Galectin-9 regulates the threshold of B cell activation and autoimmunity
Figures
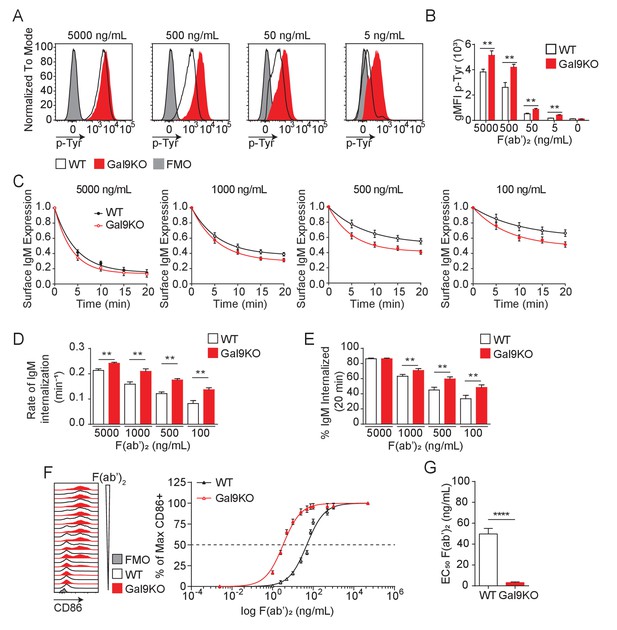
Gal9KO B cells respond to limiting concentrations of antigen.
(A) Representative histograms of total tyrosine phosphorylation (p-Tyr) in WT (black) and Gal9KO (red) B cells at 5 min post-stimulation with anti-IgM F(ab’)2, as indicated. FMO (gray shaded). (B) Summary geometric mean fluorescence intensity (gMFI) of p-Tyr shown in (A). (C) IgM internalization over 20 min for WT (black) and Gal9KO (red) B cells following stimulation with anti-IgM F(ab’)2 as indicated. (D) Rate of IgM internalization (k) of data shown in (C). (E) Proportion of total IgM internalized at 20 min for data shown in (C). (F) Representative histograms of CD86 expression on WT (black) and Gal9KO (red) B cells stimulated with increasing concentrations of anti-IgM F(ab’)2 (left). Summary statistic, proportion of CD86 expressing B cells (right) as a function of F(ab’)2 concentration. (G) EC50 of F(ab’)2 titration shown in (F). Data show mean ± SEM and are representative of nine biological replicates over three independent experiments. Statistical significance was assessed by Mann–Whitney **p≤0.01, ****p<0.0001.
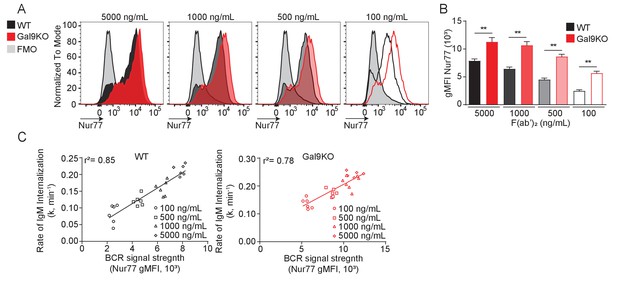
Nur77 reporter expression correlates with BCR internalization rate.
(A) Representative histograms of Nur77 expression at 16 hr in WT (black) and Gal9KO (red) B cells stimulated with anti-IgM F(ab’)2 for 20 min, as indicated. (B) Summary gMFI of Nur77 expression shown in (A), mean and SEM. (C) Correlation analysis of the internalization rate (k) of IgM-BCR over a 20 min interval relative to the extent of BCR signal transduction as measured by the intensity of Nur77 expression, WT (left), Gal9KO (right). Data are representative of six biological replicates over two experiments. Statistical significance was assessed by Mann–Whitney **p≤0.01.
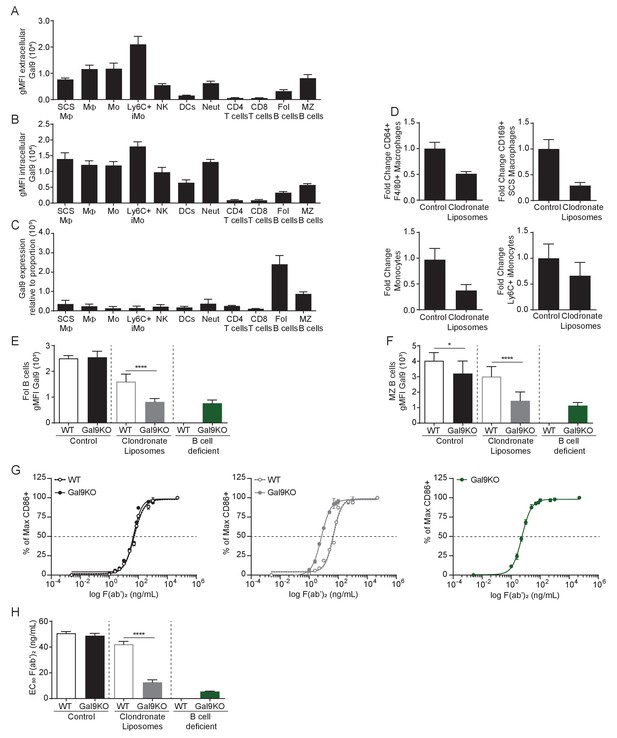
B cells and macrophage populations are a source of extrinsic Gal9, and are sufficient to alter the threshold of B cell activation.
(A) Summary gMFI of surface Gal9 expression on splenic immune cell populations in WT mice. Subcapsular sinus macrophages (SCS MΦ) were defined as CD45+, Ly6G-, CD11b+, F4/80-, CD169+. Macrophages (MΦ) were defined as CD45+, Ly6G−, CD11c+, CD64+, F4/80+. Monocytes (Mo) were defined as CD45+, Ly6G-, CD11c+, MHCII-, CD64+. Ly6C+ inflammatory Monocytes (Ly6C+ iMo) were defined as CD45+, Ly6G−, CD11c+, MHCII−, CD64+, Ly6C+. NK cells (NK) were defined as CD45+, Ly6G-,CD11b+, MHCII-, CD64-. Dendritic cells (DCs) were defined as CD45+, Ly6G-, CD11cHi, MHCIIHI. Neutrophils (Neut) were defined as CD45+, Ly6G+. CD4 T cells were defined as CD45+, B220-, CD4+, CD8-. CD8 T cells were defined as CD45+, B220-, CD4-, CD8+. Follicular B cells (Fol B cells) were defined as CD45+, B220+, CD21/35-, CD23+. Marginal zone B cells (MZ B cells) were defined as CD45+, B220+, CD21/35HI, CD23-. (B) Summary gMFI of intracellular Gal9 expression on splenic immune cell populations in WT mice. (C) Gal9 expression relative to subset abundance. Ratio of total Gal9 expression (gMFI surface Gal9 A + gMFI intracellular Gal9 B) normalized against frequency of splenic population. (D) Fold change in splenic phagocyte populations, as indicated, following treatment with control or Clodronate Liposomes. (E) Summary gMFI of surface Gal9 expression on endogenous Follicular B cells (WT) or Gal9KO Follicular B cells (Gal9KO) following adoptive transfer into recipient hosts, as indicated. (F) Summary gMFI of surface Gal9 expression as in E, gated on Marginal zone B cells. (G) Proportion of activated B cells (as in E) determined by CD86 upregulation at 16 hr of stimulation with increasing concentrations of anti-IgM F(ab’)2. (H) EC50 of F(ab’)2 titration shown in (G). Data show mean ± SEM and are representative of at least nine biological replicates. Statistical significance was assessed by Mann–Whitney *p≤0.05, ****p<0.0001.
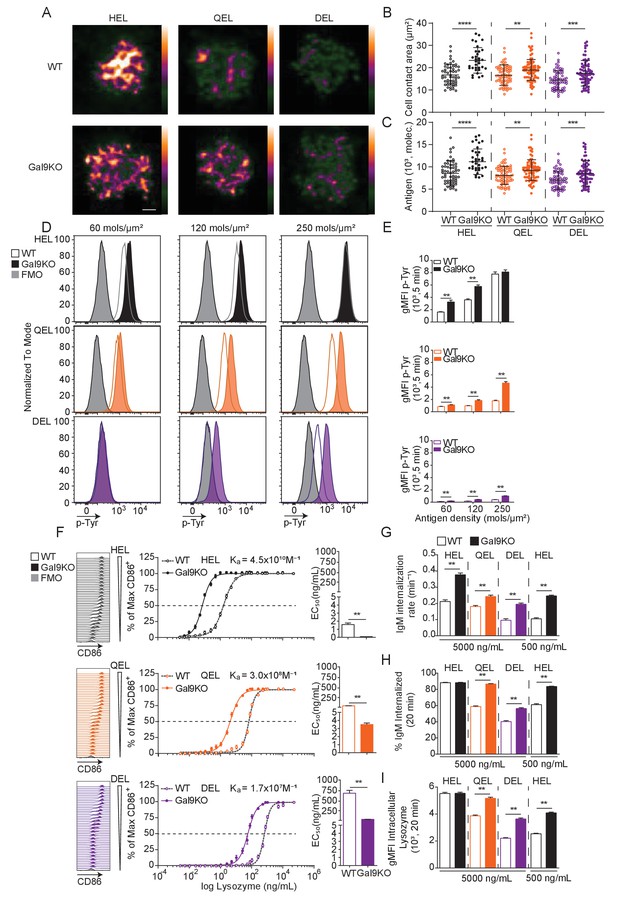
Gal9KO B cells respond more readily to low-affinity antigens.
(A) Representative images of primary WT and Gal9KO B cells fixed on planar lipid bilayers containing fluorescently conjugated antigen, as indicated, after 90 s of spreading and imaged by TIRF microscopy. Images mapped to a blue-orange ice 8-bit color scale (ImageJ). Scale bar 2 μm. (B) Quantification of cell contact area of WT (open) and Gal9KO (filled) in response to planar lipid bilayers containing HEL (black), QEL (orange), and DEL (purple) antigens. (C) Quantification of the amount of accumulated antigen as in (B). (D) Representative histograms of total tyrosine phosphorylation (p-Tyr) in WT (open) and Gal9KO (filled) B cells stimulated for 5 min with indicated antigen deposited on OP9 stromal cells, as indicated. FMO (gray shaded). (E) Summary gMFI of data shown in (D). (F) Representative histograms of CD86 expression on WT (open) and Gal9KO (filled) B cells stimulated with increasing concentrations of lysozymes, as indicated (left). Summary statistic, proportion of CD86 expressing B cells (middle). EC50 of lysozyme titration (right). (G) Internalization rate (k) of IgM; (H) Proportion of total IgM internalized; and (I) gMFI of intracellular lysozyme expression for WT (open) and Gal9KO (filled) B cells following 20 min stimulation with lysozyme, as indicated. Data show mean ± SEM and are representative of nine biological replicates over three independent experiments. Statistical significance was assessed by Mann–Whitney **p≤0.01, ***p≤0.001, **** p<0.0001.
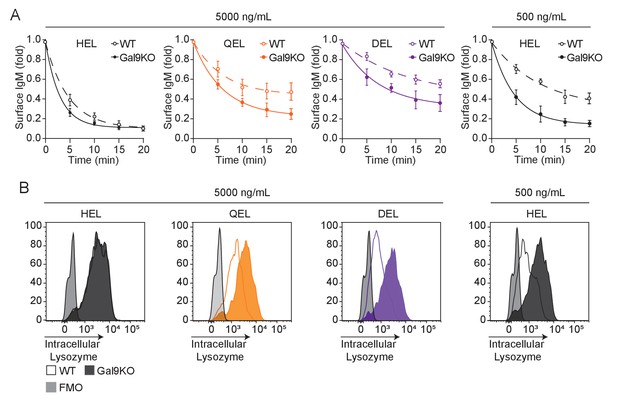
Gal9KO B cells have enhanced internalization of low-affinity antigens.
(A) Summary statistics of IgM internalization over 20 min as indicated, mean and SEM. (B) Representative histograms of intracellular lysozyme levels at 20 min as indicated. Data are representative of nine biological replicates over three independent experiments.
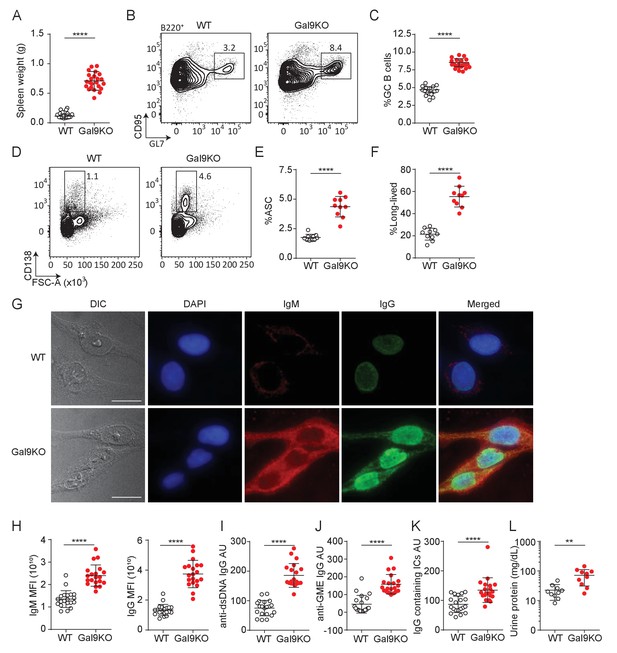
Aged Gal9KO mice develop spontaneous autoimmunity.
(A) Spleen weight of WT (black) or Gal9KO (red) mice aged >8 months. (B) Representative plots of GC B cells in the spleen of aged mice, as indicated. (C) Summary proportion of GC B cells, as in (B). (D) Representative plots of antibody secreting cells (ASC) in the spleen of aged mice. (E) Summary proportion of ASC, as in (D). (F) Proportion of ASC lacking B220 expression. (G) Representative images of HEp-2 cells stained with sera from WT (top) or Gal9KO (bottom) mice. (H) Summary mean fluorescence intensity (MFI) of IgM (left) and IgG (right) of HEp-2 staining as in (G). (I) Anti-dsDNA specific IgG titers. (J) nti-glomerulus membrane extract (GME) specific IgG titers. (K) IgG-containing immune complexes (IC) titers. (L) Proteinuria, determined by protein secretion into the urine. Data are representative of 10–20 biological replicates over at least three independent experiments. Statistical significance was assessed by Mann–Whitney **p≤0.01, ****p<0.0001.
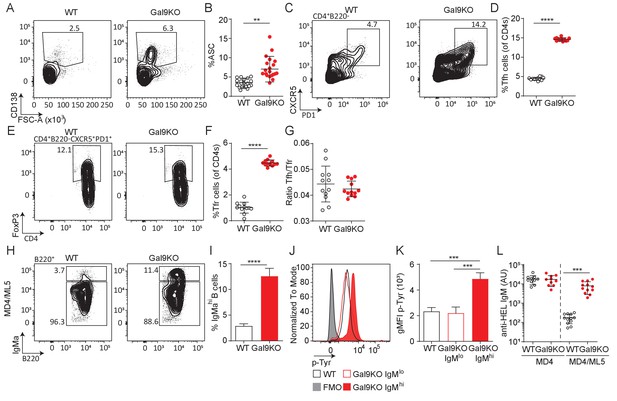
Gal9KO B cells can escape anergy.
(A) Representative plots of ASCs in the bone marrow of aged WT (left) and Gal9KO (right) mice. (B) Proportion of ASC in the bone marrow of WT (black) and Gal9KO (red), as in (A). (C) Representative plots of Tfh cells in the spleen of WT (left) and Gal9KO (right) aged mice. (D) Proportion of Tfh cells shown in (C). (E) Representative plots of FoxP3 expressing Tfr cells of Tfh cells in WT (left) and Gal9KO (right). (F) Proportion of Tfr cells. (G) Ratio of Tfh cells to regulatory Tfr cells in the spleen of aged mice. (H) Representative plots of IgM-high (hi) and IgM-low (lo) populations in WT (left) and Gal9KO (right) MD4/ML5 mice. (I) Proportion of IgMhi expressing cells shown in (H). (J) Representative histograms of p-Tyr levels in B cells 5 min post-stimulation with HEL, as indicated. (K) Summary gMFI of p-Tyr expression in (J). (L) Anti-HEL specific IgM titers in MD4 and Gal9KO-MD4 mice (left) and MD4/ML5 and Gal9KO-MD4/ML5 mice (right). Data show mean and SEM and are representative of 9–12 biological replicates over at least 3 experiments. Statistical significance for B, D, F, G, I, and L was assessed by Mann–Whitney. Statistical significance for (K) was assessed by Kruskal–Wallis. **p≤0.01, ***p≤0.001, ****p<0.0001.
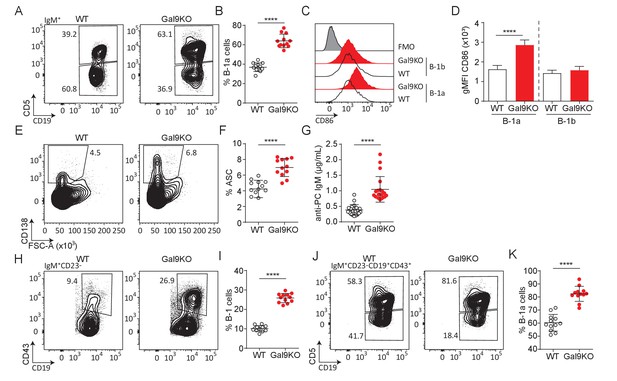
Loss of Gal9 leads to enhanced activation and accumulation of B-1a cells at steady-state.
(A) Representative plots of IgM expressing cells in the PerC of aged WT and Gal9KO mice, as indicated. (B) Summary proportion of CD5 expressing B-1a cells in the PerC shown in A. (C) Representative histograms of CD86 expression at steady state on PerC cells shown in (A). (D) Summary gMFI of CD86 expression shown in C. (E) Representative plots of ASCs in the PerC of aged mice. (F) Summary proportion of ASCs shown in (E). (G) Anti-phosphorylcholine (PC) specific IgM in sera of aged mice. (H) Representative plots of B-1 cells of IgM expressing cells in the spleen of aged mice. (I) Proportion of B-1 cells shown in H. (J) Representative plots of B-1a and B-1b cells of H. (K) Summary proportion of CD5 expressing B-1a cells in the spleen of aged mice shown in (I). Data are representative of ten biological replicates over three independent experiments. Statistical significance was assessed by Mann–Whitney ****p<0.0001.
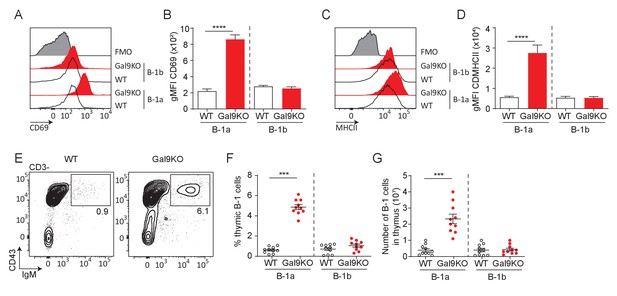
B1–a cells from Gal9KO mice are activated at steady-state.
(A) Representative histograms of CD69 expression on PerC cells in aged WT (black) and Gal9KO (red) mice, as indicated. (B) Summary gMFI of CD69 expression shown in (A). (C) Representative histograms of MHCII expression on PerC cells in aged mice, as indicated. (D) Summary gMFI of MHCII expression shown in (C). (E) Representative plots of B-1 cells in the thymus of aged WT (left) and Gal9KO (right) mice. (F) Summary proportion of thymic B-1 cells of data shown in (E). (G) Total number of B-1 cells in the thymus in aged mice, as in (F). Data represent ten biological replicates over three experiments. Statistical significance was assessed by Mann–Whitney ***p≤0.001, ****p<0.0001.
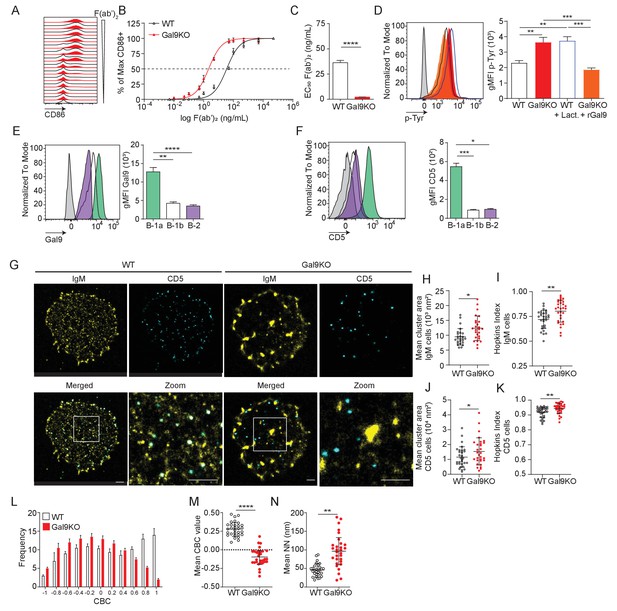
Gal9 regulates BCR signal transduction in B-1a cells through interactions with IgM and the negative co-receptor CD5.
(A) Representative histograms of CD86 expression on B-1a cells from WT (black) and Gal9KO (red) mice stimulated with titrated concentrations of anti-IgM F(ab’)2. (B) Summary proportion of CD86 expressing cells as a function of F(ab’)2 concentration as in (A). (C) EC50 of F(ab’)2 titration shown in (A). (D) Representative histogram of p-Tyr level in B-1a cells from WT (black, open) or Gal9KO (red, filled), and WT B-1a cells treated with lactose (blue, open) and Gal9KO B-1a cells treated with rGal9 (orange, filled) stimulated with anti-IgM (Fab’)2 for 5 min (left), summary gMFI of p-Tyr levels (right). (E) Representative histograms of Gal9 expression in B-1a cells (green), B-1b cells (white), and splenic B-2 cells (purple) from WT mice (left), summary gMFI of Gal9 expression (right). (F) Representative histograms of CD5 detection on rGal9-coated beads incubated with lysates from different B cell subsets, as indicated (left), summary gMFI of CD5 enrichment on beads incubated with lysates (right). (G) Reconstructed dual-dSTORM images of IgM (yellow) and CD5 (cyan) on the surface of B-1a cells from WT and Gal9KO mice, ROI (3 µm x 3 µm) used for analysis is expanded in zoom. Scale bars represent 1 µm. (H) Quantification of IgM mean cluster area from dSTORM images. (I) Quantification of IgM clustering tendency using Hopkins index. (J) Quantification of CD5 mean cluster area from dSTORM images. (K) Quantification of CD5 clustering tendency using Hopkins index. (L) Frequency distribution of coordinate-based colocalization (CBC) between IgM and CD5. (M) Mean CBC value per ROI. (N) Mean distance between nearest neighbors. Data in (A–F) are representative of nine biological replicates over three independent experiments. Data in (G–N) are representative of 30 ROIs acquired over at least three independent experiments. Data represent mean ± SEM. Statistical significance for C, H, I, J, K, M, and N was assessed by Mann–Whitney, statistical significance for D, E, and F was assessed by Kruskal–Wallis. *p≤0.05, **p≤0.01, ***p≤0.001, ****p<0.0001.
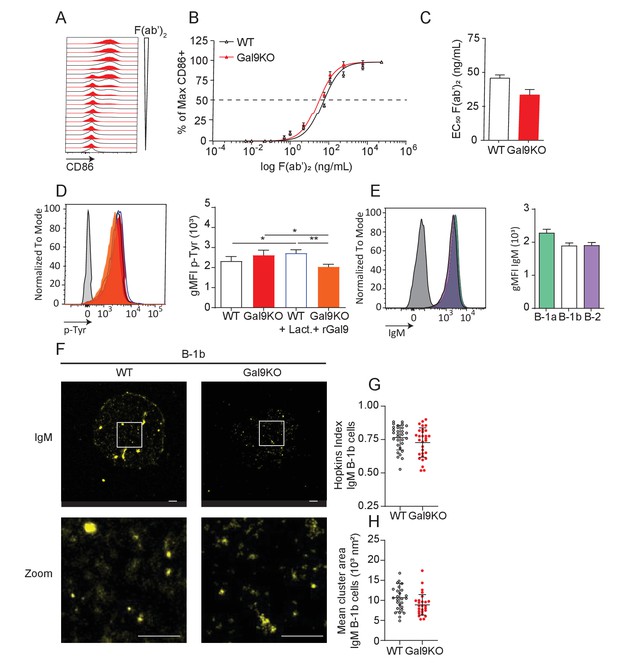
Gal9 does not alter BCR responses in B-1b cells.
(A) Representative histograms of CD86 expression on B-1b cells from WT (black) and Gal9KO (red) mice stimulated with titrated concentrations of anti-IgM F(ab’)2. (B) Summary proportion of CD86 expressing cells as in (A). (C) EC50 of F(ab’)2 titration shown in A. (D) Representative histograms of p-Tyr levels at 5 min in B-1b cells from WT (black, open) and Gal9KO (red, filled) mice, or WT B-1b cells treated with lactose (blue, open) and Gal9KO B-1b cells treated with rGal9 (orange, filled) stimulated with anti-IgM (Fab’)2 (left) and summary gMFI of p-Tyr level (right). (E) Representative histograms of IgM detection on rGal9-coated beads incubated with lysates from B-1a (green), B-1b (white), and B-2 (purple) cells (left), and summary gMFI of CD5 enrichment on beads incubated with lysates (right). (F) Reconstructed dSTORM images of IgM on the surface of B-1b cells. ROI (3 µm × 3 µm) used for analysis is expanded in zoom. Scale bars represent 1 µm. (G) Quantification of IgM clustering tendency using Hopkins index. (H) Quantification of IgM mean cluster area. Data in A–E are representative of nine biological replicates over three independent experiments. Data in G and I are representative of 30 ROIs acquired over at least three independent experiments. Data represent mean and SEM. Statistical significance for C, G, and H was assessed by Mann–-Whitney, statistical significance for D and E was assessed by Kruskal–Wallis. * p ≤ 0.05, ** p ≤ 0.01.
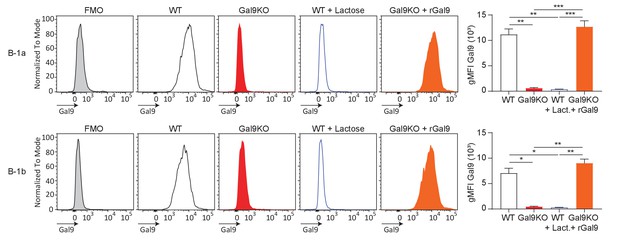
Lactose treatment inhibits Gal9 binding to B-1 cells, and treatment with recombinant Gal9 restores Gal9 expression.
Representative histograms of Gal9 expression on B-1a cells (top) and B-1b cells (bottom), as indicated (left). Summary gMFI of Gal9 expression (right). Data represent mean and SEM and are representative of at least nine biological replicates over three experiments. Statistical significance was assessed by Kruskal–Wallis. *p≤0.05, **p≤0.01, ***p≤0.001.
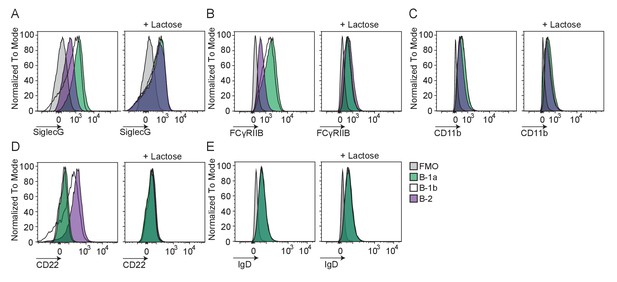
Gal9 binds only marginally to negative co-receptors Siglec G and FcγRIIB in B1 B cells.
Representative histograms of protein enrichment on Gal9 coated beads incubated with lysates from B cell populations, as indicated. Whole cell lysates were incubated with coated beads with or without the presence of the Gal9 inhibitor lactose. Beads were then washed and stained with fluorescent antibodies targeting (A) SiglecG, (B) FcγRIIB, (C) CD11b, (D) CD22, (E) IgD and measured by flow cytometry.
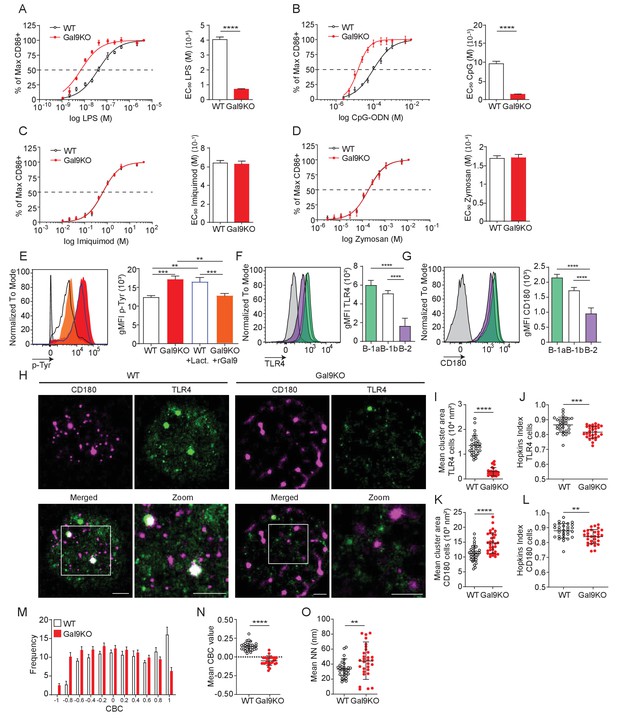
Gal9 regulates LPS-mediated activation by restraining signal transduction through interactions with TLR4 and the negative co-receptor CD180.
(A) Proportion of CD86 expressing cells of B-1a cells from WT (black) and Gal9KO (red) mice stimulated with titrated concentrations of LPS (left), EC50 (right). (B) Titration with CpG-ODN as in A (left), EC50 (right). (C) Titration with Imiquimod as in (A) (left), EC50 (right). (D) Titration with Zymosan as in (A) (left), EC50 (right). (E) Representative histograms of p-Tyr levels at 10 min in B-1a cells from WT (black, open) or Gal9KO (red, filled) mice, or WT B-1a cells treated with lactose (blue, open) or Gal9KO B-1a cells treated with rGal9 (orange, filled) and stimulated with LPS for 10 min (left) summary gMFI of p-Tyr expression (right). (F) Representative histograms (left) and summary gMFI (right) of TLR4 detection on rGal9-coated beads incubated with lysates from different B cell subsets, as indicated. (G) Representative histogram (left) and summary gMFI (right) of CD180 detection as in (F). (H) Reconstructed dual-dSTORM images of CD180 (magenta) and TLR4 (green) on the surface of B-1a cells from WT and Gal9KO mice, ROI (3 µm × 3 µm) used for analysis is expanded in zoom. Scale bars represent 1 µm. (I) Quantification of TLR4 cluster area from dSTORM images. (J) Quantification of TLR4 clustering tendency using Hopkins Index. (K) Quantification of CD180 cluster area from dSTORM images. (L) Quantification of CD180 clustering tendency using Hopkins index. (M) Frequency distribution of coordinate-based colocalization (CBC) between TLR4 and CD180. (N) Mean CBC value per ROI. (O) Mean distance between nearest neighbors. Data in A–G are representative of nine biological replicates over three independent experiments. Data in H and I are representative of 30 ROIs acquired over at least three independent experiments. Data represent mean ± SEM. Statistical significance for A, D, I, J, K, and L was assessed by Mann–Whitney, statistical significance for E, F, and G was assessed by Kruskal–Wallis **p≤0.01, *** p≤0.001, ****p<0.0001.
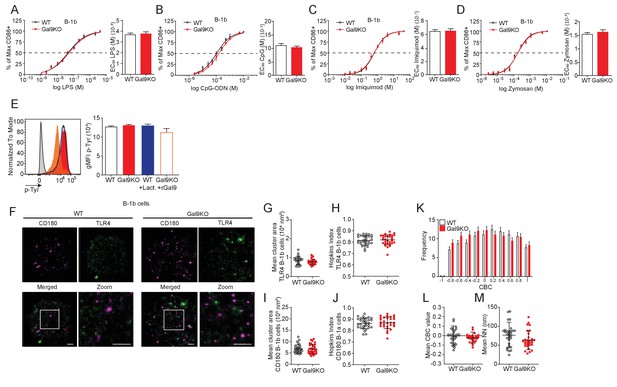
Gal9 does not alter LPS responses in B-1b cells.
B1-b cells from WT (black) or Gal9KO (red) mice were stimulated with titrated concentrations of (A) LPS, (B) CpG, (C) Imiquimod, or (D) Zymosan. Proportion of CD86 expressing B-1b cells shown on the left, and EC50 on the right. (E) Representative histogram of p-Tyr levels at 10 min in B-1b cells from WT (black, open) or Gal9KO (red, filled) mice, and WT B-1b cells treated with lactose (blue, open) or Gal9KO B-1b cells treated with rGal9 (orange, filled) and stimulated with LPS (left) and summary gMFI of p-Tyr levels (right). (F) Reconstructed dual-dSTORM images of CD180 (magenta) and TLR4 (green) on the surface of B-1b cells. ROI (3 µm × 3 µm) used for analysis is expanded in zoom. Scale bars represent 1 µm. (G) Quantification of TLR4 cluster area from dSTORM images. (H) Quantification of the clustering tendency of TLR4 using the Hopkins index. (I) Quantification of CD180 cluster area from dSTORM images. (J) Quantification of CD180 clustering tendency using the Hopkins index. (K) Frequency distribution of coordinate-based colocalization (CBC) between TLR4 and CD180. (L) Mean CBC value per ROI. (M) Mean distance between nearest neighbors. Data in A–E are representative of nine biological replicates over three independent experiments. Data in F–M are representative of 30 ROIs acquired over at least three independent experiments. Data represent mean and SEM. Statistical significance for A–D, G–I, J, L, and M was assessed by Mann–Whitney, statistical significance for E was assessed by Kruskal–Wallis.
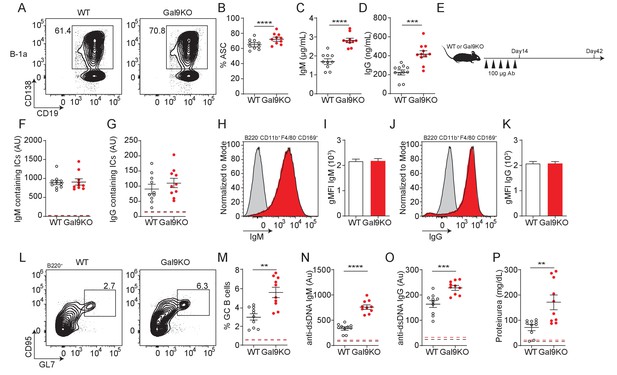
Transfer of B-1a derived antibodies leads to autoimmunity in mice.
(A) Representative plots of ASC development in sorted WT and Gal9KO B-1a cells cultured for 3 days in presence of 0.5 µg/mL LPS and 0.5 µg/mL IgM. (B) Summary proportion of ASC differentiation in sorted WT (black, open) and Gal9KO (red, filled), as in (A). (C) Secreted IgM titers, and (D) Secreted IgG titers in the culture supernatant, as in (A). (E) Schematic of in vivo B-1-derived antibody transfer experiment. (F) Titers of IgM and (G) IgG-containing ICs in circulation at day 14, mean baseline titers shown as dashed line. (H) Representative histograms and (I) summary gMFI of IgM expression on splenic subcapsular sinus macrophages at day 14. (J) Representative histograms and (K) summary gMFI of IgG expression on splenic subcapsular sinus macrophages at day 14. (L) Representative plots and (M) summary proportion of GC B cell differentiation in the spleen at day 42, mean baseline frequency shown as dashed lines. (N) Titers of dsDNA-specific IgM and (O) IgG in sera at day 42, mean baseline titers shown as dashed line. (P) Protein secretion in the urine of mice at day 42, mean baseline level shown as dashed line. Data are representative of 10 biological replicates over two independent experiments. Statistical significance was assessed by Mann–Whitney, *p≤0.05, **p≤0.01, ***p≤0.001, ****p<0.0001.
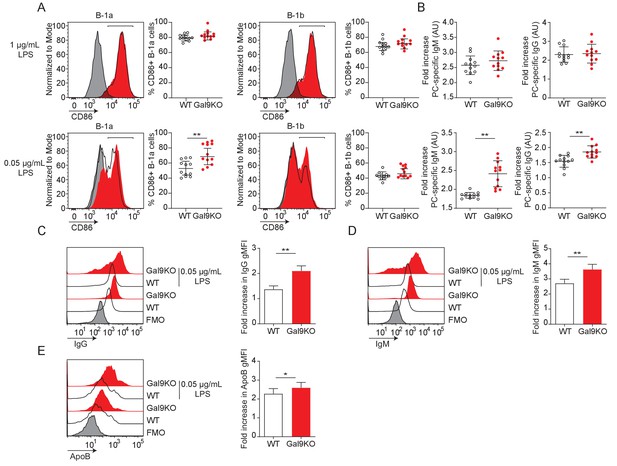
Gal9KO B-1a cells have enhanced activation to low LPS stimulation.
(A) Representative histograms of CD86 expression on B-1a and B-1b cells from WT (black) or Gal9KO (red) mice injected i.p. with 1 µg/mL (top) or 0.05 µg/mL (bottom) of LPS (left), and proportion of CD86 expressing cells (right) as indicated. (B) Fold increase in phosphorylcholine-specific IgM (left) and IgG (right) antibody titers in sera following LPS injection as in (A). (C) Representative histograms of IgG expression on splenic subcapsular sinus macrophages (left), summary fold increase in IgG gMFI following LPS injection (right). (D) Representative histograms of IgM expression on splenic subcapsular sinus macrophages (left), summary fold increase in IgG gMFI following LPS injection (right). (E) Representative histograms of apoptotic body detection on splenic subcapsular sinus macrophages (left), summary fold increase in apoptotic body gMFI following LPS injection and transfer of CFSE-labeled apoptotic bodies (right). Data are representative of twelve biological replicates over three independent experiments. Data represent mean and SEM. Statistical significance was assessed by Mann–Whitney, *p≤0.05, **p≤0.01.
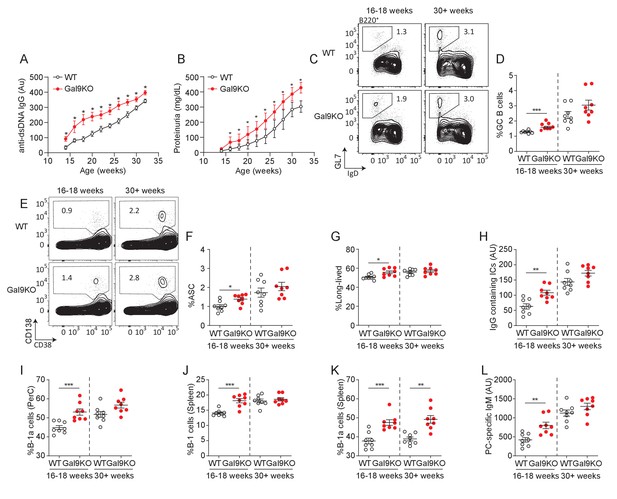
Loss of Gal9 in NZB/W mice leads to accelerated autoimmunity.
(A) Titers of dsDNA-specific IgG in sera of NZB/W mice (as indicated) monitored over time. (B) Protein secretion in the urine of NZB/W mice monitored over time. (C) Representative plots and (D) summary proportion of splenic GC B cells in NZB/W mice (as indicated). (E) Representative plots and (F) summary proportion of antibody secreting cells (ASC) in the spleen of NZB/W mice. (G) Proportion of long-lived ASC lacking B220 expression. (H) Circulating IgG-containing immune complex (ICs) titers. (I) Proportion of CD5+ B-1a cells in the peritoneal cavity of NZB/W mice. (J) Proportion of CD19+ CD43+ B-1 cells in the spleen of NZB/W mice. (K) Proportion of splenic CD5+ B-1a cells. (L) Circulating phosphoryl-choline specific IgM titers in NZB/W mice. Data in (A) and (B) are representative of at least 14 biological replicates, data in (C–L) are representative of eight biological replicates. Data represent mean and SEM. Statistical significance was assessed by Mann–Whitney, *p≤0.05, **p≤0.01, ***p≤0.001.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Mus musculus) | C57BL/6 | Charles River Laboratories | Strain code: 027 | |
| Strain, strain background (Mus musculus) | Gal9-/- | Steven Beverley (Washington University) on behalf of the Scripps Research Institute | Gal9KO | |
| Strain, strain background (Mus musculus) | Nr4a1-eGFP (Nur77-GFP) | Jackson Laboratories | Strain code: 016617 | |
| Strain, strain background (Mus musculus) | New Zealand Black (NZB/BINJ) | Jackson Laboratories | Strain code: 000684 | |
| Strain, strain background (Mus musculus) | New Zealand White (NZW/LacJ) | Jackson Laboratories | Strain code: 001058 | |
| Strain, strain background (Mus musculus) | MD4 | Jackson Laboratories | Strain code: 002595 | |
| Strain, strain background (Mus musculus) | µMT | Jackson Laboratories | Strain code: 002288 | |
| Strain, strain background (Mus musculus) | MD4/ML5 | Joan Wither (University of Toronto) | ||
| Cell Line (Homo sapiens) | Male:HEp2 | ATCC | Cat#: CCL-23 | |
| Cell Line (Mus musculus) | OP9-R7FS | Juan Carlos Zúñiga-Pflücker (University of Toronto) | ||
| Antibody | Phospho-Tyrosine (p-Tyr) | Sunnybrook Research Institute | Clone:4G10 | |
| Antibody | Anti-IgM F(ab’)2 | Jackson Immunoresearch | Cat#: 155-006-020 | Polyclonal |
| Antibody | Anti-B220-Pacific Blue | BioLegend | Cat#: 103227 | Clone: RA3-6B2 |
| Antibody | Anti-CD11b-Pacific Blue | BioLegend | Cat#: 101224 | Clone: M1/70 |
| Antibody | Anti-Ly6G-FITC | BioLegend | Cat#:127606 | Clone: 1A8 |
| Antibody | Anti-CD45- Brilliant Violet 510 | BioLegend | Cat#: 103138 | Clone: 30-F11 |
| Antibody | Anti-CD169-PE-Cyanine7 | BioLegend | Cat#: 142412 | Clone: 3D6.122 |
| Antibody | Anti-F4/80-Alexa Fluor 700 | BioLegend | Cat#: 123129 | Clone: BM8 |
| Antibody | Anti-CD11c-APC | BioLegend | Cat#: 117310 | Clone: N418 |
| Antibody | Anti-CD64-PE | BioLegend | Cat#: 139303 | Clone: X54-5/7.1 |
| Antibody | Anti-MHCII (I-A/I-E)-Brilliant Violet 605 | BioLegend | Cat#: 107639 | Clone: M5/114.15.2 |
| Antibody | Anti-CD8-APC | BioLegend | Cat#: 100712 | Clone: 53-6.7 |
| Antibody | Anti-CD4-Brilliant Violet 510 | BioLegend | Cat#: 100449 | Clone: GK1.5 |
| Antibody | Anti-CD23-APC | BioLegend | Cat#: 101620 | Clone: B3B4 |
| Antibody | Anti-CD21/35-APC-Cyanine7 | BioLegend | Cat#: 123418 | Clone: 7E9 |
| Antibody | Anti-Galectin-9-PE | BioLegend | Cat#: 137904 | Clone: 108A2 |
| Antibody | Anti-Galectin-9-Alexa Fluor 488 | BioLegend | Cat#: 137908 | Clone: 108A2 |
| Antibody | Anti-Galectin-9 | BioLegend | Cat#: 137902 | Clone: 108A2 |
| Antibody | Anti-CD86-FITC | BioLegend | Cat#: 105006 | Clone: GL-1 |
| Antibody | Anti-CD86-PE-Cyanine7 | BioLegend | Cat#: 105014 | Clone: GL-1 |
| Antibody | Anti-IgM-Alexa Fluor 488 | Jackson Immunoreasearch | Cat#: 715-547-020 | |
| Antibody | Anti-GL7-APC | BioLegend | Cat#: 144618 | Clone: GL7 |
| Antibody | Anti-CD95-FITC | BioLegend | Cat#: 152606 | Clone: SA367H8 |
| Antibody | Anti-IgD-Brilliant Violet 605 | BioLegend | Cat#: 405727 | Clone: 11-26c.2a |
| Antibody | Anti-CD138-Brilliant Violet 605 | BioLegend | Cat#: 142516 | Clone: 281-2 |
| Antibody | Anti-CD38-PE-Cyanine7 | BioLegend | Cat#: 102718 | Clone: 90 |
| Antibody | Anti-IgM-Alexa Fluor 647 | Jackson Immunoresearch | Cat#: 155-607-020 | |
| Antibody | Anti-IgG (FC)-Alexa Fluor 488 | Jackson Immunoresearch | Cat#: 155-545-008 | |
| Antibody | Anti-PD-1-PE-Cyanine7 | BioLegend | Cat#: 109110 | Clone: RMP1-30 |
| Antibody | Anti-CXCRV-Brilliant Violet 421 | BioLegend | Cat#: 145512 | Clone: L138D7 |
| Antibody | Anti-FoxP3-PE | BioLegend | Cat#: 126404 | Clone: MF-14 |
| Antibody | Anti-IgMa-PE | BioLegend | Cat#: 408608 | Clone: MA-69 |
| Antibody | Anti-B220-Alexa Fluor 647 | BioLegend | Cat#: 103226 | Clone: RA3-6B2 |
| Antibody | Anti-CD5-PE-Cyanine 5 | BioLegend | Cat#: 100610 | Clone: 53-7.3 |
| Antibody | Anti-CD19-Pacific Blue | BioLegend | Cat#: 115523 | Clone: 6D5 |
| Antibody | Anti-CD43-APC | BioLegend | Cat#: 143208 | Clone: S11 |
| Antibody | Anti-CD23-PE | BioLegend | Cat#: 101607 | Clone: B3B4 |
| Antibody | Anti-CD69-Brilliant Violet 510 | BioLegend | Cat#: 104531 | Clone: H1.2F3 |
| Antibody | Anti-CD5-Alexa Fluor 647 | BioLegend | Cat#: 100614 | Clone: 53-7.3 |
| Antibody | Anti-SiglecG-APC | Fisher Scientific | Cat#: 501123146 | Clone: SH2.1 |
| Antibody | Anti-CD22-Alexa Fluor 647 | BioLegend | Cat#: 126108 | Clone: OX-97 |
| Antibody | Anti-CD180 | BioLegend | Cat#:117710 | Clone:RP/14 |
| Antibody | Anti-TLR4-Alexa Fluor 488 | Fisher Scientific | Cat#: 5016829 | Clone: UT41 |
| Antibody | Anti-C3 | BioLegend | Cat#: 518106 | Clone: K13/16 |
| Recombinant Proteins | Recombinant Galectin-9 | R&D Systems | Cat#: 3535-GA-050 | |
| Chemical compound/drug | Calf Thymus DNA | ThermoFisher Scientific | Cat#: 15633019 | |
| Chemical compound/drug | Phosphorylcholine | Sigma Aldrich | Cat#: P0378 | |
| Chemical compound/drug | Hen Egg Lysozyme | Sigma Aldrich | Cat#: 10837059001 | |
| Chemical compound/drug | β-lactose | Sigma Aldrich | Cat#: L3750 | |
| Chemical compound/drug | Streptavidin-Alexa Fluor 647 | Thermo Fisher Scientific | Cat#: S21374 | |
| Chemical compound/drug | CFSE | Thermo Fisher Scientific | Cat#: C34554 | |
| Chemical compound/drug | Cell tracker eFluor 450 | Thermo Fisher Scientific | Cat#: 65-0842-85 | |
| Chemical compound/drug | Puromycin Dihydrochloride | Thermo Fisher Scientific | Cat#: A1113803 | |
| Chemical compound/drug | LPS | Invivogen | Cat#: tlrl-eklps | |
| Chemical compound/drug | Imiquimod | Invivogen | Cat#: tlrl-imq | |
| Chemical compound/drug | Zymosan | Invivogen | Cat#: tlrl-zyn | |
| Chemical compound/drug | CpG-ODN | Invivogen | Cat#: tlrl-1585 | |
| Chemical compound/drug | M2 Beads | Sigma Aldrich | Cat#: M8823 | |
| Commercial assay/kit | B cell Isolation kit | Stemcell Technologies | Cat#: 19854 | |
| Commercial assay/kit | Mouse IgM Quantitation set | Bethyl Laboratories | Cat#: E90-101 | |
| Commercial assay/kit | Mouse IgG Quantitation set | Bethyl Laboratories | Cat#: E90-131 | |
| Commercial assay/kit | Clophosome-A and control liposome kit | FormuMax | Cat#: F70101C-AC | |
| Commercial assay/kit | Fluorescence quantitation beads | Bangs Labs | Cat#: 817 | |
| Commercial assay/kit | BCA kit | Sigma Aldrich | Cat#: A53225 | |
| Software, algorithm | Flowjo v10.6 | Tree Star | ||
| Software, algorithm | ImageJ | NIH/LOCI | ||
| Software, algorithm | ThunderSTORM | Github | zitmen | |
| Software, algorithm | Rstudio | Rstudio PBC | ||
| Software, algorithm | SMoLR | Github | ErasmusOIC | |
| Software, algorithm | Spatstat | Github | spatstat |
Additional files
-
Source data 1
Source data for figures and figure supplements.
- https://cdn.elifesciences.org/articles/64557/elife-64557-data1-v1.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/64557/elife-64557-transrepform-v1.pdf





