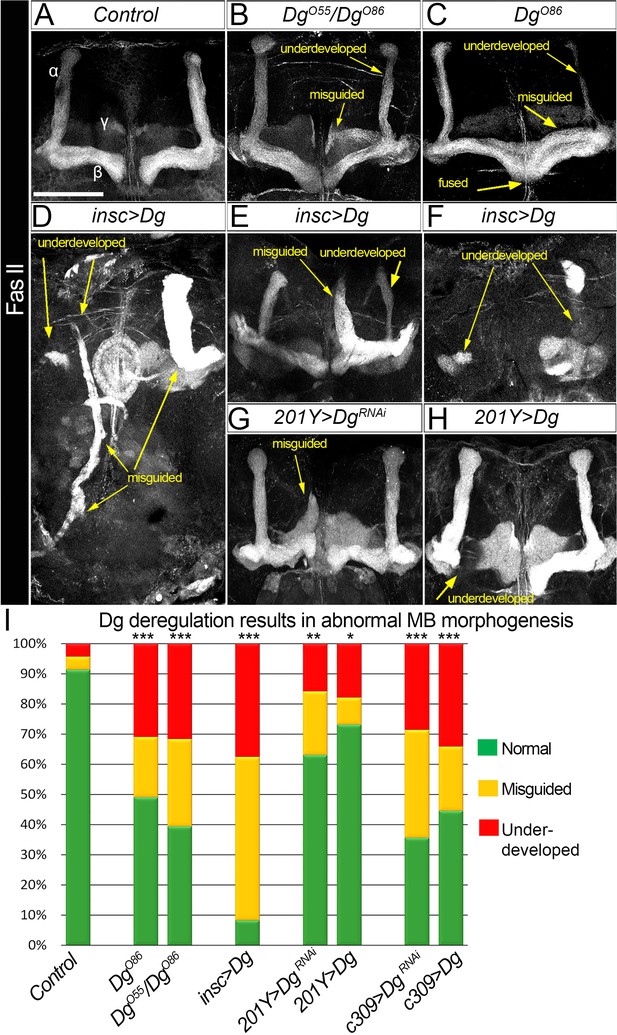
Dystroglycan (Dg) is required for proper mushroom body (MB) fasciculation.
(A–H) Fasciclin II (FasII) staining of adult mutant brains reveals various morphological α/β lobe defects upon Dg deficiency (B: DgO55/DgO86; C: DgO86), pan-neuronal Dg upregulation (insc>Dg, D–F), and γ and α/β MB neuron-specific Dg down- or upregulation (201Y>DgRNAi, G, and 201Y>Dg, H). In Dg mutants, axons of α lobe neurons stop migration prematurely or abnormally project into β lobe space, forming underdeveloped α lobes. Axons of β lobe neurons are improperly clustered and misguided, projecting into γ lobe space or overshooting the midline to form a fused β lobe. Note that misguided and underdeveloped α/β lobe phenotypes are more dramatic when Dg is overexpressed in all neuronal cells. (I) Quantification of the observed MB phenotypes (see also Supplementary file 1). For comparison of the observed phenotypes, χ2 test was used. ***p≤0.001; **p≤0.01; *p≤0.05; n.s.: not significantly different.
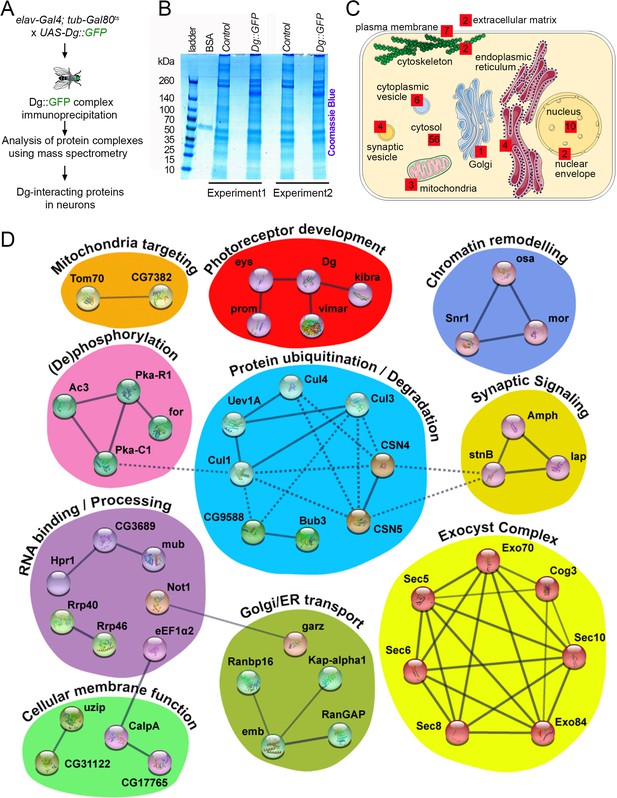
Neuronal dystroglycan (Dg)-associated components identified through proteomics approach.
(A) Scheme represents experimental techniques carried out to identify neuronal proteins that interact with Dg. GFP-tagged full-length Dg was expressed specifically in neurons by driving expression of UAS-Dg::GFP with elav-Gal4 using the Gal4/Gal80ts system. Dg::GFP protein was immunoprecipitated with GFP-Trap beads containing anti-GFP antibodies. Proteins that form complexes with Dg in neuronal tissue were detected by mass spectrometry analysis. (B) Coomassie blue-stained gel confirms increased protein levels in samples immunoprecipitated from protein extracts from Dg-overexpressing adult animal heads. Experiments were performed in duplicate. (C) Cartoon represents neuronal cell with subcellular compartments, where red squares and numbers indicate identified Dg-associated proteins and their reported subcellular localization. See also Supplementary files 2 and 4. (D) Dg-associated components placed into a protein interaction network. Colored shapes outline functional groups. Nodes symbolize identified proteins, lines show previously reported associations, and line thickness represents confidence of association. Non-dashed lines show protein complexes identified by Markov clustering algorithm. See also Supplementary file 5. Source data file. Mass spec data for neuronal Dg interactome https://doi.org/10.5061/dryad.8sf7m0cmf.
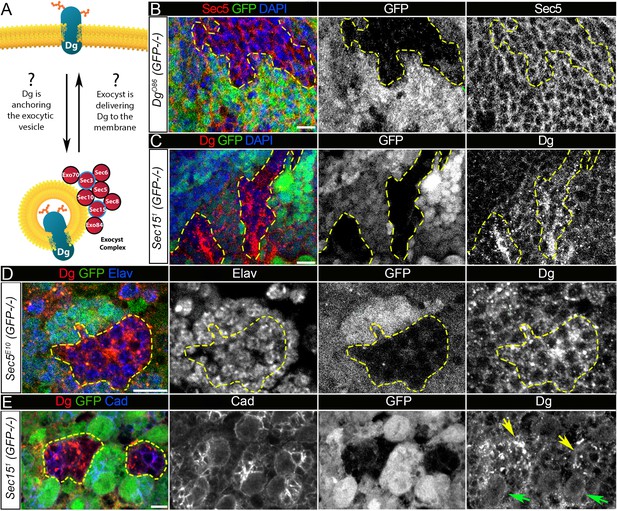
Exocyst mediates dystroglycan (Dg) trafficking in neuronal cells.
(A) Scheme of a potential Dg–exocyst interaction hypothesized based on the reported data on functions and subcellular localizations of Dg and exocyst complex proteins. The hypothesis predicts either Dg’s function in mediating membrane-targeting of exocytic vesicles through interaction with the exocyst at the membrane site or exocyst-mediated Dg delivery to the cell membrane. Black circles outline proteins of the exocyst complex found to interact with Dg in the mass spectrometry screen. (B) Larval brain with GFP-negative Dg loss-of-function clones (outlined with yellow) immunostained with anti-Sec5. GFP and Sec5 are shown in separate channels. No obvious changes are observed in Sec5 protein levels or localization in Dg mutant clones when compared to GFP-positive control cells. Sec5 (red), GFP (green), and DAPI (blue). (C) GFP-negative Sec151 mutant clones in larval brains show altered Dg localization when compared to neighboring control cells (GFP-positive). Dg (red), GFP (green), and DAPI (blue). (D) GFP-negative Sec5E10 mutant clones show an impaired Dg expression pattern in neuronal cells marked with the neuron-specific marker Elav. Dg (red), GFP (green), and Elav (blue). Elav, GFP, and Dg are also shown in separate channels. Yellow dashed line outlines Dg-/Dg- clonal area. (E) The surface of the larval brain showing control and Sec15 clonal neuronal stem cells and their progeny. Note that in mutant cells (GFP-negative, yellow arrows) Dg protein is enriched in cytoplasmic puncta, more randomly distributed, and not properly delivered to the membrane in comparison to controls (green arrows). Dg (red), GFP (green), and Cad (blue). Scale bar 5 µm.
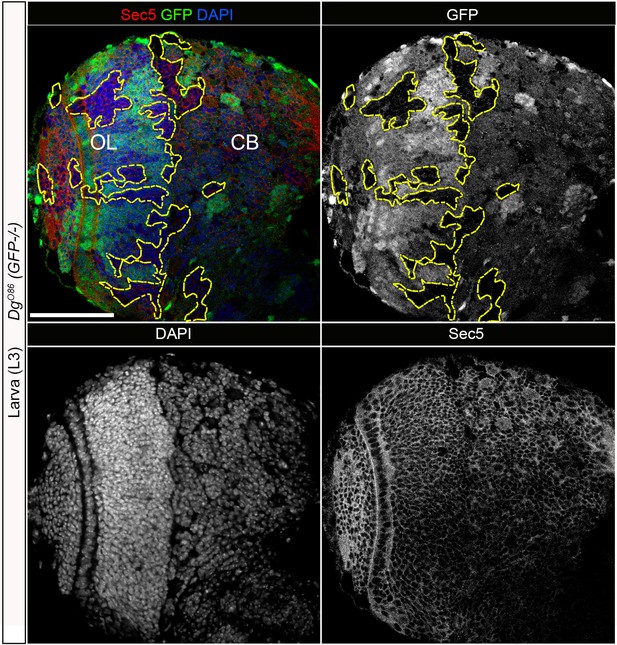
Dystroglycan (Dg) loss in neural cells does not have a significant effect on Sec5 protein.
Larval brain with GFP-negative Dg loss-of-function clones (outlined with yellow) immunostained with anti-Sec5. GFP, Sec5, and DAPI shown in separate channels. No obvious changes exist in Sec5 protein levels or localization in Dg mutant clones when compared to GFP-positive control cells. Scale bar 50 µm.
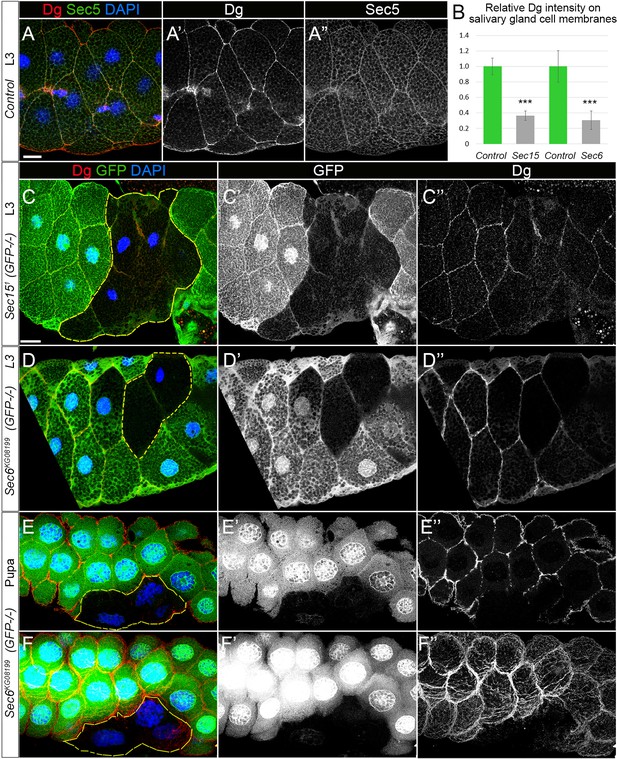
Perturbed exocyst causes downregulated dystroglycan (Dg) at the salivary gland cell membrane.
(A) Larval salivary gland stained with anti-Dg (A’) and anti-Sec5 (A”) shows co-localization of both proteins at the membrane. (B) The bar graph shows relative intensities of Dg staining at the membranes of control (GFP+) and Sec151 and Sec6KG08199 mutant (GFP-) salivary gland cells. n = 20 and 15 for control and Sec151, respectively, p=8.4×10−18 and n = 64 and 25 for control and Sec6KG08199, respectively, p=1.4×10−20. Two-tailed Student’s t-test was applied for statistical analyses. (C) Single z-section image of larval salivary gland with cells mutant for exocyst subunit Sec15 (GFP-negative clone outlined with yellow). (C’) GFP signal shown in a separate channel. (C’’) Dg staining reveals strong downregulation of Dg protein at the membranes of Sec15 mutant cells when compared to control (GFP-positive) cells. (D, E) Single z-section image of larval and pupal salivary glands with cells mutant for exocyst subunit Sec6 (GFP-negative clone outlined with yellow). (D’, E’) GFP signal shown in a separate channel. (D’’, E”) Dg staining shown in separate channel reveals strong downregulation of Dg protein at the membranes of Sec6 mutant cells when compared to control (GFP-positive cells). (F, F’), Maximum intensity projection image of the salivary gland shown in (F). Scale bar 25 µm.
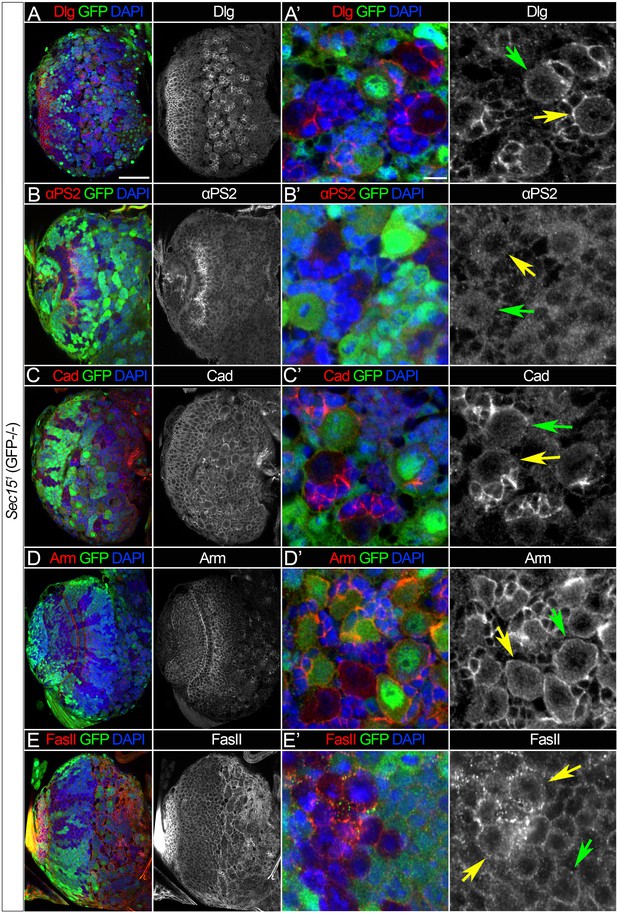
Sec15-mediated trafficking shows protein specificity.
(A–E) Analyses of the expression patterns of various cell adhesion proteins in brains containing sec15 mutant clones: Discs large 1 (Dlg, A, A’); αPS2 integrin (αPS2, B, B’); DE-Cadherin (Cad, C, C’); its binding partner Armadillo (Arm; D, D’); and Fasciclin II (FasII, E, E’). Note that at the level of light microscopy the protein localization of Dlg, αPS2, Cad, and Arm was not altered upon sec15 deficiency. (E, E’) FasII protein was enriched in cytoplasmic puncta, more randomly distributed, and not properly delivered to the membrane. Compare protein distribution in controls (green arrows) and sec15 mutant cells (yellow arrows). (A–E) show larval brain hemispheres, scale bar 25 µm. (A’–E’) show the surface of the larval brain, scale bar 5 µm.
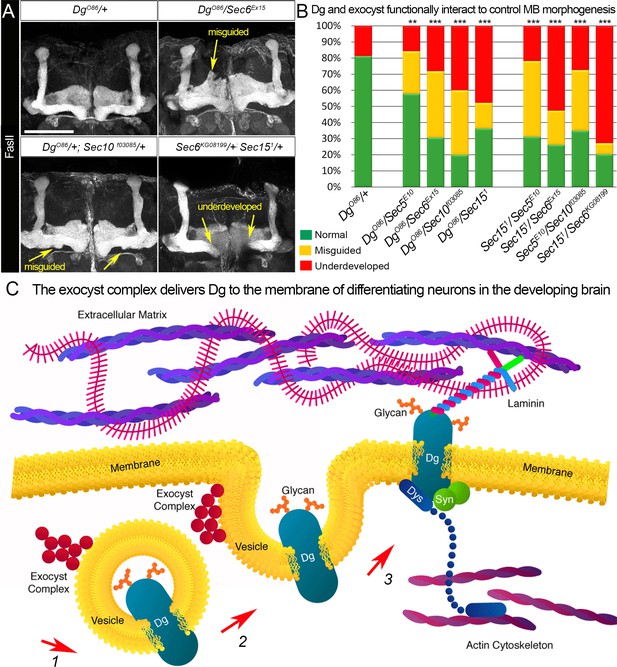
Proper patterning of the mushroom body (MB) neuropil is guaranteed by joint exocyst–dystroglycan (Dg) function.
(A) Abnormal MB lobe architecture phenotypes are observed in trans-heterozygous animals carrying only one copy of Dg and one copy of Sec (DgO86/+; Sec10f03085/+ and DgO86/Sec6Ex15), confirming a Dg–exocyst functional interaction in the process of MB morphogenesis (compare to Figure 3G–H). Viable combination of mutations in different exocyst subunits (Sec6KG08199/+; Sec151/+) results in similar phenotypes. The MB lobes are marked with Fasciclin II (FasII). Scale bar 50 µm. For more phenotypes, see also Figure 6—figure supplement 3. (B) Bar graph presents the quantification of the phenotypes observed in the genetic interaction analysis (see also Supplementary file 1). Reduction by one copy of any two of the analyzed genes significantly affects MB morphogenesis. For comparison of MB phenotypes, χ2 test was used. ***p≤0.001; **p≤0.01. See also Supplementary file 1. (C) In differentiating neurons, the Dg protein is loaded into the exocyst-positive secretory vesicle where it may be glycosylated (1). Then, the vesicle transports the glycoprotein Dg to the membrane. Upon vesicle fusion with the membrane (2), Dg is localized at the membrane where it acts as the extracellular matrix receptor (3). Exocyst-mediated delivery of Dg is necessary for the establishment of brain compartments and proper neuronal networking.
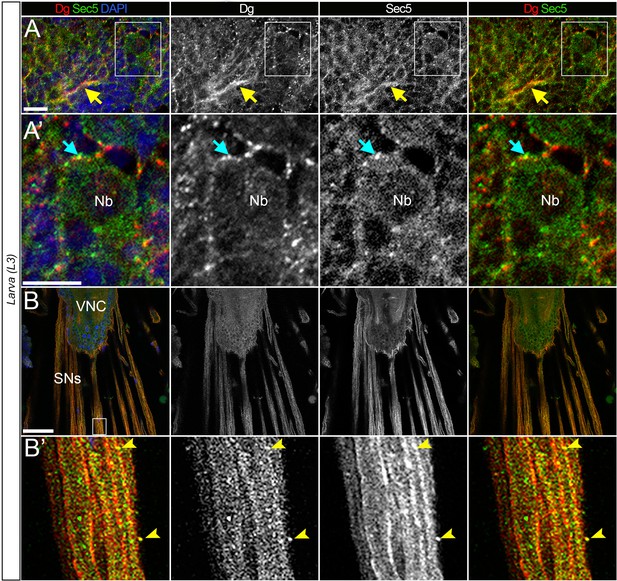
Dystroglycan (Dg) co-localizes with the exocyst in the differentiating neurons.
(A) Enlarged image of L3 brain shows co-localization of Dg and Sec5. Yellow arrow points to axonal extensions, where enriched Dg and Sec5 levels are detected. (A’) Zoomed-in region from A. In differentiating neural cells (cyan arrow), Dg can be found in speckles located in very close proximity to Sec5-positive puncta. Nb: neuroblast. (B) Image of fully differentiated segmental nerves (SNs) connected to ventral nerve cord (VNC) of third instar larva stained with anti-Dg and anti-Sec5. (B’) Enlarged view of area marked by white rectangle in B. SNs have a strong Dg expression and rarer scattered Sec5-positive speckles. Arrow points to a speckle positive for both proteins. From the enlarged images, it is evident that a gross pool of Dg protein is scattered in Sec5-positive puncta and enriched at the cell body periphery of differentiating neural cells (cyan arrows) and along projecting axonal tracts (yellow arrows). Scale bar 10 µm.
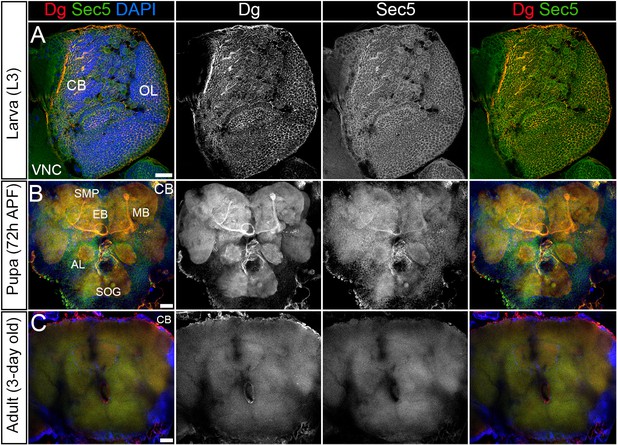
Dystroglycan (Dg) co-localizes with the exocyst in the developing brain.
(A) Brain lobe of a third instar larva (L3) immunostained with antibodies against Dg and Sec5, a subunit of the octameric exocyst complex, showing similar expression patterns. CB: central brain; OL: optic lobe. (B) Brain at 72 hr after puparium formation. Co-localization of Dg and Sec5 is seen in the midbrain neuropils: antennal lobe (AL), mushroom body (MB), superior medial protocerebrum (SMP), subesophageal ganglion (SOG), and the ellipsoid body (EB). (C) In adult brains, both proteins’ expression is strongly reduced compared to pre-adult stages, and strong enrichment in the neuropils is no longer observed. Scale bar 25 µm.
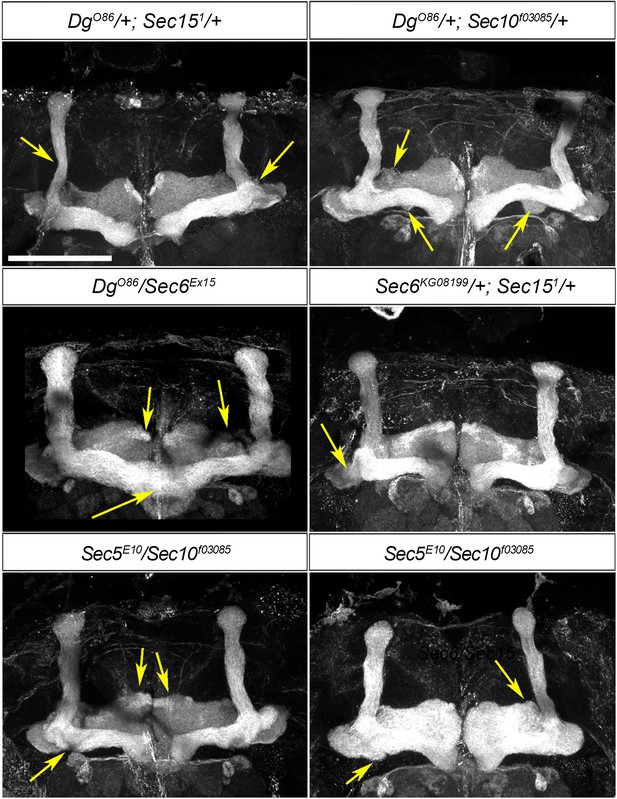
Mushroom body (MB) neuropil formation depends on exocyst–dystroglycan (Dg) interaction.
Viable combination of mutations in different exocyst subunits (Sec6KG08199/+; Sec151/+ and Sec5E10/Sec10f03085) results in abnormal MB lobe architecture (compare to controls in Figures 3A and 6A). Similar phenotypes are observed in trans-heterozygous animals carrying one copy of Dg and one copy of Sec (DgO86/+; Sec151/+ and DgO86/Sec6Ex15), confirming a functional interaction between Dg and the exocyst in the process of MB morphogenesis. Arrows point to the observed morphological abnormalities of MB lobes. The MB lobes are marked with Fasciclin II. Scale bar 50 µm.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | Anti-Dg (rabbit polyclonal) | Gift from Hannele Ruohola-Baker (Deng et al., 2003) | Dg | IF(1:1000) |
| Antibody | Anti-Sec5 (mouse monoclonal) | Gift from Thomas Schwarz (Langevin et al., 2005) | Sec5 | IF(1:50) |
| Antibody | Anti-GFP (chicken polyclonal) | Abcam | Cat# ab, 13970 | IF(1:5000) |
| Antibody | Anti-FAsII (mouse monoclonal) | Developmental Studies Hybridoma Bank | Cat# 1D4 | IF(1:50) |
| Antibody | Anti-Dlg (mouse monoclonal) | Developmental Studies Hybridoma Bank | Cat# 4F3 | IF(1:20) |
| Antibody | Anti-Elav (mouse monoclonal) | Developmental Studies Hybridoma Bank | Cat# 9F8A9 | IF(1:20) |
| Antibody | Anti-Arm (mouse monoclonal) | Developmental Studies Hybridoma Bank | Cat# N2 7A1 | IF(1:20) |
| Antibody | Anti-integrin alphaPS2 (mouse monoclonal) | Developmental Studies Hybridoma Bank | Cat# CF.2C7 | IF(1:50) |
| Antibody | Anti-DE-Cad (rat monoclonal) | Developmental Studies Hybridoma Bank | Cat# DCAD2 | IF(1:50) |
| Antibody | Anti-mouse IG1 Cy3 (goat polyclonal) | Jackson ImmunoResearch | Cat# 115-165-205 | Secondary antibody IF(1:500) |
| Antibody | Anti-rabbit Alexa 568 (goat polyclonal) | Thermo Fisher Scientific | Cat# A-11011 | Secondary antibody IF(1:500) |
| Antibody | Anti-rabbit Alexa 488 (goat polyclonal) | Thermo Fisher Scientific | Cat# A-11039 | Secondary antibody IF(1:500) |
| Genetic reagent (Drosophila melanogaster) | w[1118] | Bloomington Drosophila Stock Center | BDSC: 5905 FBgn0003996 | Wild-type strain |
| Genetic reagent (D. melanogaster) | Oregon-R-C | Bloomington Drosophila Stock Center | BDSC: 5 FBgn0003996 | Wild-type strain |
| Genetic reagent (D. melanogaster) | insc-Gal4 | Bloomington Drosophila Stock Center | BDSC: 8751 | w[*]; P{w[+mW.hs]=GawB}insc[Mz1407] |
| Genetic reagent (D. melanogaster) | C305a-Gal4 | Bloomington Drosophila Stock Center | BDSC: 30829 | w[*]; P{w[+mW.hs]=GawB}Cka[c305a] |
| Genetic reagent (D. melanogaster) | C309-Gal4 | Bloomington Drosophila Stock Center | BDSC: 6906 | w[*]; P{w[+mW.hs]=GawB}c309 |
| Genetic reagent (D. melanogaster) | 201Y-Gal4 | Bloomington Drosophila Stock Center | BDSC: 4440 | w[1118]; P{w[+mW.hs]=GawB}Tab2[201Y] |
| Genetic reagent (D. melanogaster) | elav-Gal4 | Bloomington Drosophila Stock Center | BDSC: 458 | w[1118], elavGal4; tubGal80ts (temperature sensitive) |
| Genetic reagent (D. melanogaster) | FRT40A GFP | Bloomington Drosophila Stock Center | BDSC: 5629 | hsFlp; Ubi GFP FRT 40A/CyO (clone induction line) |
| Genetic reagent (D. melanogaster) | FRT G13 GFP | Bloomington Drosophila Stock Center | BDSC: 5826 | hsFlp; FRTG13 GFP/CyO (clone induction line) |
| Genetic reagent (D. melanogaster) | FRT 82B GFP | Bloomington Drosophila Stock Center | BDSC: 5827 | hsFlp; +; FRT 82B GFP/TM3 (clone induction line) |
| Genetic reagent (D. melanogaster) | UAS-Dg::GFP | Gift from Marie-Laure Parmentier (Bogdanik et al., 2008) | Dg | UAS-Dg::GFP (Dg tagged GFP under control of UAS promoter) |
| Genetic reagent (D. melanogaster) | UAS-Dg | Gift from Hannele Ruohola-Baker (Deng et al., 2003) | Dg | UAS-Dg (Dg gene under control of UAS promoter) |
| Genetic reagent (D. melanogaster) | UAS-DgRNAi | Gift from Hannele Ruohola-Baker (Deng et al., 2003) | Dg | UAS-DgRNAi (Dg RNAi construct under control of UAS promoter) |
| Genetic reagent (D. melanogaster) | DgO86/CyO | Gift from Robert Ray (Christoforou et al., 2008) | Dg | DgO86/CyO (premature stop codon at 87 aa, strong hypomorph or null) |
| Genetic reagent (D. melanogaster) | DgO55/CyO | Gift from Robert Ray (Christoforou et al., 2008) | Dg | DgO55/CyO (premature stop codon at 653 aa, strong hypomorph or null) |
| Genetic reagent (D. melanogaster) | FRT G13 DgO55/CyO | Gift from Robert Ray (Christoforou et al., 2008) | Dg | w[*]; FRT G13 DgO86/SM6a (line for Dg mutant clone induction line) |
| Genetic reagent (D. melanogaster) | FRT40A, Sec5E10/CyO | Gift from Yohanns Bellaiche (Langevin et al., 2005) | Sec5 | w[*]; FRT40A, Sec5E10/CyO (null) |
| Genetic reagent (D. melanogaster) | FRT82B Sec151/TM3 | Gift from Yohanns Bellaiche (Langevin et al., 2005) | Sec15 | w[*]; FRT82B Sec151/TM3 (premature stop codon, strong hypomorph or null) |
| Genetic reagent (D. melanogaster) | FRT G13, Sec6KG08199/CyO | Gift from Yohanns Bellaiche (Langevin et al., 2005) | Sec6 | w[*]; FRT G13, Sec6KG08199/CyO (P-element insertion, null) |
| Genetic reagent (D. melanogaster) | FRT 82B, Sec10f03085/TM6, Tb | Gift from Yohanns Bellaiche (Langevin et al., 2005) | Sec10 | FRT 82B, Sec10f03085/TM6, Tb (PBac-element insertion, null) |
| Genetic reagent (D. melanogaster) | FRT G13 Sec6Ex15/CyO | Gift from Mark Metzstein (Jones et al., 2014) | Sec6 | w[*]; FRT G13 Sec6Ex15/CyO, Act-GFP (null) |
| Software, algorithm | Adobe Photoshop | Adobe | Adobe CC | |
| Software, algorithm | Zen 2011 | Carl Zeiss | Zen 2011 | |
| Software, algorithm | MaxQuant software 1.3.0.5 | Cox and Mann, 2008 | MaxQuant | |
| Software, algorithm | Markov clustering algorithm | https://micans.org/mcl/ | MLC | |
| Software, algorithm | Human disease-association enrichment analysis | http://ctdbase.org/tools | Disease Association | |
| Software, algorithm | Protein domain structure analysis | http://smart.embl-heidelberg.de | SMART | |
| Software, algorithm | Functional protein-association network clustering | https://string-db.org/ (Szklarczyk et al., 2015) | String v.10 | |
| Chemical compound, drug | Brilliant Blue R | Sigma Aldrich | Cat# 27816-25G | |
| Chemical compound, drug | Acetic acid | Sigma Aldrich | Cat# 27225-1 L-M | |
| Chemical compound, drug | Chloroform | Sigma Aldrich | Cat# 288306–2L | |
| Chemical compound, drug | Glycerol | Sigma Aldrich | Cat# G6279-1L | |
| Chemical compound, drug | Sodium azide | Sigma Aldrich | Cat# S2002-25G | |
| Chemical compound, drug | Formaldehyde, 16% | Polysciences Inc | Cat# 18814-20 | Methanol free, ultra pure |
| Commercial assay or kit | RealTime ready Cell Lysis Kit | Roche | Cat# 06366 821001 | |
| Commercial assay or kit | GFP-Trap A Kit | Chromotek | Cat# 5062685 | |
| Other | DAPI stain | Sigma Aldrich | Cat# D9542-10MG | IF concentration used: 1 µg/mL |
| Other | Normal Goat Serum | Abcam | Cat# ab7481 | |
| Other | Trans-Blot Turbo Mini PVDF Transfer Packs 0.2 µm | Bio-Rad | Cat# 1704156 | |
| Other | Immun-Blot PVDF/Filter Paper Sandwiches | Bio-Rad | Cat# 1620218 | |
| Other | Precision Plus Protein Kaleidoscope Prestained Protein Standard | Bio-Rad | Cat# 1610375 | |
| Other | 10× Tris/Glycine/SDS Running Buffer | Bio-Rad | Cat# 1610772 | |
| Other | NuPAGE Novex 4–12% Protein Gels | Thermo Fisher Scientific | Cat# NP0321PK2 | |
| Other | PicoFrit Columns | New Objective | Cat# PF360-75-15-N | |
| Other | Paraplast Plus | Sigma Aldrich | Cat# 76258-1KG | |
| Other | Casein Blocking Buffer 10x | Sigma Aldrich | Cat# B6429-500ML | |
| Other | Hematoxylin Solution, Mayer’s | Sigma Aldrich | Cat# MHS16-500ML | |
| Other | Eosin Y solution, aqueous | Sigma Aldrich | Cat# HT110232 | |
| Other | DPX Mountant for histology | Sigma Aldrich | Cat# 06522-100ML | |
| Other | PBS buffer (10× Dulbecco's) | AppliChem | Cat# A0965,9010 | |
| Other | LSM700 confocal laser-scanning microscope | Carl Zeiss | LSM700 | |
| Other | Hyrax M25 microtome | Carl Zeiss | Hyrax M25 | |
| Other | ReproSil-Pur analytical column 120 C18-AQ | Dr. Maisch GmbH | ReproSil-Pur | |
| Other | Nanoflow liquid chromatography system EASY n-LC 1000 | Thermo Scientific | Nanoflow | |
| Other | Q Exactive Hybrid Quadrupole-Orbitrap | Thermo Scientific | Orbitrap |
Additional files
-
Supplementary file 1
Mushroom body (MB) morphology is affected by deregulation of dystroglycan (Dg) and the exocyst.
*Misguided = fused β lobes or β lobe neurons projecting into γ lobe space.
**Underdeveloped = smaller α or β lobes due to α lobe neurons projecting into β lobe space or vice versa.
aCompared to Oregon (Control).
bCompared to DgO86/+ (Control).
cCompared to DgO86/Sec6Ex15.
dCompared to DgO86/Sec10f03085.
eCompared to DgO86/Sec151.
For comparison of the observed phenotypes, two-way tables and χ2 test were used.
- https://cdn.elifesciences.org/articles/63868/elife-63868-supp1-v2.docx
-
Supplementary file 2
Dystroglycan (Dg)-interacting proteins in neurons.
- https://cdn.elifesciences.org/articles/63868/elife-63868-supp2-v2.docx
-
Supplementary file 3
Human disease enrichment in the dystroglycan (Dg) neuronal interactome network.
For human disease-association enrichment analysis, the entry of human orthologs of identified Dg-interacting components (Table S2) was examined with Comparative Toxicogenomics Database (CTD) Disease Tool http://ctdbase.org/tools. The corrected threshold value of p<0.001 was used.
- https://cdn.elifesciences.org/articles/63868/elife-63868-supp3-v2.docx
-
Supplementary file 4
Functional enrichments in the dystroglycan (Dg) neuronal interactome network based on cellular component (GO) terms.
- https://cdn.elifesciences.org/articles/63868/elife-63868-supp4-v2.docx
-
Supplementary file 5
Dystroglycan (Dg)-associated components placed into the protein-interaction network identified by Markov clustering algorithm (MCL).
STRING database (https://string-db.org) was used to identify functional networks with the interaction score of high confidence (0.700). Interacting protein are clustered by the MCL with the inflation parameter (3). For the schematic representation of the obtained interaction network, see Figure 4D.
- https://cdn.elifesciences.org/articles/63868/elife-63868-supp5-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/63868/elife-63868-transrepform-v2.docx





