Globally defining the effects of mutations in a picornavirus capsid
Figures
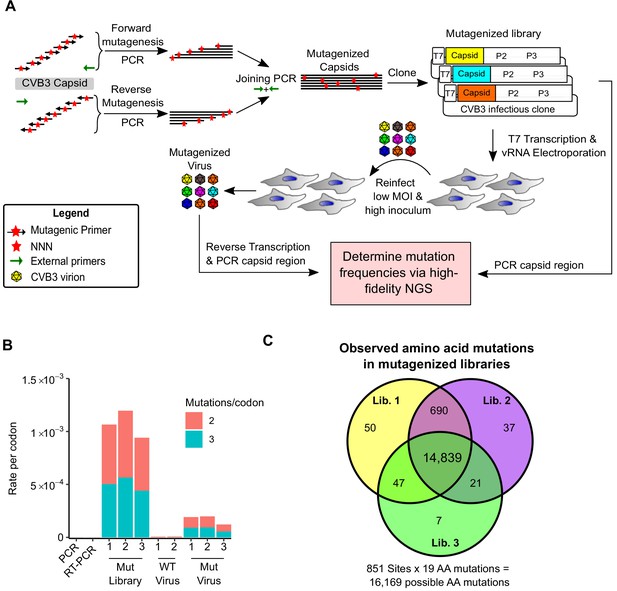
Deep mutational scanning (DMS) of the CVB3 capsid.
(A) Overview of the deep mutational scanning experimental approach. A mutagenesis PCR was used to introduce all possible single amino acid mutations across the CVB3 capsid region (Mut Library 1–3). Viral genomic RNA (vRNA) produced from the mutant libraries was then electroporated into cells to generate high diversity CVB3 populations (Mut Virus 1–3). The frequency of each mutation relative to the WT amino acid was then determined in both the mutagenized libraries and the resulting virus populations via high-fidelity duplex sequencing. (B) The average rate of double or triple mutations per codon observed in the mutagenized libraries (Mut Library 1–3), the resulting mutagenized virus (Mut Virus 1–3), as well as controls for the error rate of the amplification and sequencing process (PCR and RT-PCR) or the WT unmutagenized virus (WT Virus 1–2). Single mutations per codon were omitted from the analysis to increase the signal-to-noise ratio. (C) Venn diagram showing the number of amino acid mutations observed in the mutagenized libraries. MOI: multiplicity of infection. NGS: next-generation sequencing.
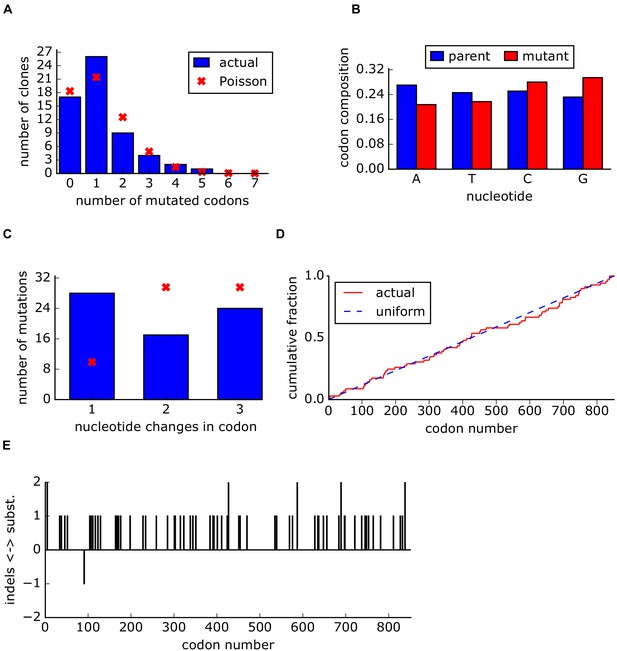
Sanger analysis of DMS libraries.
(A) The number of mutated codons per clone. (B) Original and mutated base for each mutation. (C) The number of nucleotide changes per codon. (D) Cumulative fraction of mutations versus the codon position. (E) Location of both mutations and indels across the capsid sequence.
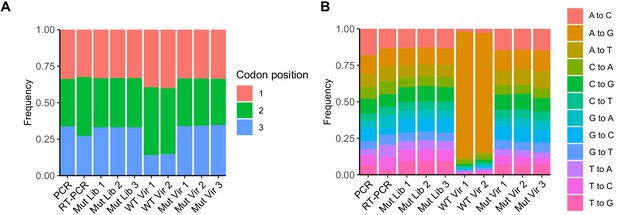
Results of high-fidelity duplex sequencing.
(A) The relative frequency of the mutated base within each mutated codon. (B) The relative frequency of each mutation type.
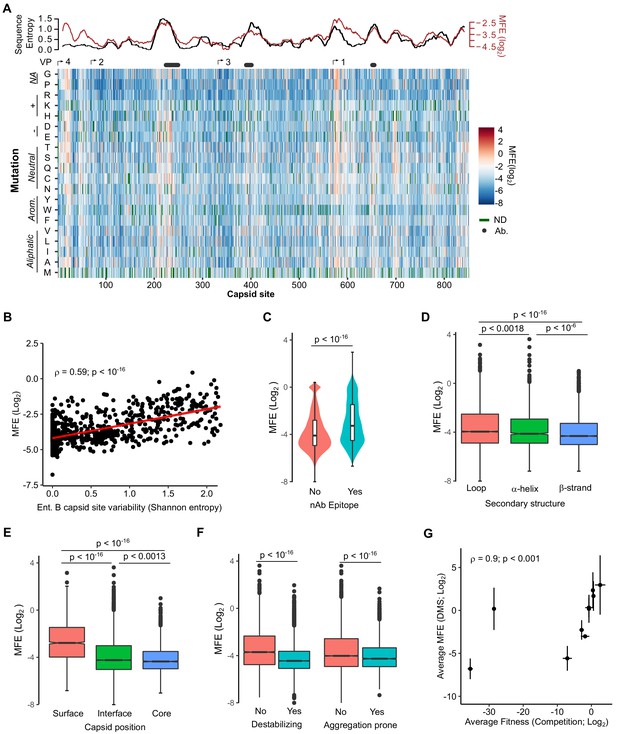
Mutational fitness effects (MFE) across the CVB3 capsid and their correlation with structural, evolutionary, and immunological attributes.
(A) Overview of the MFE observed across the CVB3 capsid. Bottom: A heatmap representing the MFE of all mutations observed at each capsid site. Green indicates no data available (ND), and the positions of the mature viral proteins (VP1–4) or antibody neutralization sites (nAb) are indicated above. Top: A 21 amino acid sliding window analysis of the average sequence variation in enterovirus B genomes (Shannon entropy; black line) or a 21 amino acid sliding window of the average MFE observed at each capsid site (red line). (B) Correlation between the average MFE observed at each capsid site and variation in enterovirus B sequence alignments (Shannon entropy). (C) Violin plot of MFE in antibody neutralization sites versus other capsid sites. (D–F) Boxplots of MFE as a function of secondary structure (D), position in the capsid (E), or the predicted effect of mutations on stability or aggregation propensity (F). (G) Validation of the MFE obtained by DMS using a competition assay. For each mutant, the average and standard deviation of the MFE obtained by DMS (n = 3) is plotted against the average and standard deviation of the fitness derived using the competition assay (n = 4). A two-sided Mann–Whitney test was used for two- category comparisons.
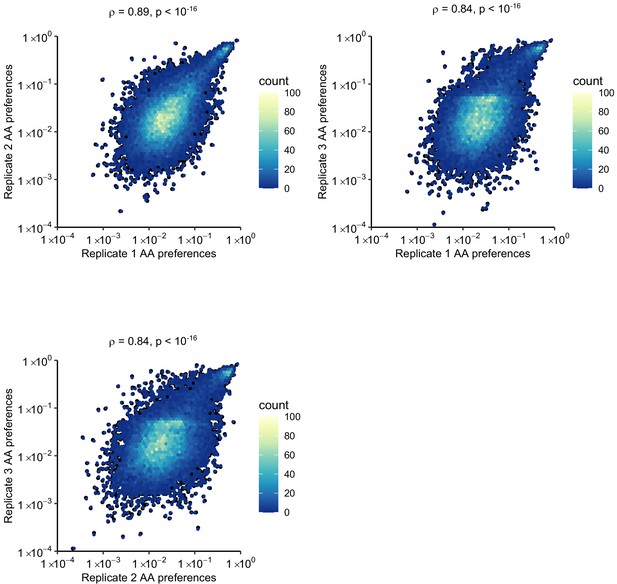
Correlation of amino acid preferences observed in experimental replicates.
Hexagonal bin plots showing the correlation of amino acid preferences between the three experimental replicates. Spearman’s correlation coefficient and p-value are shown above each plot.
Prediction of MFE based on structural and sequence information.
(A) The top 10 predictors identified in a random forest model for explaining MFE in the CVB3 capsid based on the percent of mean squared error (MSE) increase. (B) Hexagonal plot showing the correlation between MFE predicted using a random forest algorithm trained on the top five variables versus observed MFE. The random forest model was trained on 70% of the data and then tested on the remaining 30% (shown). RSA, relative surface area.
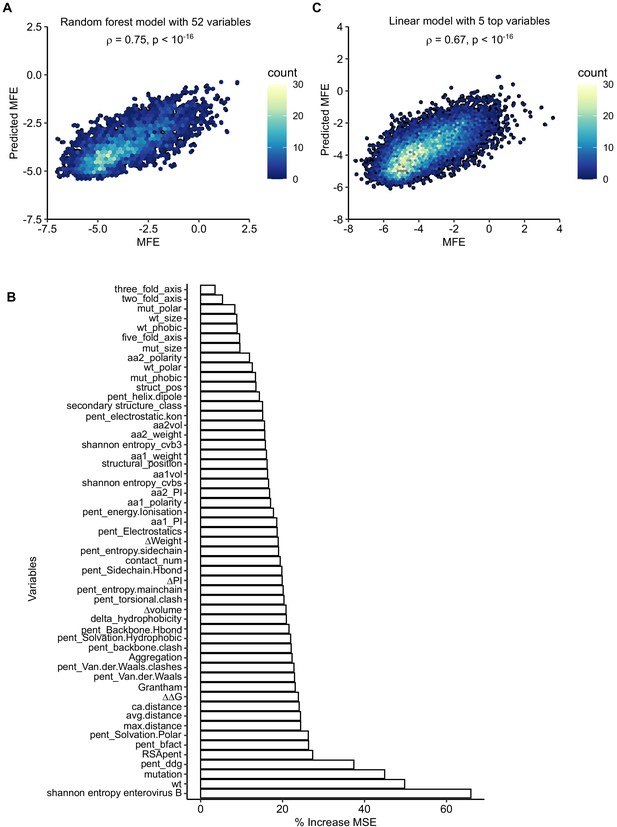
Prediction of mutational fitness effects using random forest or linear models.
(A) Hexagonal bin plot showing the correlation between actual and predicted MFE derived from a random forest model using all 52 variables. The model was trained on 70% of the data and tested on the remaining 30% of the data (shown). (B) Variable importance was obtained from the random forest model. (C) Linear model using the top five parameters of the random forest model. See supplementary file 6 for parameter description.
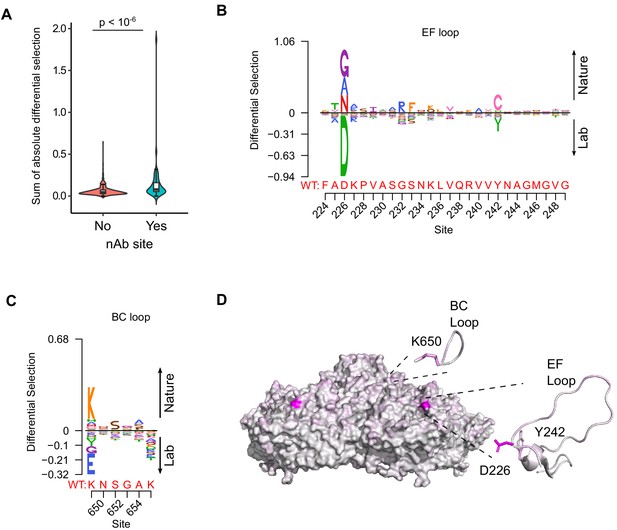
Antibody neutralization sites show differential selection between laboratory conditions and nature.
(A) Violin plot showing the sum of absolute differential selection observed at capsid sites comprising antibody neutralization epitopes (nAb) versus all other capsid sites. (B–C) Logoplots showing the observed differential selection of sites in the EF loop or BC loop. The WT sequence is indicated in red. (D) The CVB3 capsid pentamer (PDB:4GB3), colored according to the amount of differential selection. The BC and EF loops are shown next to the structure together with the side chains for sites showing the highest differential selection.
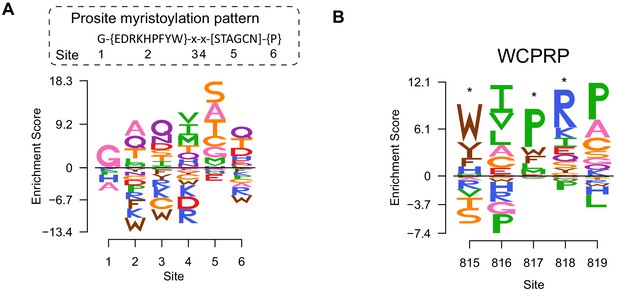
Sequence preferences of capsid-encoded motifs.
(A) Amino acid preferences of the CVB3 myristoylation motif. The canonical Prosite myristoylation motif is indicated above, with curly brackets indicating disfavored amino acids and square brackets indicating tolerated amino acids. (B) WCPRP motif required for 3CDpro cleavage of P1. Asterisks indicate analogous positions in FMDV shown to be essential for viability (Kristensen and Belsham, 2019).
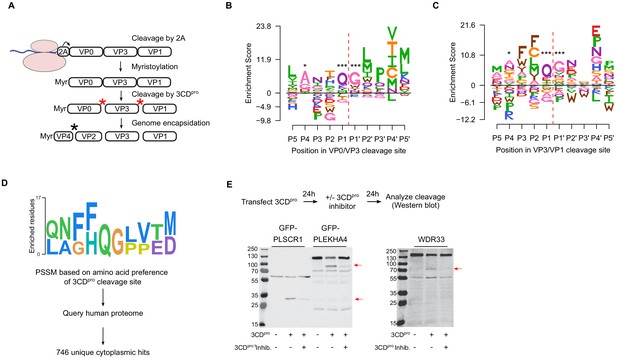
Sequence preference of capsid 3CDpro cleavage sites and their use for the identification of novel cellular targets of the viral protease.
(A) Overview of the CVB3 capsid maturation pathway. The CVB3 capsid precursor P1 is co-translationally cleaved by the viral 2A protease. P1 is then myristoylated and cleaved by the viral 3CDpro to generate the capsid proteins VP0, VP3, and VP1. Finally, upon assembly and genome encapsidation, VP0 is further cleaved into VP4 and VP2 in a protease-independent manner to generate the mature capsid. Red and black asterisks indicated 3CDpro or protease-independent cleavage events, respectively. (B,C) Logoplots showing amino acid preferences for the 10 amino acid regions spanning the 3CDpro cleavage sites (P1–P’1) of both VP0/VP3 and VP3/VP1 in the DMS dataset. (D) Overview of the bioinformatic pipeline for identification of novel 3CDpro cellular targets using the amino acid preferences for the capsid cleavage sites from our DMS study. A position-specific scoring matrix (PSSM) was generated based on the amino acid preferences for the 10 amino acid regions spanning the two 3CDpro cleavages sites. This PSSM was then used to query the human genome for potential cellular targets, and non-cytoplasmic proteins were filtered out, yielding 746 proteins. (E) The cellular proteins PLSCR1, PLEKHA4, and WDR33 are cleaved by 3CDpro. Western blot analysis of cells cotransfected with 3CDpro and GFP-PLSCR1 or GFP-PLEKHA4 and probed with a GFP antibody or transfected with 3CDpro and probed using a WDR33 antibody. When indicated, the 3CDpro inhibitor rupintrivir was included to ensure cleavage was mediated by the viral protease. Red arrows indicate cleavage products of the expected size (GFP-PLSCR1 full length = 64 kDa, cleaved N-terminus = 36 kDa; GFP-PLEKHA4 full length = 118 kDa, cleaved N-terminus = 72 kDa; WDR33 full length = 146 kDa, cleaved N-terminus = 72 kDa). *p<0.05, ***p<0.001.
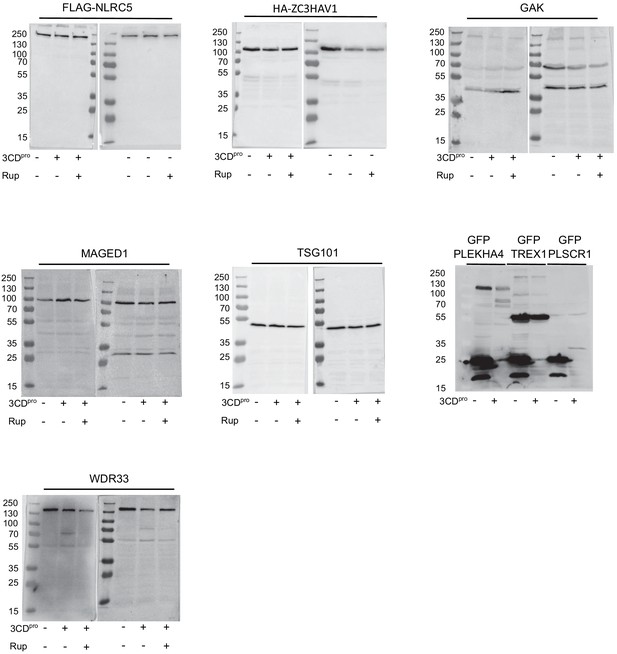
Evaluation of select hits identified as potential 3CDpro target proteins.
Western blots of cells transfected with 3CDpro and probed for the indicated endogenous protein or cotransfected with 3CDpro and the indicated fusion protein and blotted for the tag. Each experiment was performed twice. When indicated, the 3Cpro inhibitor rupintrivir was added.
Tables
Incorporation of DMS results in evolutionary models better describes natural CVB3 evolution compared to standard codon models.
| Model | ΔAIC | Log-likelihood | Parameters | Parameter values |
|---|---|---|---|---|
| ExpCM | 0.00 | −14,580.51 | 6 | Beta = 2.18, kappa = 7.47, omega = 0.16 |
| Goldman-Yang M5 | 4187.56 | −16,668.29 | 12 | Alpha_omega = 0.30, beta_omega = 10.00, kappa = 7.15 |
| Averaged ExpCM | 4303.74 | −16,732.38 | 6 | Beta = 0.61, kappa = 7.55, omega = 0.02 |
| Goldman-Yang M0 | 4371.26 | −16,761.14 | 11 | Kappa = 7.14, omega = 0.02 |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (coxsackievirus B3) | pCVB3-XhoI-P1-Kpn21 | 10.1016/j.celrep.2019.09.014 | Infectious CVB3 clone based on the Nancy strain (Taxon identifier 103903) | |
| Strain, strain background (coxsackievirus B3) | pCVB3-XhoI-∆P1-Kpn21 | This paper | Infectious CVB3 clone without P1 region | |
| Strain, strain background (coxsackievirus B3) | Marked reference CVB3 virus | 10.1038/nmicrobiol.2017.88 | Infectious CVB3 clone with silent mutations in the polymerase region used as a reference for fitness assays | |
| Strain, strain background (Escherichia coli) | NZY5α | NZY Tech | MB004 | Competent cells, standard cloning |
| Strain, strain background (Escherichia coli) | MegaX DH10B T1R Electrocomp cells | ThermoFisher | C6400-03 | Electrocompetent cells, library cloning |
| Cell line (Homo sapiens) | HeLa-H1 | ATCC | CRL-1958; RRID:CVCL_3334 | Cell line for CVB3 infection and DMS library production |
| Cell line (Homo sapiens) | HEK293 | ATCC | CRL-1573; RRID:CVCL_0045 | Cell line used for production of CVB3 mutants and for protease cleavage |
| Antibody | Anti-GFP (Mouse monoclonal) | SantaCruz | Sc-9996 | Western blot (1:2000) |
| Antibody | Anti-FLAG (Mouse monoclonal) | SantaCruz | Sc-166335 | Western blot (1:2000) |
| Antibody | Anti-HA (Mouse monoclonal) | SantaCruz | Sc-7392 | Western blot (1:2000) |
| Antibody | Anti-WDR33 (Mouse monoclonal) | SantaCruz | Sc-374466 | Western blot (1:1000) |
| Antibody | Anti-TSG101 (Mouse monoclonal) | SantaCruz | Sc-136111 | Western blot (1:1000) |
| Antibody | Anti-GAK (Mouse monoclonal) | SantaCruz | Sc-137053 | Western blot (1:1000) |
| Antibody | Anti-MAGED1 (Mouse monoclonal) | SantaCruz | Sc-393291 | Western blot (1:1000) |
| Recombinant DNA reagent | DMS libraries (1–3) | This paper | CVB3 infectious clone libraries with mutagenized capsid region | |
| Recombinant DNA reagent | pUC19-HiFi-P1 (plasmid) | This paper | CVB3 capsid region used as template for DMS cloned into SalI digested pUC19 vector. Used for site-directed mutagenesis | |
| Recombinant DNA reagent | T7 encoding plasmid (plasmid) | 10.1128/jvi.02583–14 | RRID:Addgene_65974 | Plasmid encoding T7 polymerase for transfection |
| Recombinant DNA reagent | pIRES-3CDpro (plasmid) | This paper | CVB3 3 CD protease region cloned into XhoI and NotI pIRES plasmid (Clonetech) | |
| Recombinant DNA reagent | peGFP_PLEKHA4 | 10.1016/j.celrep.2019.04.060 | Kind gift from Dr. Jeremy Baskin GFP-PLEKHA4 expression plasmid | |
| Recombinant DNA reagent | peGFP_PLSCR1 | 10.1371/journal.pone.0005006 | Kind gift from Dr. Serfe Benichou GFP-PLSCR1 expression plasmid | |
| Recombinant DNA reagent | pAcGFP-C1 WDR33 | https://doi.org/10.1016/j.molcel.2018.11.036 | Kind gift from Dr. Matthias Altmeyer pAcGFP-C1 WDR33 expression plasmid | |
| Recombinant DNA reagent | FLAG-NLCR5 | Addgene | RRID:Addgene_37521 | NLCR5 expression plasmid |
| Recombinant DNA reagent | HA-ZC3HAV1 | Addgene | RRID:Addgene_45907 | HA-ZC3HAV1 expression plasmid |
| Recombinant DNA reagent | Fluc-eGFP | Addgene | RRID:Addgene_90170 | Fluc-eGFP expression plasmid |
| Sequence-based reagent | HiFi_F | IDT | PCR primer | For generating PCR to clone libraries and sequencing: CTTTGTTGGGTTTATACCACTTAGCTCGAGAGAGG |
| Sequence-based reagent | HiFi_R | IDT | PCR primer | For generating PCR to clone libraries and sequencing: CCTGTAGTTCCCCACATACACTGCTCCG |
| Sequence-based reagent | DMS primers | IDT | PCR primer | Primers spanning the full coding region of the CVB3 capsid to perform codon mutagenesis. Listed in Supplementary file 1. |
| Sequence-based reagent | 2045_F | IDT | PCR primer | Primer used for Sanger sequencing. TCGAGTGTTTTTAGTCGGACG |
| Sequence-based reagent | 2143_R | IDT | PCR primer | Primer used for Sanger sequencing. TCGAGTGTTTTTAGTCGGACG |
| Sequence-based reagent | 3450_RT | IDT | PCR primer | Primer used for Sanger sequencing and RT-PCR. TCGAGTGTTTTTAGTCGGACG |
| Sequence-based reagent | qPCR_F | 10.1038/nmicrobiol.2017.88 | PCR primer | qPCR primer for competition assays. GATCGCATATGGTGATGATGTGA |
| Sequence-based reagent | qPCR_R | 10.1038/nmicrobiol.2017.88 | PCR primer | qPCR primer for competition assays. AGCTTCAGCGAGTAAAGATGCA |
| Sequence-based reagent | MGB_CVB3_wt | 10.1038/nmicrobiol.2017.88 | TaqManProbe | qPCR probe for competition assays. 6FAM-CGCATCGTACCCATGG-TAMRA |
| Sequence-based reagent | MGB_CVB3_Ref | 10.1038/nmicrobiol.2017.88 | TaqManProbe | qPCR probe for competition assays. HEX-CGCTAGCTACCCATGG-TAMRA |
| Sequence-based reagent | Q8D_F | IDT | PCR primer | Primer for site-directed mutagenesis: gtatcaacgGATaagactggg |
| Sequence-based reagent | Q8D_R | IDT | PCR primer | Primer for site-directed mutagenesis: ttgagctcccattttgctgt |
| Sequence-based reagent | K829L_F | IDT | PCR primer | Primer for site-directed mutagenesis: gagaaggcaCTAaacgtgaac |
| Sequence-based reagent | K829L_R | IDT | PCR primer | Primer for site-directed mutagenesis: gtattggcagagtctaggtgg |
| Sequence-based reagent | K235D_F | IDT | PCR primer | Primer for site-directed mutagenesis: gggtccaacGATttggtacag |
| Sequence-based reagent | K235D_R | IDT | PCR primer | Primer for site-directed mutagenesis: ggatgcgaccggtttgtccgc |
| Sequence-based reagent | R16G_F | IDT | PCR primer | Primer for site-directed mutagenesis: catgagaccGGActgaatgct |
| Sequence-based reagent | R16G_R | IDT | PCR primer | Primer for site-directed mutagenesis: tgccccagtcttttgcgttg |
| Sequence-based reagent | K827G_F | IDT | PCR primer | Primer for site-directed mutagenesis: caatacgagGGGgcaaagaac |
| Sequence-based reagent | K827G_R | IDT | PCR primer | Primer for site-directed mutagenesis: gcagagtctaggtggtctagg |
| Sequence-based reagent | Q566M_F | IDT | PCR primer | Primer for site-directed mutagenesis: atttcgcagATGaactttttc |
| Sequence-based reagent | Q566M_R | IDT | PCR primer | Primer for site-directed mutagenesis: gaaaggagtgtccttcaatag |
| Sequence-based reagent | T315P_F | IDT | PCR primer | Primer for site-directed mutagenesis: attacggtcCCCatagcccca |
| Sequence-based reagent | T315P_R | IDT | PCR primer | Primer for site-directed mutagenesis: tgggacgtacgtggtgga |
| Sequence-based reagent | N395H_F | IDT | PCR primer | Primer for site-directed mutagenesis: gagaaggtcCATtctatggaa |
| Sequence-based reagent | N395H_R | IDT | PCR primer | Primer for site-directed mutagenesis: tccaacattttggactgggac |
| Sequence-based reagent | T849A_F | IDT | PCR primer | Primer for site-directed mutagenesis: actacaatgGTCaatacgggc |
| Sequence-based reagent | T849A_R | IDT | PCR primer | Primer for site-directed mutagenesis: gatgctttgcctagtagtgg |
| Sequence-based reagent | K235D_F | IDT | PCR primer | Primer for site-directed mutagenesis: gggtccaacGATttggtacag |
| Sequence-based reagent | K235D_R | IDT | PCR primer | Primer for site-directed mutagenesis: ggatgcgaccggtttgtccgc |
| Sequence-based reagent | 3C_For | IDT | PCR primer | Primer for cloning CVB3 3 CD into pIRES: TATTCTCGAGACCATGGGCCCTGCCTTTGAGTTCG |
| Sequence-based reagent | 3D_Rev | IDT | PCR primer | Primer for cloning CVB3 3 CD into pIRES: TATTGCGGCCGCCTAGAAGGAGTCCAACCATTTCCT |
| Commercial assay or kit | NEBuilder HiFi DNA Assembly kit | NEB | E2621X | Seamless cloning |
| Commercial assay or kit | TranscriptAid T7 High Yield Transcription Kit | ThermoFisher Scientific | K0441 | T7 in vitro transcription kit |
| Commercial assay or kit | Quick-RNA Viral kit | Zymo Research | R1035 | RNA purification |
| Commercial assay or kit | DNA Clean andConcentrator-5 | Zymo Research | D4013 | DNA purification, gel purification |
| Commercial assay or kit | Luna Universal Probe One-Step RT-qPCR kit | NEB | E3006X | One-step qPCR master mix |
| Chemical compound, drug | Rupintivir | Tocris Biosciences | Cat. #: 6414 | CVB3 3C protease inhibitor |
| Software, algorithm | CodonTilingPrimers | https://doi.org/10.1016/j.chom.2017.05.003 | Software to design primers for mutagenesis (https://github.com/jbloomlab/CodonTilingPrimers) | |
| Software, algorithm | Sanger Mutant Library Analysis | Dr. Jesse Bloom | Software to assess library mutagenesis by Sanger sequencing (https://github.com/jbloomlab/SangerMutantLibraryAnalysis) | |
| Software, algorithm | Samtools | http://www.htslib.org/ | version 1.5 | Suite of programs for interacting with high-throughput sequencing data |
| Software, algorithm | Fastp | 10.1093/bioinformatics/bty560 | Software for NGS read trimming and QC | |
| Software, algorithm | PicardTools, FastqToSam | https://broadinstitute.github.io/picard/ | Version 2.2.4 | Used to generate Bam files from Fastq files |
| Software, algorithm | Duplex pipeline | https://github.com/KennedyLabUW/Duplex-Sequencing; Kennedy et al., 2014 | Version 3.0 | Analysis pipeline for duplex sequencing (UnifiedConsensusMaker.py) |
| Software, algorithm | VariantBam | 10.1093/bioinformatics/btw111 | Software to filter Bam files | |
| Software, algorithm | BWA | https://sourceforge.net/projects/bio-bwa/files/ | Version 0.7.16 | Software to align NGS reads |
| Software, algorithm | Fgbio | http://fulcrumgenomics.github.io/fgbio/ | version 1.1.0 | Software used to hard-clip NGS reads |
| Software, algorithm | VirVarSeq | 10.1093/bioinformatics/btu587 | version 1.1.0 | Software used to identify codons in each NGS read |
| Software, algorithm | Custom R scripts | This paper | Custom R scripts to process output of VirVarSeq script. Available athttps://github.com/RGellerLab/CVB3_Capsid_DMS | |
| Software, algorithm | DMS_tools2 | 10.1186/s12859-015-0590-4 | Software to determine amino acid preferences and mutational fitness effects | |
| Software, algorithm | TANGO | 10.1038/nbt1012 | Software to determine the effect of mutations on aggregation | |
| Software, algorithm | FoldX | 10.1093/nar/gki387 | Software to determine the effect of mutations on stability | |
| Software, algorithm | DSSP | http://swift.cmbi.ru.nl/gv/dssp/ | Software used to obtain secondary structure and RSA within DMS_tools2 | |
| Software, algorithm | ViprDB | http://viperdb.scripps.edu/ Carrillo-Tripp et al., 2009 | Software used to obtain structural information on capsid sites | |
| Software, algorithm | DECIPHER Package | 10.32614/RJ-2016–025 | R package for performing codon alignments | |
| Software, algorithm | PhyDMS | doi:10.7717/peerj.3657 | For phylogenetic and differential selection analyses. https://jbloomlab.github.io/phydms/index.html | |
| Software, algorithm | Custom R scripts | This paper | Custom R script to generate in silico peptides spanning 10AA 3 CD protease cleavage site. Available at https://github.com/RGellerLab/CVB3_Capsid_DMS | |
| Software, algorithm | PSSMSearch | 10.1093/nar/gky426 | Used to generate position-specific scoring matrix and search human proteome for hits. http://slim.icr.ac.uk/pssmsearch/ | |
| Software, algorithm | Peptides R package | ISSN 2073–4859 | Version 2.4.2 | R package to predict molecular weight of proteins |
| Software, algorithm | RandomForest R package | 10.1023/A:1010933404324 | Version 4.6–16 | R package for random forest prediction |
| Software, algorithm | Logolas | 10.1186/s12859-018-2489-3 | Package to generate logo plots in R |
Additional files
-
Supplementary file 1
Primers for next-generation sequencing.
- https://cdn.elifesciences.org/articles/64256/elife-64256-supp1-v2.csv
-
Supplementary file 2
Next-generation sequencing statistics.
- https://cdn.elifesciences.org/articles/64256/elife-64256-supp2-v2.xlsx
-
Supplementary file 3
Mutations observed by Sanger sequencing.
- https://cdn.elifesciences.org/articles/64256/elife-64256-supp3-v2.txt
-
Supplementary file 4
Mutational fitness effects of the mutagenized viral populations.
- https://cdn.elifesciences.org/articles/64256/elife-64256-supp4-v2.csv
-
Supplementary file 5
Results of qPCR validation of MFE.
- https://cdn.elifesciences.org/articles/64256/elife-64256-supp5-v2.xlsx
-
Supplementary file 6
Data used for random forest model and parameter explanation.
- https://cdn.elifesciences.org/articles/64256/elife-64256-supp6-v2.xlsx
-
Supplementary file 7
Differential selection results.
- https://cdn.elifesciences.org/articles/64256/elife-64256-supp7-v2.csv
-
Supplementary file 8
PSSMsearch results.
- https://cdn.elifesciences.org/articles/64256/elife-64256-supp8-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/64256/elife-64256-transrepform-v2.docx





