Basis of specificity for a conserved and promiscuous chromatin remodeling protein
Figures
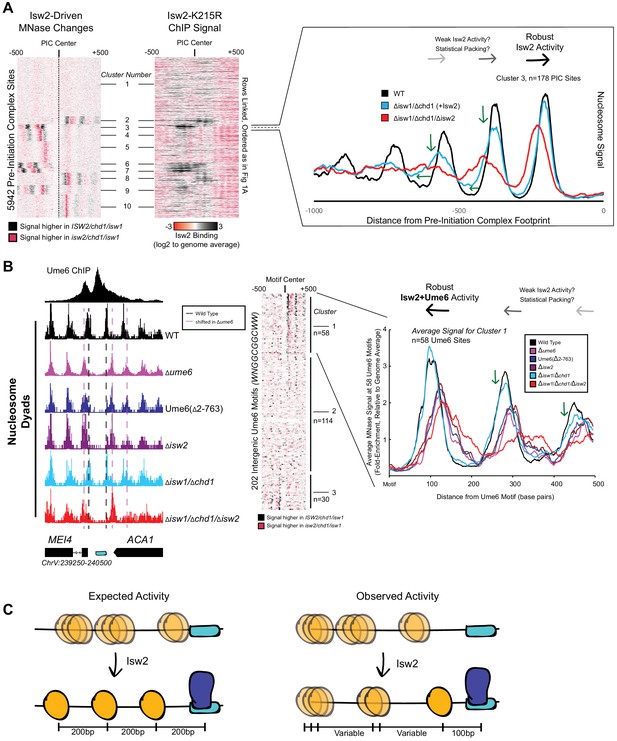
Isw2 is a specialist remodeler that positions single nucleosomes at target sites.
(A) (Left) Clustered heatmap showing differences in nucleosome dyad signal between isw2/isw1/chd1 and ISW2/isw1/chd1 strains at 5942 pre-initiation complex sites (PICs). Black indicates positions where Isw2 preferentially positions nucleosomes compared to the strain lacking Isw2. (Middle) Heatmap of ISW2(K215R) ChIP signal, with rows linked to the PIC data on the left, shows that Isw2-dependent nucleosome changes overlap with regions where Isw2 is present. (Right) Average nucleosome dyad signal for wild type (WT) (black), isw1/chd1 (cyan), and isw2/isw1/chd1 (red) strains for the 178 PIC sites in cluster 3. Black arrows denote Isw2-driven nucleosome shifts. Green arrows indicate rapid decay of positioning at PIC-distal nucleosomes in the ISW2/isw1/chd1 mutant. (B) (Left) Genome Browser image showing nucleosome dyad signal at a unscheduled meiotic gene expression (Ume6) motif (cyan rectangle) for indicated strains. Vertical gray dashed line denotes the motif-proximal WT nucleosome positions while vertical pink dashed line indicates the nucleosome positions in the absence of Ume6 or Isw2. (Center) Clustered heatmap showing the difference in nucleosome dyad signal between isw2/isw1/chd1 and ISW2/isw1/chd1 strains at 202 intergenic Ume6 motifs. Black indicates positions where Isw2 preferentially positions nucleosomes compared to strains lacking Isw2. (Right) Average nucleosome dyad signal for indicated strains at Ume6 motifs in cluster 1. Black arrows indicate direction of nucleosome positioning by Isw2. Green arrows signify decreased positioning of motif-distal nucleosomes in the ISW2/isw1/chd1 strain (cyan) compared to WT (black). (C) (Left) Cartoon depicting the expected activity of Isw2 at barrier elements according to current biochemical data and nucleosome positioning models. Isw2 is thought to move nucleosomes away from bound factors and space nucleosomes with an approximately 200 base pair repeat length. (Right) Cartoon of the observed activity of Isw2 at target sites where only a motif-proximal single nucleosome is precisely positioned but distal nucleosomes are not well-spaced by Isw2.
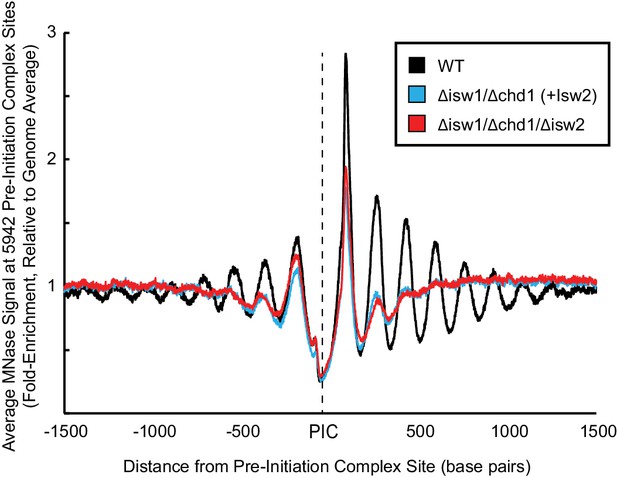
Isw2 is a precise specialist at target nucleosomes.
Meta-analysis nucleosome dyad signal at all 5942 pre-initiation complex sites (PICs) shows that Isw2 confers no global nucleosome organizing activity throughout yeast cells. WT: wild type.
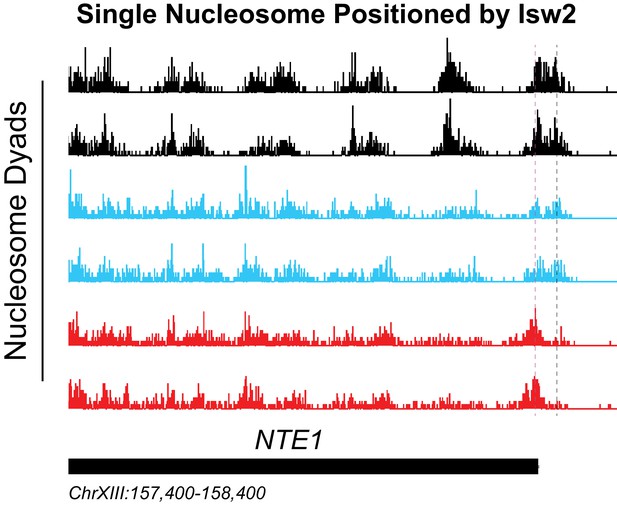
Isw2 is a precise specialist at target nucleosomes.
Example Isw2 target locus where addition of Isw2 in the absence of Isw1 and Chd1 leads to positioning of a single motif-proximal nucleosome at the NTE1 locus. Vertical gray dashed lines indicate the wild-type nucleosome locations while vertical dashed pink lines indicate an alternate isw2 deletion strain position at the motif-proximal nucleosome. The ISW2/chd1/isw1 strain can position the motif-proximal nucleosome, but all distal nucleosomes are disorganized much like the isw2/chd1/isw1 strain.
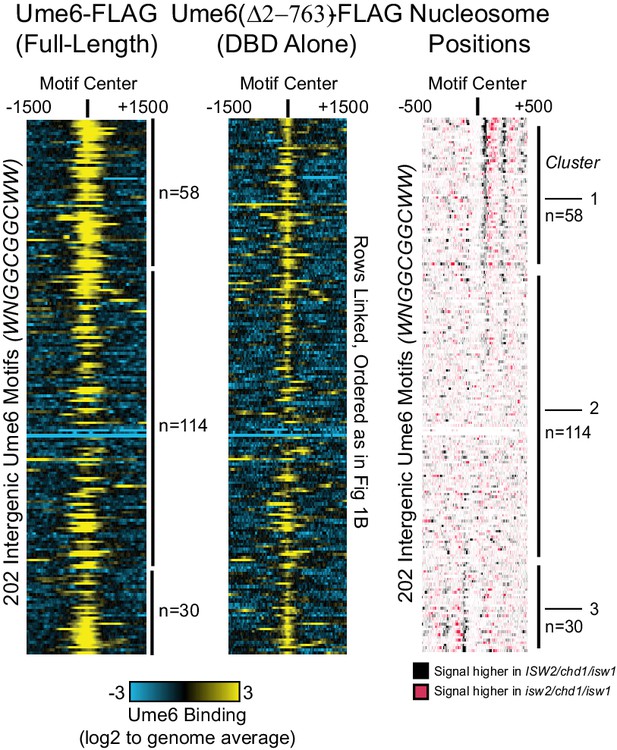
Isw2 is a precise specialist at target nucleosomes.
Heatmaps showing that full-length unscheduled meiotic gene expression (Ume6)-FLAG (left) and Ume6(∆2–763)-FLAG (middle) associate with similar targets. (Right) Clustered heatmap from Figure 1B showing the difference in nucleosome dyad signal between isw2/isw1/chd1 and ISW2/isw1/chd1 strains at 202 intergenic Ume6 motifs. Black indicates positions where Isw2 preferentially positions nucleosomes compared to strains lacking Isw2. All rows are linked, and all 202 intergenic Ume6 motifs are displayed.
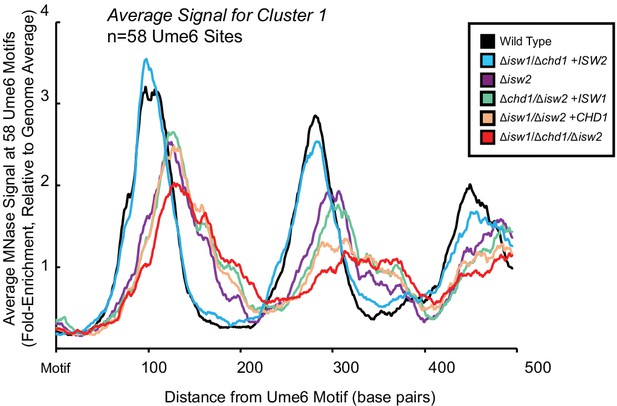
Isw2 is a precise specialist at target nucleosomes.
Meta-analysis of nucleosome dyad signal for indicated strains at the 58 unscheduled meiotic gene expression (Ume6) target loci associated with cluster 1 in Figure 1B demonstrates that Isw2 but not Chd1 or Isw1 is necessary and sufficient to position nucleosomes at Ume6 target loci.
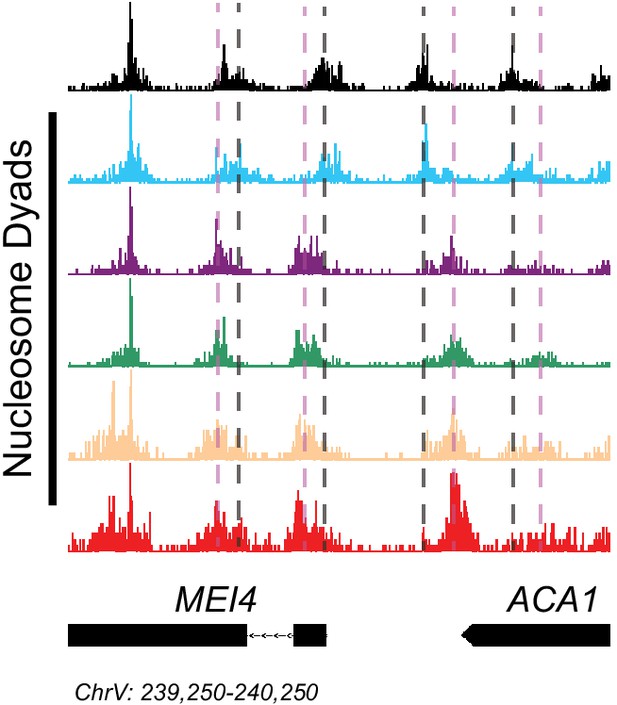
Isw2 is a precise specialist at target nucleosomes.
Genome Browser image showing nucleosome dyad signal for indicated strains at the MEI4-ACA1 locus, a representative unscheduled meiotic gene expression (Ume6) target locus. The color schemes are shared between Figure 1—figure supplement 1D and E according to the key between figures. Vertical gray lines indicate wild-type nucleosome positions while vertical pink lines indicate ectopic nucleosome positions associated with loss of Isw2 activity.
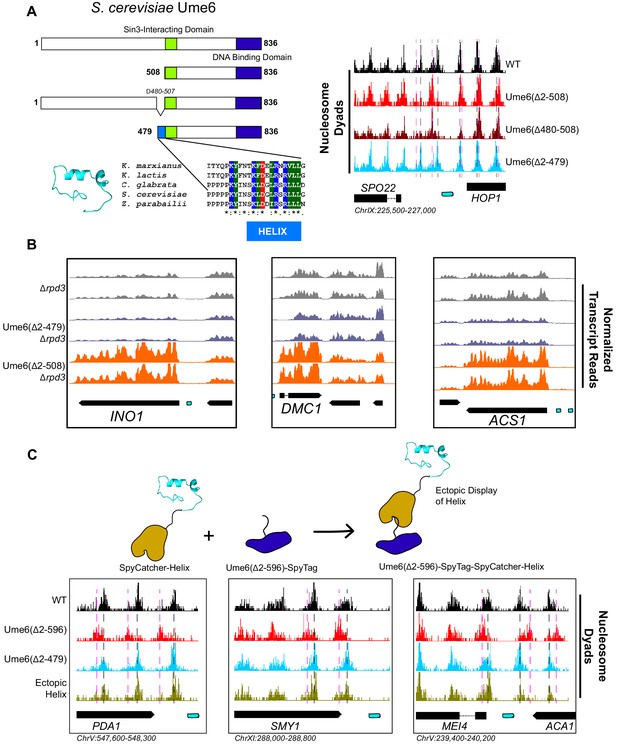
A small predicted helix is the Isw2-recruitment epitope in unscheduled meiotic gene expression (Ume6).
(A) (Top left) Schematic diagram of Ume6 truncation and deletion constructs used to identify the Isw2-recruitment epitope, with the known Sin3-interacting domain depicted as a green square, the DNA binding domain as a dark blue rectangle, and the putative Isw2-recruitment helix as a light blue rectangle. (Bottom left) Modeled helical peptide (by Phyre2) and sequence conservation of the identified Isw2-recruitment motif in Ume6 constructs from other yeasts. Asterisks denote invariant residues. (Right) Nucleosome dyad signal for Ume6 truncation and deletion strains indicates deletion of the region from residues 480 to 507 completely abrogates nucleosome positioning by Isw2 at Ume6 target sites. Vertical dashed gray lines denote wild-type (WT) positions of nucleosomes while vertical dashed pink lines indicate isw2 or ume6-deficient positions of nucleosomes. (B) Genome Browser image showing transcript abundance at three Ume6 target sites for yeast strains lacking Rpd3 with WT Ume6 (gray), Ume6(∆2–479) (blue), and Ume6(∆2–508) (orange). Grossly increased transcription is seen when residues 480–507 are deleted, consistent with expected transcriptional increase associated with loss of Isw2 and Rpd3. Upstream repression sequence (URS) sites are indicated as cyan rectangles. No significant increase in transcription is detected when Ume6 residues 2–479 are deleted. Biological replicates are shown to highlight reproducibility. (C) (Top) Cartoon schematic for ectopic display of the Isw2-recruiting helix (residues 480–507) to the C-terminus of a truncated Ume6 construct lacking Isw2-directed nucleosome positioning. A short SpyTag is appended to the C-terminus of the Ume6 construct and residues 480–507 are fused to the SpyCatcher domain and introduced on a yeast expression vector. (Bottom) Nucleosome dyad signal demonstrating recovery of Isw2-directed nucleosome positions at a subset of Ume6 target genes by the ectopically displayed helical element. Vertical dashed gray lines denote WT positions of nucleosomes while vertical dashed pink lines indicate isw2 or ume6-deficient positions of nucleosomes. URS sites are indicated as cyan rectangles. Individual biological replicates for nucleosome positions after ectopic display of the recruitment helix are provided in Figure 2—figure supplement 4.
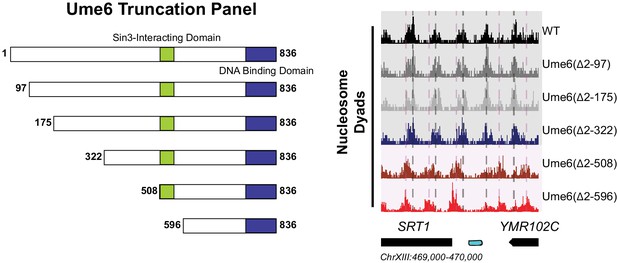
The unscheduled meiotic gene expression (Ume6) helix between residues 479 and 508 recruits Isw2 to Ume6 targets.
(Left) Schematic representation of the truncation panel initially used to identify the region of Ume6 required for Isw2 recruitment. Green square indicates the part of Ume6 known to recruit SIN3 while the blue rectangle indicates the Ume6 DNA binding domain. (Right) Genome Browser image for a representative locus showing nucleosome dyad signal for the indicated strains. Vertical gray lines denote wild-type (WT) nucleosome positions while vertical pink lines indicate ectopic nucleosome positions associated with truncations beyond 322 N-terminal amino acids. The Isw2-recruitment domain was thus determined to be between residues 322 and 508 in Ume6.
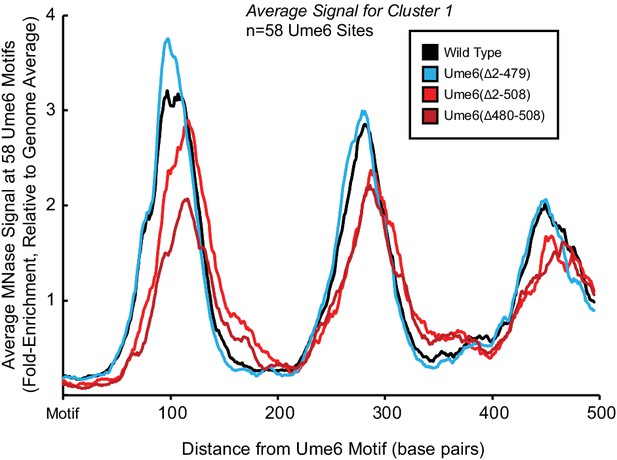
The unscheduled meiotic gene expression (Ume6) helix between residues 479 and 508 recruits Isw2 to Ume6 targets.
Meta-analysis of nucleosome dyad signal at the 58 Ume6 target loci associated with cluster 1 in Figure 1B showing loss of residues 479–508 from Ume6 results in ectopic nucleosome positioning while N-terminal deletion of residues 2–479 preserves wild-type (WT) nucleosome positioning at Ume6 sites.
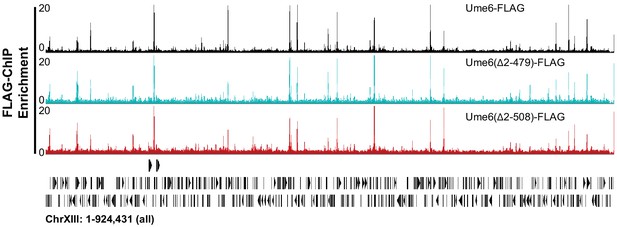
The unscheduled meiotic gene expression (Ume6) helix between residues 479 and 508 recruits Isw2 to Ume6 targets.
Genome Browser image demonstrating no loss in Ume6-ChIP signal for relevant Ume6 truncations across all of chromosome XIII. Signal is read-corrected enrichment, relative to genome average.
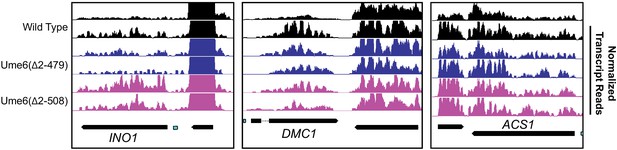
Transcription data support a role of unscheduled meiotic gene expression (Ume6) residues 479–508 for Isw2 recruitment and not Rpd3 activity.
Genome Browser images showing transcript abundance for the three representative Ume6 target loci for indicated Ume6 truncation strains in the presence of functional Rpd3, demonstrating the expected very subtle increase in transcription associated with loss of Isw2 at the INO1 locus. URS sites are indicated as cyan rectangles. Biological replicates for each strain are shown.
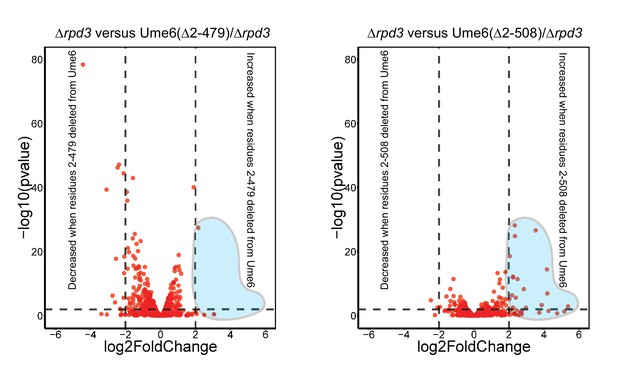
Transcription data support a role of unscheduled meiotic gene expression (Ume6) residues 479–508 for Isw2 recruitment and not Rpd3 activity.
Volcano plots for genes containing Ume6 motifs showing log2 change in transcription and associated statistical significance for indicated strains. Horizontal dashed line indicates a p value of 0.01. Vertical dashed lines indicate a change in transcription of ±4-fold. Retention of residues 479–508 prevents large-scale increases in transcription associated with deletion of Rpd3 and Isw2 (see blue shaded region).
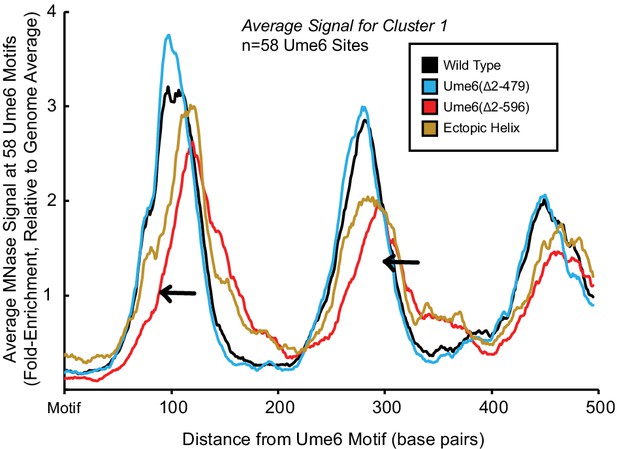
Ectopic display of the unscheduled meiotic gene expression (Ume6) helical element can rescue Isw2 activity at Ume6 targets.
Meta-analysis of nucleosome dyad signal at the 58 Ume6 target loci associated with cluster 1 in Figure 1B showing partial rescue of nucleosome positioning when residues 479–508 are ectopically displayed on the C-terminus of Ume6 ∆2–596 through SpyTag-SpyCatcher pairs. Black arrows indicate wild-type signal gained by ectopic display of the recruitment helix.
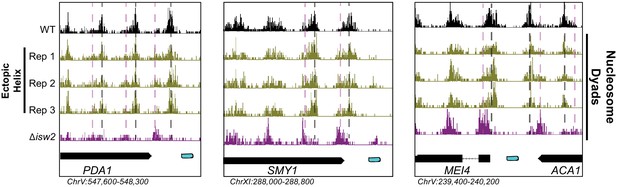
Ectopic display of the unscheduled meiotic gene expression (Ume6) helical element can rescue Isw2 activity at Ume6 targets.
Genome Browser image showing additional replicates where SpyTag/SpyCatcher-mediated ectopic tethering of Ume6 residues 479–508 recovers wild-type nucleosome positions at Ume6 targets (as in Figure 2C).
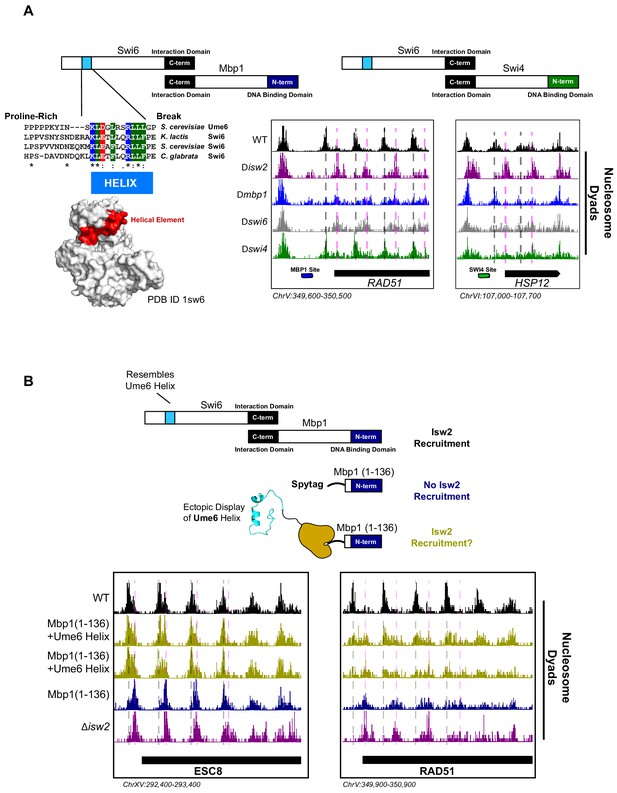
The cell cycle regulator Swi6 contains a similar helical element and recruits Isw2 to MBF and SBF target genes.
(A) (Top left) Schematic representation of the Swi6-Mbp1 MBF complex. Swi6 interacts with Mbp1 through the C-terminal domain (black rectangle). Mbp1 has an N-terminal DNA binding domain (dark blue rectangle). The putative Isw2-recruitment helix is in the Swi6 N-terminus (light blue rectangle). (Center left) Conserved residues in the putative Isw2-recruitment helix in Swi6 for three yeast species compared to the Isw2-recruitment helix in Ume6 for S. cerevisiae. (Bottom left) Crystal structure (Protein Data Bank [PDB] ID 1sw6) showing the location of the surface-exposed, conserved helical element from Swi6 in red. (Top right) Schematic representation of the Swi6-Swi4 SBF complex. Swi6 interacts with Swi4 through the C-terminal domain (black rectangle). Swi4 has an N-terminal DNA binding domain (green rectangle). Putative Isw2-recruitment helix is shown (small blue rectangle). (Bottom center) Genome Browser image showing nucleosome dyad signal for indicated strains at the RAD51 locus, an MBF target gene with an indicated Mbp1 binding motif (blue rectangle). Wild-type (WT) nucleosome positions are indicated by vertical dashed gray lines while ectopic positions associated with isw2, mbp1, and swi6 deletion strains are indicated by vertical dashed pink lines. (Bottom right) Genome Browser image showing nucleosome dyad signal for indicated strains at the HSP12 locus, an SBF target gene with an indicated Swi4 binding motif (green rectangle). WT positions are denoted by vertical gray dashed lines while ectopic nucleosome positions associated with isw2, swi6, and swi4 deletion strains are indicated with vertical pink dashed lines. (B) (Top) Schematic representation of constructs used to determine if ectopic display of an Isw2-recruitment helix on the Mbp1 N-terminus could recover Isw2-positioned nucleosomes at Mbp1 target genes. Either WT Mbp1, a C-terminal deletion of Mbp1 leaving only the DNA binding domain and an appended SpyTag, or a C-terminal deletion of Mbp1 leaving the DNA binding domain and SpyTag with constitutively expressed SpyCatcher fused to the Isw2-recruitment helix from Ume6 was examined. (Bottom) Genome Browser image showing nucleosome dyad signal for indicated strains at the ESC8 (left) or RAD51 (right) loci. Gray vertical dashed lines indicate WT nucleosome positions while vertical dashed pink lines indicate ectopic nucleosome positions associated with inactive Isw2 or Mbp1/Swi6. Biological replicates for ectopic display of the recruitment helix are provided as two separate tracks (gold) to emphasize reproducibility.
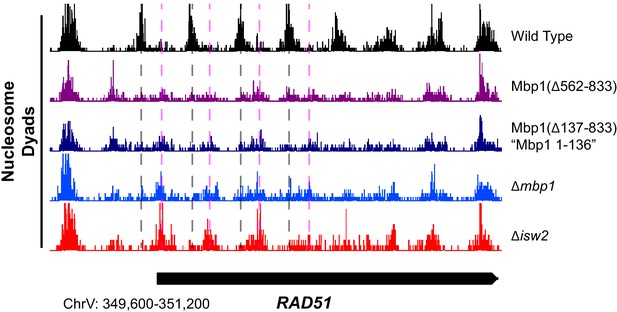
Truncation of the Mbp1 C-terminus eliminates Isw2-directed nucleosome positioning at Mbp1 targets.
Genome Browser image showing nucleosome positioning at the RAD51 locus for indicated strains. Dashed vertical gray lines denote positions of nucleosomes in the wild-type strain while dashed vertical pink lines show positions of nucleosomes in the absence of functional Isw2.
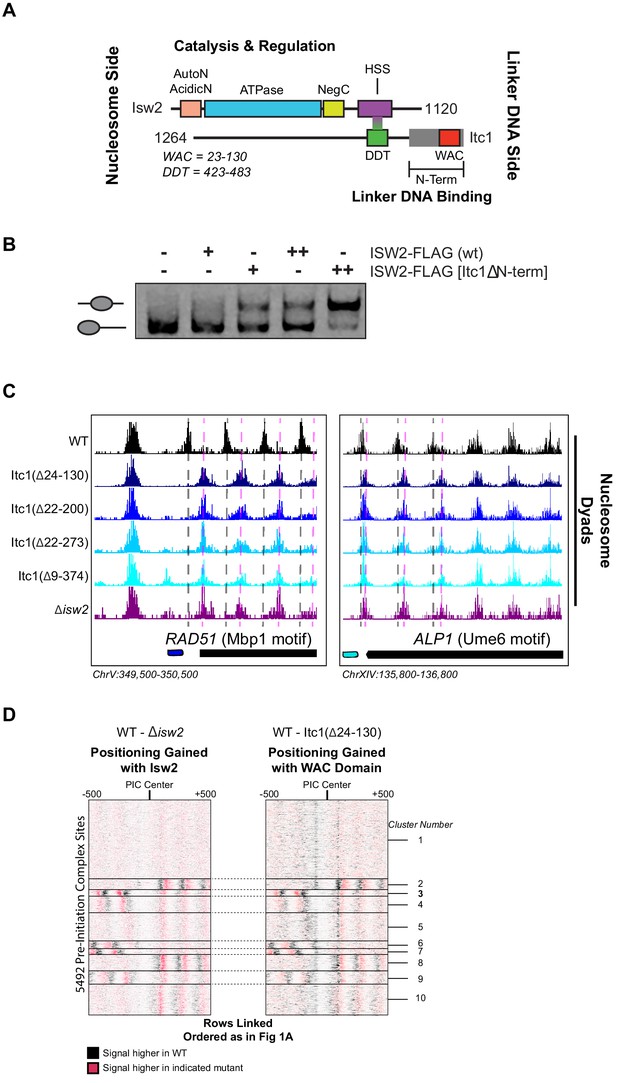
The N-terminal WAC domain in Itc1 couples Isw2 biochemical activity to all Isw2 genomic targets.
(A) Cartoon representation the Isw2 and Itc1 subunits of the yeast ISW2 complex. Isw2 possesses autoregulatory domains on either side of the catalytic ATPase domain (AutoN and NegC). The HAND-SANT-SLIDE (HSS) domain of Isw2 interacts with the DDT domain of Itc1 for complex formation. Itc1 has an N-terminal region thought to act as a length-sensing domain (gray rectangle) and an N-terminal WAC domain with putative nonspecific linker DNA binding ability. (B) Nucleosome sliding assay demonstrating that deletion of the N-terminal domain (∆9–374) from Itc1 does not impair nucleosome sliding in vitro by the Isw2 complex. Higher electrophoretic mobility indicates end-positioned (unslid) nucleosomes while lower electrophoretic mobility indicates centrally positioned (slid) nucleosomes. Isw2-FLAG complexes were purified from exponentially growing yeast cells as described in 'Materials and methods'. Amount of Isw2 added was 1 µl (+) or 1.5 µl (++). Sliding assays were performed three independent times with similar results. (C) Genome Browser images showing nucleosome dyad positions for indicated strains at RAD51 and ALP1, two representative Isw2 targets. Only wild-type (WT) cells display the proper nucleosome positions (vertical gray dashed lines) while all Itc1 truncations and isw2 deletion display similar ectopic nucleosome positions (vertical pink dashed lines). (D) Heatmap comparing difference in nucleosome positions at 5942 PIC locations for isw2 deletion versus WT strains (left) and Itc1(∆24–130) versus WT strains (right). Black indicates where nucleosomes are shifted by functional Isw2 while red indicates where nucleosomes shift when Isw2 complex is perturbed. All rows are linked and ordered identically to Figure 1A.
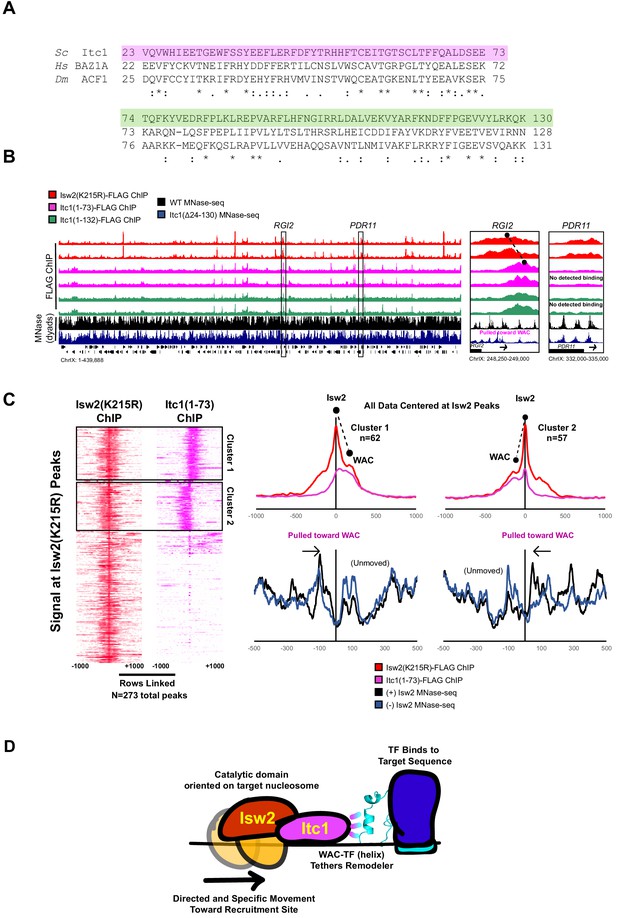
The Itc1 WAC domain associates with genomic Isw2 targets and orients Isw2 on the proper nucleosomes.
(A) Sequence conservation for regions of Itc1 examined by ChIP. Itc1(1–73)-FLAG incorporates the pink highlighted region while Itc1(1–132)-FLAG incorporates the pink and green highlighted regions. Sequence conservation is shown relative to human BAZ1A and Drosophila melanogaster Acf1, two widely studied Itc1 orthologs. (B) (Left) Full view of yeast chromosome IX showing Isw2(K215R)-FLAG ChIP (red), Itc1(1–73)-FLAG ChIP (pink), Itc1(1–132)-FLAG ChIP (green), nucleosome dyad signal from wild-type (WT) yeast (black), and nucleosome dyad signal from Itc1(∆24–130) yeast (blue). Regions indicated by black rectangles are shown with higher resolution on the right. (Right) Zoomed-in view of a locus where Isw2-ChIP and Itc1 truncation ChIP overlap (RGI2) or where only Isw2 binding is detected (PDR11). Black circles indicate center of ChIP peaks and are connected by a dashed black line to highlight offset of indicated peaks. (C) (Left) Heatmap showing 273 detected Isw2 ChIP peaks (red) clustered by associated Itc1(1–73)-FLAG ChIP (pink). The two clusters (right-side Itc1 and left-side Itc1) are shown on the right. (Right) Meta-analysis of Isw2(K215R)-FLAG ChIP signal at 62 cluster 1 peaks or 57 cluster 2 peaks (from left) with associated Itc1(1–73)-FLAG signal. The offset between Isw2 and Itc1 is indicated by two circles connected by a dashed line. Associated nucleosome positions for WT and isw2 deletion strains for each cluster are shown below in black and blue, respectively. All data are centered at called Isw2 peaks. (D) Cartoon representation for how the N-terminal WAC domain of Itc1 interacts with a helical element in a sequence-specific DNA-associated transcription factor to orient Isw2 on the proper motif-proximal nucleosome for directional movement toward the recruitment site.
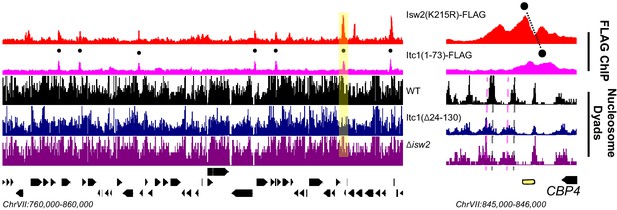
The WAC domain orients the Isw2 catalytic domain at nearly half of detected Isw2 targets in yeast.
(Left) Genome Browser image showing Isw2(K215R)-FLAG and Itc1(1–73)-FLAG ChIP signal and wild type (WT), Itc1(∆24–130), and ∆isw2 nucleosome dyad signal for a 100 kb section of chromosome VII. Black circles indicate genomic loci where Isw2(K215R) and Itc1(1–73) ChIP signal overlap. (Right) Zoomed Genome Browser image of the section highlighted in yellow showing offset nature of the Isw2 peak and Itc1 peak. Black circles connected by a black line indicate the offset nature of the Isw2 and Itc1 ChIP peaks. The shifted nucleosome is to the left of the Itc1-Isw2 axis.
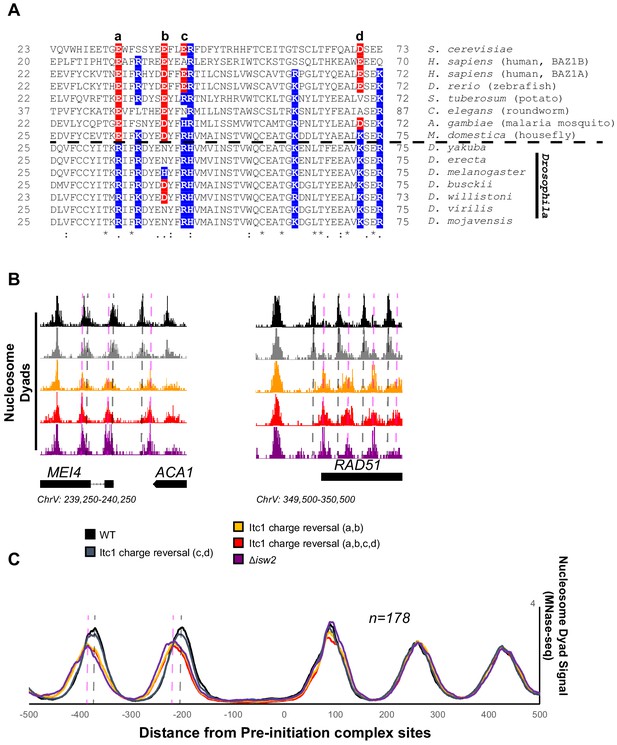
Essential targeting-specific charged residues in the conserved WAC domain is lost in Drosophila.
(A) Sequence conservation of the N-terminal region (22–73) of Itc1 across various organisms with key charged residues highlighted in blue or red for positive and negative charge, respectively. Horizontal dashed line indicates separation of all other species from members of the Drosophila genus. (B) Genome Browser image showing nucleosome dyad signal at two representative Isw2 target loci. Wild-type (WT) nucleosome positions are indicated by gray vertical dashed lines while ectopic nucleosome positions associated with isw2 deletion or indicated charge reversal mutations are denoted by vertical pink dashed lines. (C) Meta-analysis of nucleosome dyad signal at 178 PIC sites associated with cluster 3 (from Figure 1A). Only WT and charge reversal c, d display proper nucleosome positions while charge reversal a, b or a, b, c, and d display ectopic positions identical to deletion of ISW2 completely.
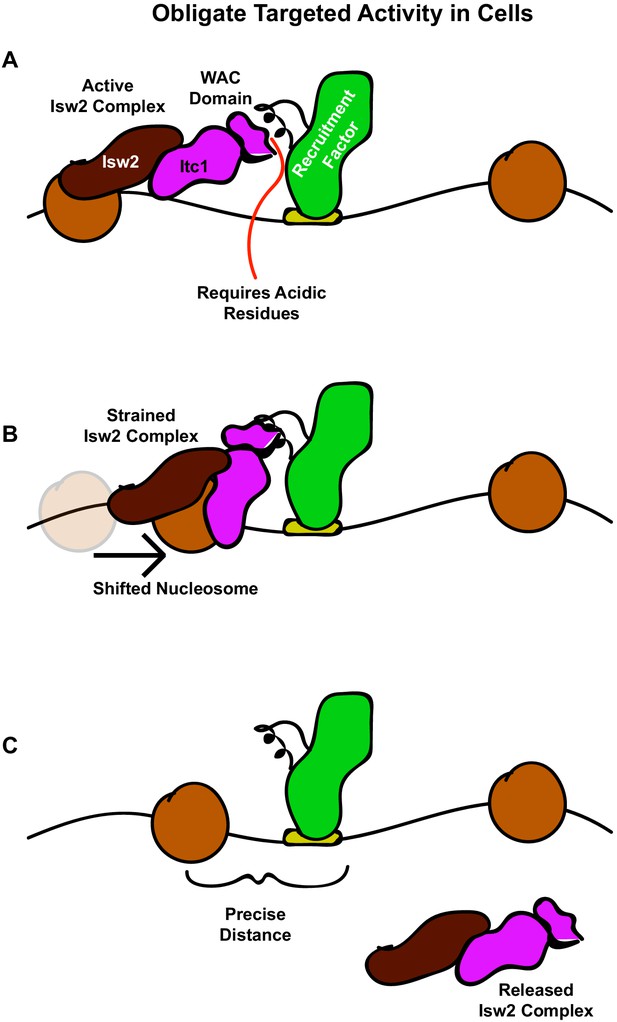
The interacting barrier model for specific nucleosome placement by Isw2.
(A) A DNA binding factor with an Isw2-recruitment helix (or other epitope) associates with DNA. The WAC domain of Itc1 engages with the recruitment epitope to proximally align the catalytic subunit of Isw2 with the proper nucleosome. (B) Nucleosome sliding by Isw2 creates a nucleosome position that is too close to the recruitment epitope for proper alignment of the recruitment epitope – Itc1 WAC – Isw2 catalytic subunit axis, leading to a ‘strained complex’. (C) The precise distance between the DNA-bound Isw2-recruitment factor and proximal nucleosome after nucleosome positioning by Isw2 is no longer a good substrate for Itc1 WAC interaction and further remodeling, so the Isw2 complex diffuses to new target loci.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Saccharomyces cerevisiae) | W303-1A | Laboratory ofRodney Rothstein | American Type Culture Collection (ATCC): 208352 | |
| Strain, strainbackground (S. cerevisiae) | For additional strains, see Supplementary file 1 | |||
| Antibody | Anti-FLAG M2 (mouse monoclonal) | Sigma | Cat# F1084;RRID:AB_262044 | 5 μl antibody per 30 μl beads |
| Antibody | Anti-FLAG M2 magneticbeads (mouse monoclonal) | Sigma | Cat# M8823;RRID:AB_2637089 | |
| Recombinant DNA reagent | Plasmid | For plasmid informatio,n see Supplementary file 2 | ||
| Peptide, recombinant protein | Dynabeads Protein G | Thermo Fisher Scientific | Cat# 10004D | 200 μl |
| Peptide, recombinant protein | 3x FLAG | Sigma | Cat# F4799-4MG | MDYKDHDGDYKDHDIDYKDDDDK |
| Commercial assay or kit | MinElute PCR purification kit | Qiagen | Cat# 28006 | |
| Commercial assay or kit | Ovation Ultralow System V2 | NuGEN | Cat# 0344NB-32 | |
| Commercial assay or kit | Universal Plus mRNA-Seq | NuGEN | Cat# 0508-32 | |
| Commercialassay or kit | Quant-IT PicoGreen dsDNA assay kit | Invitrogen | Cat# P7589 | |
| Chemical compound, drug | Proteoloc protease inhibitor cocktail (EDTA-free) | Expedeon | Cat# 44214 | |
| Chemical compound, drug | Nuclease, micrococcal | Worthington Biochemical | Cat# LS004798 | |
| Chemical compound, drug | Zymolyase (R) 100T | AMS Bio | Cat# 120493-1 | |
| Chemical compound, drug | Agencourt AMPure XP beads | Beckman Coulter | Cat# A63881 | |
| Chemical compound, drug | Oligo(dT)25 beads | NEB | Cat# S1408S | |
| Software, algorithm | Trimmomatic | PubMed ID (PMID):24695404 | RRID:SCR_011848 | |
| Software, algorithm | STAR (V.2.5.3) | PMID:23104886 | RRID:SCR_015899 | |
| Software, algorithm | HTSeq (V.0.9.1) | PMID:25260700 | RRID:SCR_005514 | |
| Software, algorithm | DESeq2 (V.1.22.2) | PMID:25516281 | RRID:SCR_015687 | |
| Software, algorithm | ggplot2 | ISBN 978-0-387-98140-6 | RRID:SCR_014601 | |
| Software, algorithm | Bowtie 2 | PMID:22388286 | RRID:SCR_005476 | |
| Software, algorithm | Integrated Genome Browser | PMID:27153568 | RRID:SCR_011792 |
Additional files
-
Supplementary file 1
Yeast strains used in this study.
- https://cdn.elifesciences.org/articles/64061/elife-64061-supp1-v2.docx
-
Supplementary file 2
Plasmids used in this study.
- https://cdn.elifesciences.org/articles/64061/elife-64061-supp2-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/64061/elife-64061-transrepform-v2.docx





