A structure of substrate-bound Synaptojanin1 provides new insights in its mechanism and the effect of disease mutations
Figures
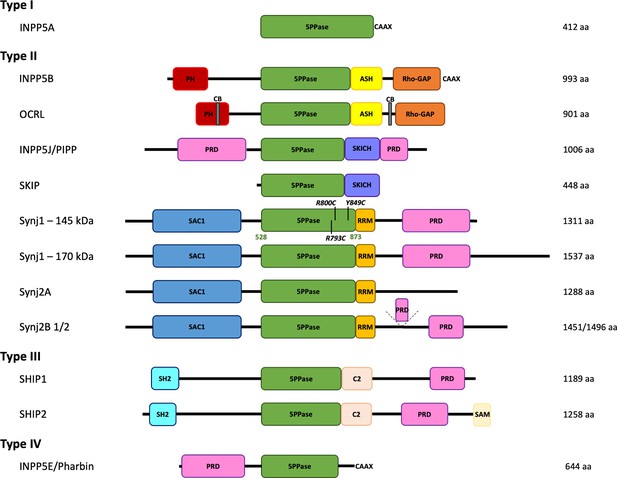
Domain organization of the inositol polyphosphate 5-phosphatase family (5PPases).
The domain organization of the ten human 5PPases, subdivided in four groups (type I–IV), is shown schematically. The different splice forms for Synaptojanin 1 (Synj1) and 2 (Synj2) are also shown. The domain boundaries of the 5PPase domain of Synj1 145 kDa used in this study and the disease mutations under study are indicated. 5PPase = 5-phosphatase domain; PH = Pleckstrin homology domain; ASH = ASPM, SPD-2, Hydin domain; Rho-GAP = Rho GTPase-activating protein domain; CB = clathrin-binding domain; PRD = proline-rich domain; SKICH = SKIP carboxyl homology domain; SAC1 = suppressor of actin 1-like domain; RRM = RNA recognition motif; SH2 = Src homology two domain; SAM = sterile alpha motif.
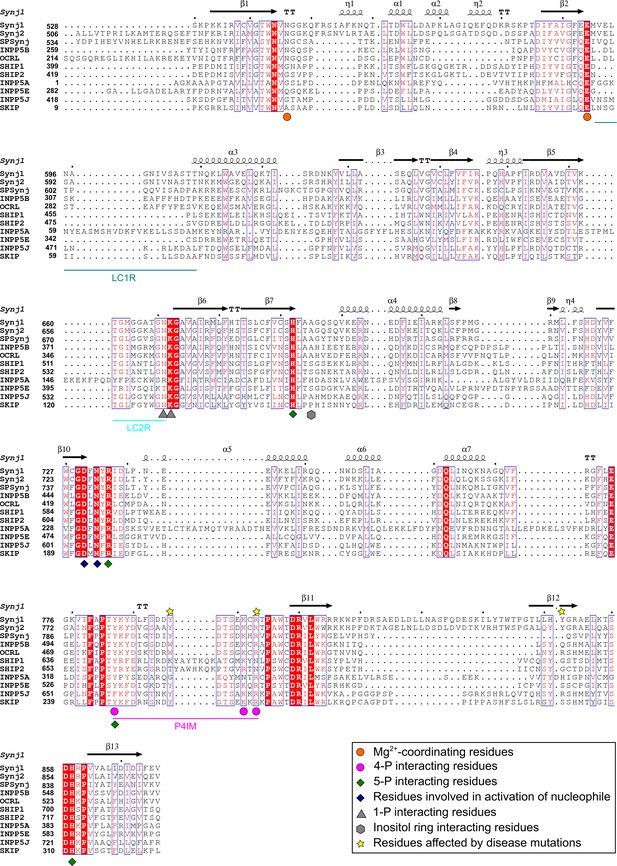
Sequence alignment of the 5PPase domains of ten human 5-phosphatases and Schizosaccaromyces pombe Synaptojanin (SPSynj).
The sequence alignment of the 5PPase domain of all human 5PPases and SPSynj was obtained via Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/). Secondary structure assignment was performed using ESPript (http://espript.ibcp.fr/ESPript/ESPript/). Residues important for substrate or metal-ion interactions are indicated as described in the figure legend. The loop containing the P4-interacting-motif (P4IM) and the lipid chain 1 (LC1R) and 2 (LC2R) recognition motifs are indicated by magenta, green-blue, and cyan lines, respectively. Completely conserved residues are indicated by a red background and residues that show similarity across the 5PPases are written in red. Similar sequence motifs are indicated with a blue square. α-helices and 310-helices are displayed as helices and indicated by α or η respectively. β-strands are displayed as arrows and strict β-turns as TT.
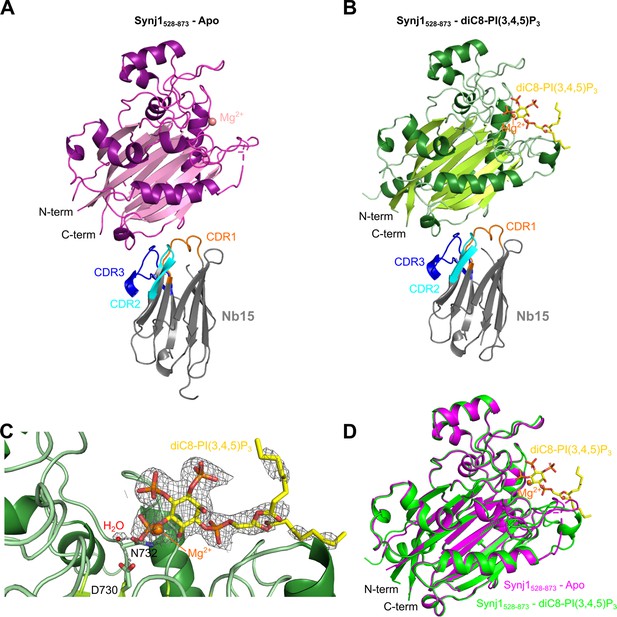
Structure of the Nb15-Synj1528–873 complex in presence or absence of the substrate diC8-PI(3,4,5)P3.
(A) Apo-structure of the Nb15-Synj1528–873 complex. Synj1528–873 (chain E) is represented in different shades of magenta, while the Nb (chain F) is represented in grey with indication of the different CDR regions. The Mg2+-ion is shown as a salmon sphere. (B) The Nb15-Synj1528–873 complex bound to diC8-PI(3,4,5)P3. Synj1528–873 (chain A) is represented in different shades of green, while the Nb (chain B) is represented similar as in (A). The Mg2+-ion is shown as an orange sphere and diC8-PI(3,4,5)P3 is shown as yellow sticks. (C) Zoom-in on the active site region of Synj1528–873 with bound diC8-PI(3,4,5)P3 (yellow sticks), Mg2+ (orange sphere) and the nucleophilic water molecule (red sphere) shown with their corresponding 2FO-FC-map contoured at 1σ. Residues D730 and N732, which play a role in the activation of the nucleophilic water, are shown as green sticks. (D) Superposition of the apo (magenta) and diC8-PI(3,4,5)P3-bound (green) Synj1528–873 structure.
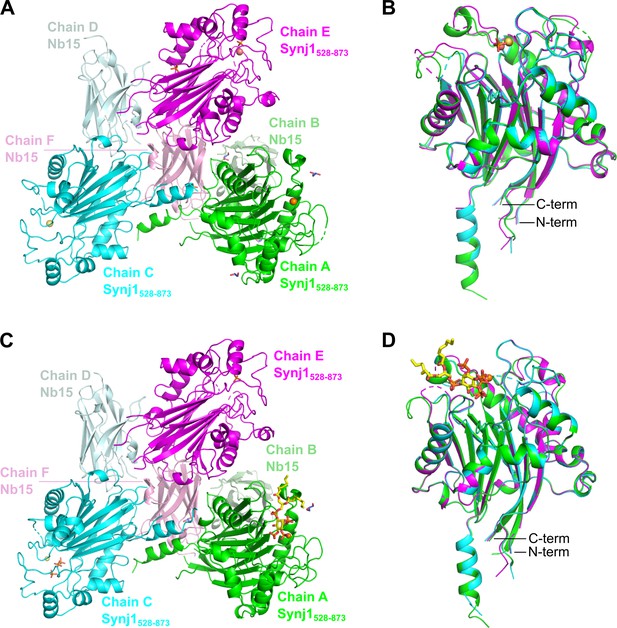
Content of the asymmetric unit (AU) of the apo and the diC8-PI(3,4,5)P3-bound Nb15-Synj1528–873 complex.
The asymmetric unit (AU) of both structures contains three Nb15-Synj1528–873 complexes. (A) AU of the apo-structure. (B) Superposition of the three Synj1528–873 chains from the apo-structure. (C) AU of the diC8-PI(3,4,5)P3-bound structure. (D) Superposition of the three Synj1528–873 chains from the diC8-PI(3,4,5)P3-bound structure. Mg2+-ions are represented as orange, yellow, and salmon spheres for chain A, C, and E, respectively. diC8-PI(3,4,5)P3, free phosphates, and glycerol are shown as sticks in yellow, orange, and purple, respectively.
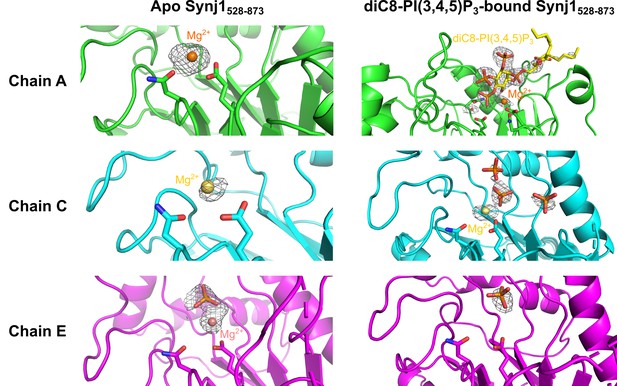
Close-up view on the active site of different Synj1528–873-chains of the apo- and diC8-PI(3,4,5)P3-bound Nb15-Synj1528–873 structure.
The close-up views of the different Synj1528–873 chains of the apo-structure are show on the left, while the same chains of the diC8-PI(3,4,5)P3-bound structure are shown on the right. The Mg2+-ions are shown as orange, yellow or salmon spheres for chain A (green), chain C (cyan) of chain E (magenta), respectively. The Mg2+-interacting residues, N543 and E591, are shown as sticks in every chain. In chain A of the diC8-PI(3,4,5)P3-bound Synj1528–873 structure (top right panel) residues D730 and N732, which play a direct role in activation of the nucleophilic water (red sphere), are also shown as sticks. Free orthophosphates and the substrate diC8-PI(3,4,5)P3 are shown as orange and yellow sticks, respectively. The omit map, contoured at 3 σ, is shown as a grey mesh around the phosphates, Mg2+-ions, diC8-PI(3,4,5)P3 and the nucleophilic water.
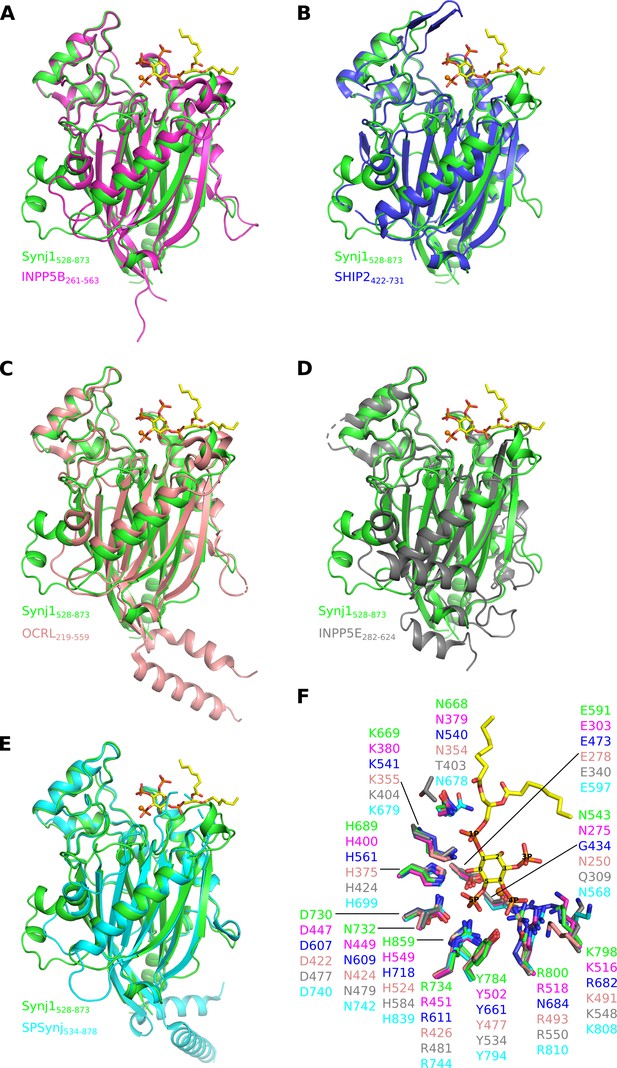
Superposition of the catalytic (5PPase) domain of human Synj1528–873 on the available structures of the other human 5-phosphatases and SPSynj.
Superposition of Synj1528–873 (green, PDB 7A17) on the 5PPase domain of (A) INPP5B (magenta, PDB 4CML), (B) SHIP2 (blue, PDB 4A9C), (C) OCRL (salmon, PDB 4CMN), (D) INPP5E (grey, PDB 2XSW) and (E) SPSynj (cyan, PDB 1I9Z). The Mg2+-ion and the diC8-PI(3,4,5)P3 substrate of the Synj1528–873 structure are shown as an orange sphere and yellow sticks, respectively. (F) Zoom-in on the active site of the superposed 5PPases described in (A to E). For clarity, only the Mg2+-ion and the diC8-PI(3,4,5)P3 substrate of the Synj1528–873 structure are shown as an orange sphere and yellow sticks, respectively.
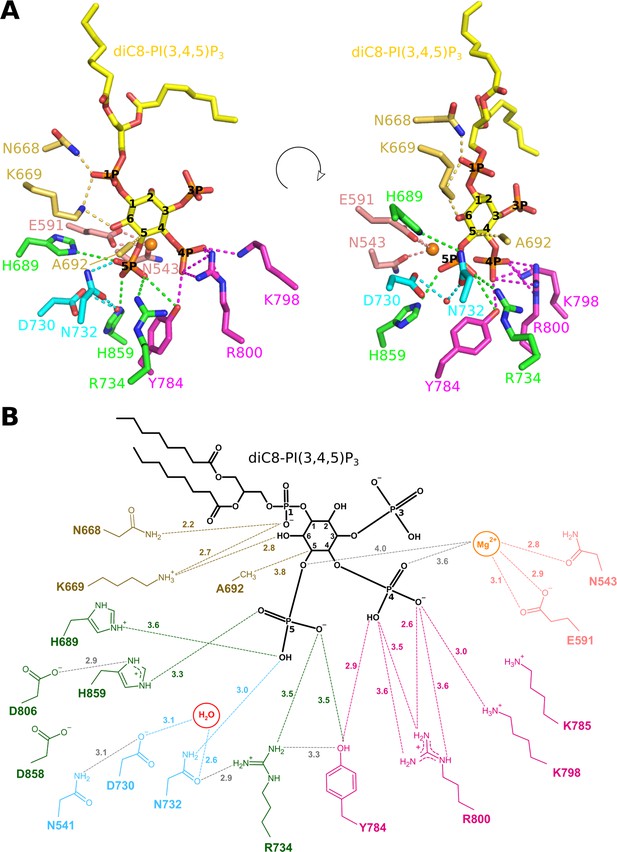
Enzyme-substrate interactions in the Synj1528–873-diC8-PI(3,4,5)P3 complex.
(A) Zoom-in on the active site where Synj1528–873 forms interactions with different groups of the diC8-PI(3,4,5)P3 substrate (yellow sticks). The residues coloured in gold are forming interactions with the 1 P group or inositol ring of the PIP (gold dashes), residues coloured in magenta form interactions with the 4 P group (magenta dashes), and residues shown in green are interacting with the 5 P group (green dashes). Residues important for activation of the nucleophilic water (red sphere) are shown in cyan, while two residues shown in salmon are interacting with the Mg2+-ion (orange sphere). (B) Schematic representation of the interactions between Synj1528–873 and diC8-PI(3,4,5)P3. The same colour-code as in (A) was used, except for the substrate that is shown in black. Distances (in Å) between interacting atoms are indicated.
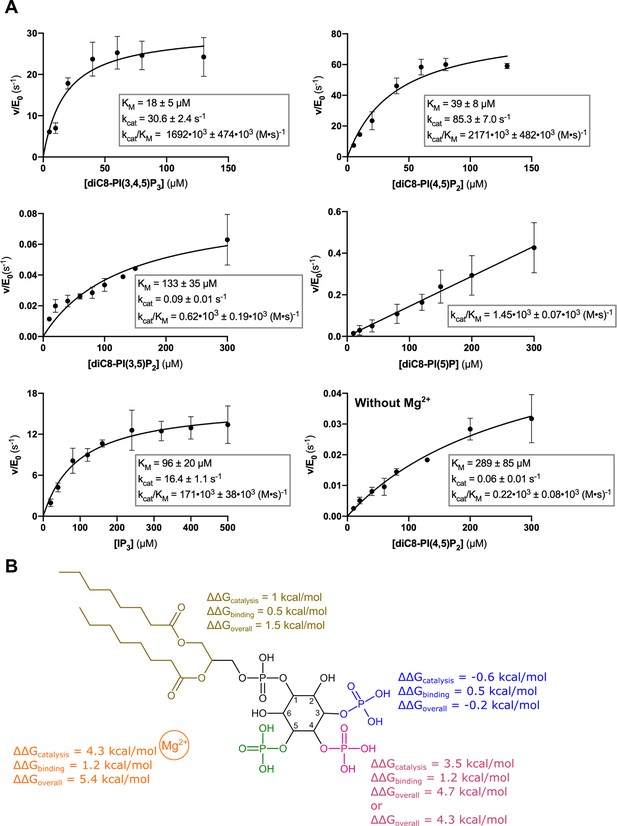
Kinetic analysis of the contribution of different substrate groups to the Synj1528–873 5-phosphatase activity.
(A) Michaelis-Menten curves obtained for Synj1528–873 with different substrates: diC8-PI(3,4,5)P3, diC8-PI(4,5)P2, diC8-PI(3,5)P2, diC8-PI(5)P, IP3, and diC8-PI(4,5)P2 in the absence of Mg2+. The turnover number (kcat), the Michaelis-Menten constant (KM) and specificity constant (kcat/KM) are given for every measurement, together with the standard error. Each datapoint is the average of three independent measurements with the error bars representing the standard deviation. (B) Schematic overview of the contribution of the different groups of the diC8-PI(3,4,5)P3-substrate to the Synj1528–873 mechanism, with the acyl chains coloured in gold, the 3 P group in blue, the 4 P group in magenta, the 5 P group in green and the Mg2+-ion in orange. The ΔΔG value shows the contribution of the acyl chains, 3 P, 4 P, and Mg2+-ion to catalysis (kcat), binding (KM) and overall catalytic efficiency (kcat/KM). The more positive the ΔΔG value, the larger the contribution of the respective group to either catalysis, binding or overall catalytic efficiency.
-
Figure 4—source data 1
Steady-state enzyme kinetics data of Synj1528–873 wild-type in combination with different substrates.
- https://cdn.elifesciences.org/articles/64922/elife-64922-fig4-data1-v2.xlsx
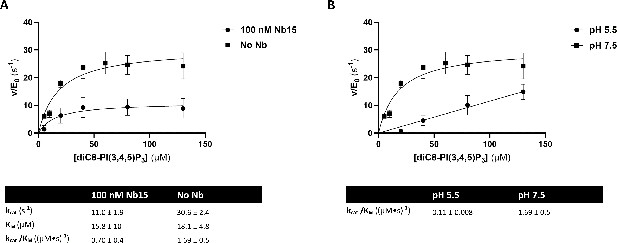
The 5-phosphatase activity of Synj1528–873 is affected by Nb15 and acidic pH.
(A) Michaelis-Menten curves obtained by using the Malachite green assay with 2.5 nM of human Synj1528–873 and various concentrations of diC8-PI(3,4,5)P3 in absence or presence of 100 nM Nb15. (B) Michaelis-Menten curves obtained by using the Malachite green assay with 2.5 nM of human Synj1528–873 and various concentration of diC8-PI(3,4,5)P3 in assay buffer containing 25 mM HEPES at pH 7.5 or in assay buffer containing 25 mM sodium citrate at pH 5.5. The turnover number (kcat), the Michaelis-Menten constant (KM), and specificity constant (kcat/KM) are given for the different measurements, together with the standard error. Each datapoint is the average of three independent measurements with the error bars representing the standard deviation.
-
Figure 4—figure supplement 1—source data 1
Steady-state enzyme kinetics data of Synj1528–873 wild-type using diC8-PI(3,4,5)P3 as substrate at pH 5.5 and in presence of an excess of Nb15 (comparable to the crystallization conditions).
- https://cdn.elifesciences.org/articles/64922/elife-64922-fig4-figsupp1-data1-v2.xlsx
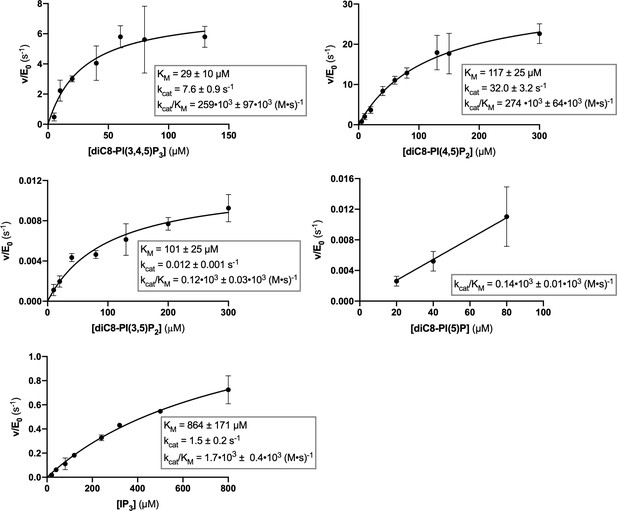
Steady-state enzyme kinetics of the Synj1528–873 Y793C mutant in combination with different substrates.
Michaelis-Menten curves obtained for Synj1528–873 Y793C with different substrates: diC8-PI(3,4,5)P3, diC8-PI(4,5)P2, diC8-PI(3,5)P2, diC8-PI(5)P, and IP3. The turnover number (kcat), the Michaelis-Menten constant (KM) and specificity constant (kcat/KM) are given for the different measurements, together with the standard error. Each datapoint is the average of three independent measurements with the error bars representing the standard deviation.
-
Figure 4—figure supplement 2—source data 1
Steady-state enzyme kinetics data of the Synj1528–873 Y793C mutant in combination with different substrates.
- https://cdn.elifesciences.org/articles/64922/elife-64922-fig4-figsupp2-data1-v2.xlsx
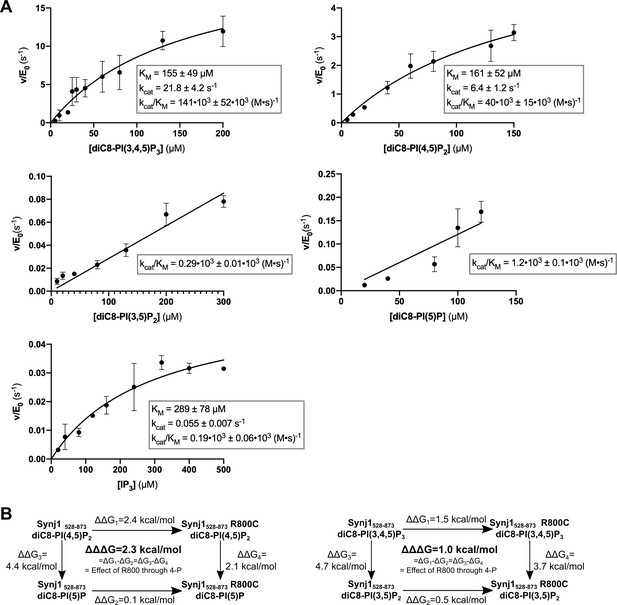
Steady-state enzyme kinetics of the Synj1528–873 R800C mutant in combination with different substrates.
(A) Michaelis-Menten curves obtained for Synj1528–873 R800C with different substrates: diC8-PI(3,4,5)P3, diC8-PI(4,5)P2, diC8-PI(3,5)P2, diC8-PI(5)P, and IP3. The turnover number (kcat), the Michaelis-Menten constant (KM) and specificity constant (kcat/KM) are given for the different measurements, together with the standard error. Each datapoint is the average of three independent measurements with the error bars representing the standard deviation. (B) Thermodynamic squares with corresponding ΔΔG values calculated from kcat/KM. Each ΔΔG value corresponds to the contribution of either the substituted R800 side chain or the deleted 4 P group according to ΔΔG = -R.T.ln[(kcat/KM)2 / (kcat/KM)1]. ΔΔΔG shows the difference between ΔΔG1 and ΔΔG2 or ΔΔG3 and ΔΔG4, indicating that R800 performs its role in the Synj1528–873 mechanism through the 4 P group.
-
Figure 4—figure supplement 3—source data 1
Steady-state enzyme kinetics data of the Synj1528–873 R800C mutant in combination with different substrates.
- https://cdn.elifesciences.org/articles/64922/elife-64922-fig4-figsupp3-data1-v2.xlsx
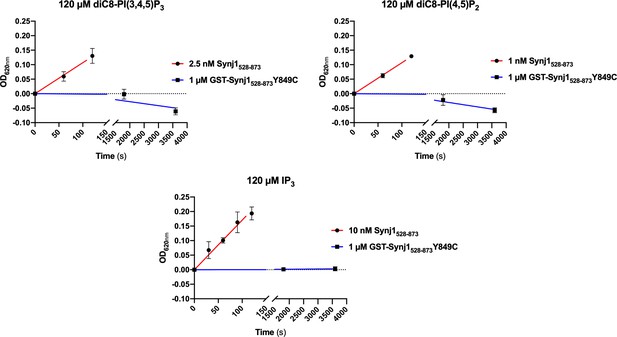
The (GST-tagged) Synj1528–873 Y849C mutant shows no 5-phosphatase activity.
Time traces monitoring the activity (change in OD620nm) of 1 µM GST-Synj1528–873 Y849C using either 120 µM diC8-PI(3,4,5)P3, diC8-PI(4,5)P2 or IP3 are shown. After measuring 1 hr, the OD620nm in function of time curve was flat, indicating that no activity could be measured. These curves were compared to the corresponding curves of the wild-type Synj1528–873 at 100- to 1000-fold lower enzyme concentration (1–10 nM).
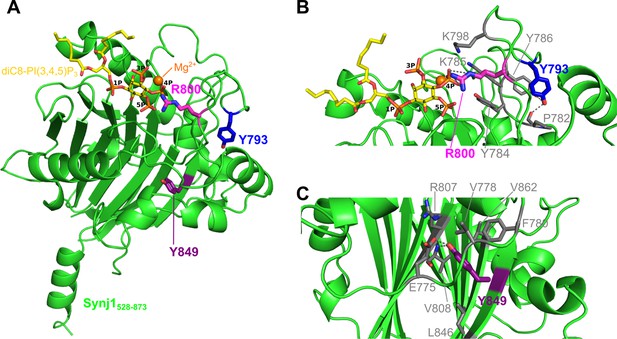
Localization of the Y793C, R800C, and Y849C disease mutations in the Synj1528–873 structure.
(A) Overall structure of Synj1528–873 (green) with the Y793, R800, and Y849 residues represented as blue, magenta, and purple sticks, respectively. The Y793 residue is present in a loop close to the active site, while R800 is present in the active site. The Y849 residue, on the other hand, is buried in the core of the 5PPase domain. The Mg2+-ion is represented as an orange sphere and the substrate, diC8-PI(3,4,5)P3, as yellow sticks. (B) Close-up view on Y793 and R800 and their surrounding residues. Y793 forms a hydrogen bond with Y786 and with the main chain of P782 (grey dashes) to potentially stabilize the conformation of the loop. R800 forms multiple hydrogen bonds with the 4 P group of diC8-PI(3,4,5)P3 (grey dashes). (C) Close-up view on Y849 and its surrounding residues. Y849 is buried in the hydrophobic core, where it forms a hydrogen bond with E775 and with the main chain NH of V808 (grey dashes).
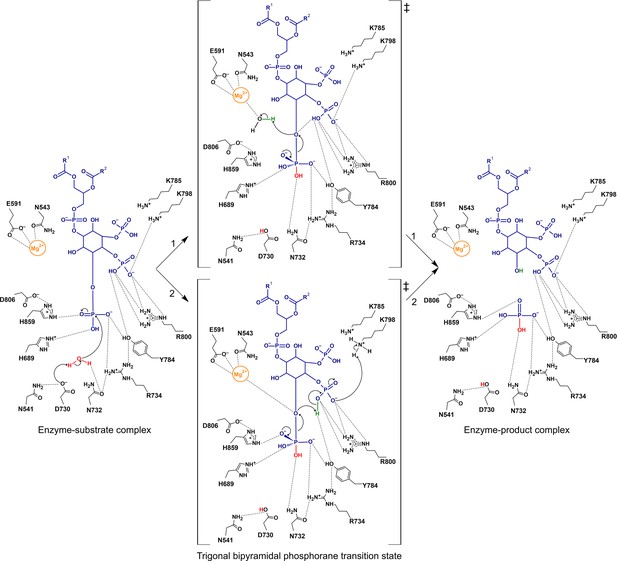
Proposed catalytic mechanism(s) of the 5-phosphatase reaction of Synj1.
The nucleophilic water is activated by proton transfer to the catalytic base D730, allowing attack on the scissile phosphate (P5), and resulting in a phosphorane transition state with excess negative charge that is stabilized by several surrounding residues. Two routes for leaving group activation are envisioned. In route 1 (upper pathway) a Mg2+-activated water molecule acts as general acid by donating a proton to the leaving hydroxylate. Route 2 (lower pathway) corresponds to a mechanism of substrate-assisted catalysis, where the adjacent 4 P group acts as general acid, potentially assisted by transfer of a proton from K798.
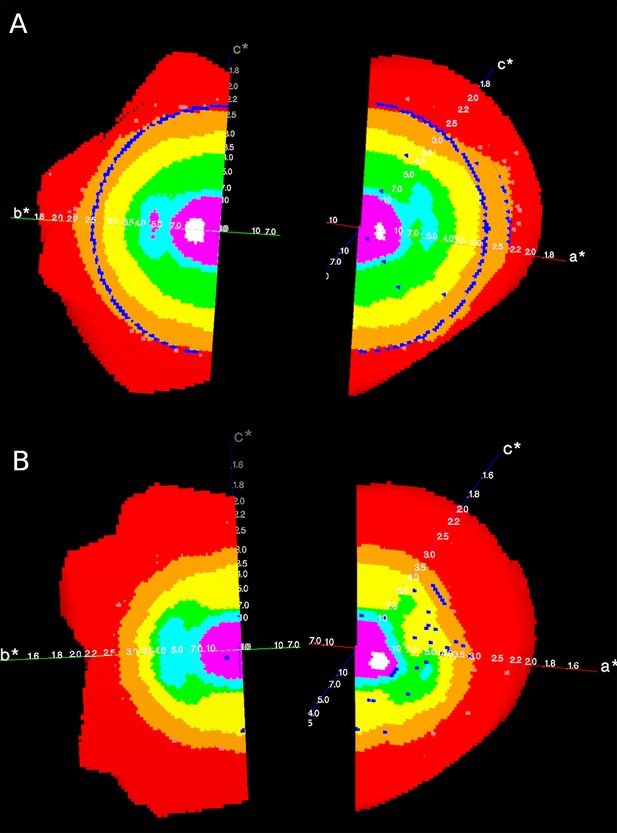
Reciprocal lattices (axes a*, b*, c*) colour coded by mean I/σ(I) as given by STARANISO, showing the anisotropic diffraction of the crystal of (A) the apo Synj1528-873 and (B) the diC8-PI(3,4,5)P3-bound Synj1528-873.
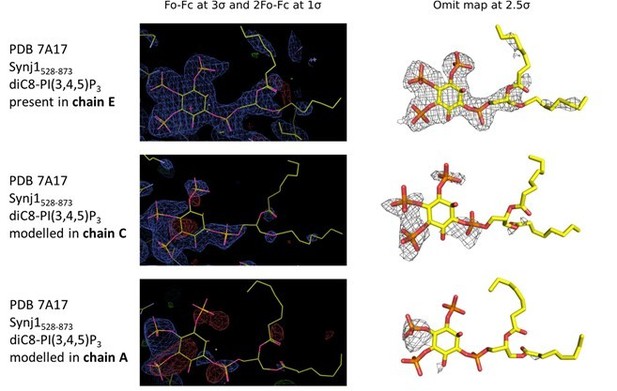
Electron density in the active sites of protein molecules corresponding to chain E (top), C (middle) and A (bottom) of the Synj1528‐873 ‐ diC8‐PI(3,4,5)P3 structure.
The panels on the left show the Fo‐Fc and 2Fo‐Fc map contoured at 3σ and 1σ respectively, while the panels on the right show the omit map contoured at 2.5σ.
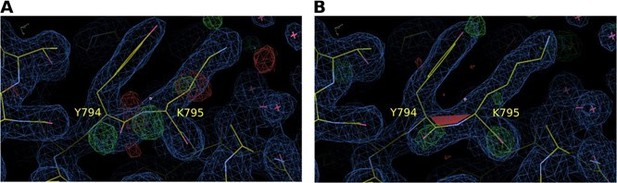
Electron density map around residues Y794 and K795 of S. pombe Synj (SPSynj).
2Fo‐Fc map at 1σ and Fo‐Fc map at 3σ around residues Y794 and K795 of SPSynj with (A) a trans‐peptide bond and (B) a cis‐peptide bond.
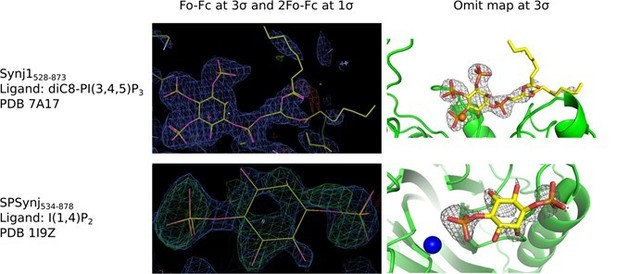
Electron density around diC8‐PI(3,4,5)P3 in the Synj1528‐873 structure (PDB 7A17) and around inositol‐(1,4)bisphosphate in the SPSynj structure (1I9Z).
The panels on the left show the Fo‐Fc and 2Fo‐Fc map contoured at 3σ and 1σ respectively, while the panels on the right show the omit map contoured at 3σ.
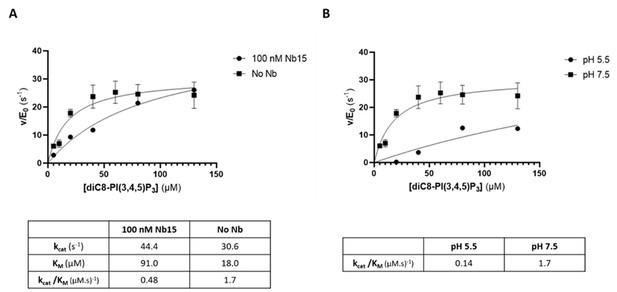
The 5‐phosphatase activity of Synj1528‐873 is affected by Nb15 and acidic pH.
(A) Michaelis‐Menten curves obtained by using the Malachite green assay with 2.5 nM of human Synj1528‐873 and various concentrations of diC8‐PI(3,4,5)P3 in absence or presence of 100 nM Nb15. (B) Michaelis‐Menten curves obtained by using the Malachite green assay with 2.5 nM of human Synj1528‐873 and various concentration of diC8‐PI(3,4,5)P3 in assay buffer containing 25 mM HEPES at pH 7.5 or in assay buffer containing 25 mM sodium citrate at pH 5.5.
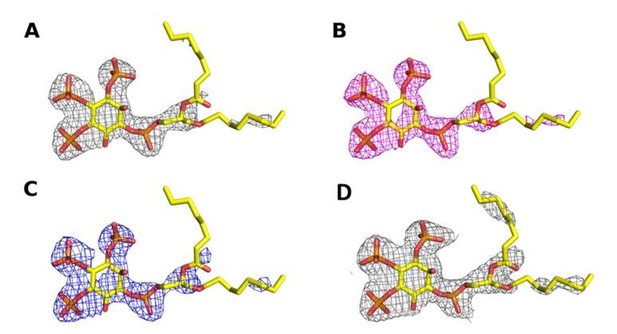
Omit map contoured at 3σ around the diC8‐PI(3,4,5)P3 substrate of a structure (A) processed with STARANISO to 2.73 Å, (B) processed without STARANISSO to 2.73 Å and (C) processed without STARANISO to 3.0 Å.
Panel (D) Refined 2Fo‐Fc map contoured at 1σ around the diC8‐PI(3,4,5)P3 substrate of a structure processed with STARANISO to 2.73 Å.
Tables
Data collection and refinement statistics.
| Synj1528–873 - Apo | Synj1528–873 - diC8-PI(3,4,5)P3 | |
|---|---|---|
| PDB code | 7A0V | 7A17 |
| Data collection | ||
| Synchrotron | Diamond | Soleil |
| Beamline | i03 | Px2a |
| Wavelength (Å) | 0.98 | 0.98 |
| Resolution range (Å)* | 87.06–2.30 (2.43–2.30) | 87.39–2.73 (3.02–2.73) |
| Space group | C121 | C121 |
| Unit cell dimensions (Å) | a = 168.87 | a = 169.32 |
| b = 108.79 | b = 109.21 | |
| c = 100.97 | c = 100.90 | |
| Unit cell angles (°) | α = 90.00 | α = 90.00 |
| β = 120.72 | β = 120.62 | |
| γ = 90.00 | γ = 90.00 | |
| Spherical completeness (%)* | 77.1 (26.4) | 76.2 (22.8) |
| Ellipsoidal completeness (%)* | 92.3 (95.0) | 91.5 (57.3) |
| Unique reflections | 53789 | 32149 |
| Mean (I)/SD(I)* | 11.2 (1.4) | 5.3 (1.4) |
| CC(1/2)* | 0.997 (0.512) | 0.964 (0.474) |
| Multiplicity* | 7.0 (5.7) | 3.5 (3.6) |
| Rmeas (%)* | 14.5 (125.1) | 28.2 (123.7) |
| Refinement | ||
| Resolution range (Å) | 86.81–2.30 | 86.83–2.73 |
| Rwork (%) | 19.64 | 19.88 |
| Rfree (%)† | 25.23 | 25.74 |
| Model content | ||
| Molecules per AU | 6 | 6 |
| Protein atoms per AU | 10574 | 10624 |
| Ligand atoms per AU | 39 | 85 |
| Metal atoms per AU | 3 | 2 |
| Water molecules per AU | 359 | 71 |
| Wilson B factors (Å2) | 38.08 | 43.28 |
| Average B factors (Å2) | ||
| Protein atoms | 47.35 | 44.88 |
| Ligand atoms | 65.62 | 67.11 |
| Metal atoms | 60.08 | 42.62 |
| Water molecules | 42.68 | 22.93 |
| Rmsd bonds (Å) | 0.002 | 0.006 |
| Rmsd angles (°) | 0.456 | 1.15 |
| Ramachandran plot (%) (favored, outliers) | 95.79, 0.23 | 97.00, 0.46 |
-
* Values in parentheses are for the high-resolution shell.
† Rfree is based on a subset of 5% of reflections omitted during refinement.
-
AU, asymmetric unit.
Steady-state kinetic parameters of Synj1528–873 and the Synj1528–873 Y793C, R800C and Y849C mutants in combination with different substrates.
| Synj1528–873 | Synj1528–873 Y793C | Synj1528–873 R800C | Synj1528–873 Y849C | ||
|---|---|---|---|---|---|
| diC8-PI(3,4,5)P3 | kcat (s−1) | 30.6 ± 2.4 | 7.6 ± 0.9 | 21.8 ± 4.2 | NMA |
| KM (µM) | 18 ± 5 | 29 ± 10 | 155 ± 49 | NMA | |
| kcat/KM (•103 (M•s)−1) | 1692 ± 474 | 259 ± 97 | 141 ± 52 | NMA | |
| diC8-PI(4,5)P2 | kcat (s−1) | 85.3 ± 7.0 | 32.0 ± 3.2 | 6.4 ± 1.2 | NMA |
| KM (µM) | 39 ± 8 | 117 ± 25 | 161 ± 52 | NMA | |
| kcat/KM (•103 (M•s)−1) | 2171 ± 482 | 274 ± 64 | 40 ± 15 | NMA | |
| IP3 | kcat (s−1) | 16.4 ± 1.1 | 1.5 ± 0.2 | 0.055 ± 0.007 | NMA |
| KM (µM) | 96 ± 20 | 864 ± 171 | 289 ± 78 | NMA | |
| kcat/KM (•103 (M•s)−1) | 171 ± 38 | 1.7 ± 0.4 | 0.19 ± 0.06 | NMA | |
| diC8-PI(3,5)P2 | kcat (s−1) | 0.09 ± 0.01 | 0.012 ± 0.001 | ND | ND |
| KM (µM) | 133 ± 35 | 101 ± 25 | ND | ND | |
| kcat/KM (•103 (M•s)−1) | 0.62 ± 0.19 | 0.12 ± 0.03 | 0.29 ± 0.01 | ND | |
| diC8-PI(5)P | kcat (s−1) | ND | ND | ND | ND |
| KM (µM) | ND | ND | ND | ND | |
| kcat/KM (•103 (M•s)−1) | 1.45 ± 0.07 | 0.14 ± 0.01 | 1.2 ± 0.1 | ND | |
| diC8-PI(4,5)P2 without Mg2+ | kcat (s−1) | 0.06 ± 0.01 | ND | ND | ND |
| KM (µM) | 289 ± 85 | ND | ND | ND | |
| kcat/KM (•103 (M•s)−1) | 0.22 ± 0.08 | ND | ND | ND |
-
ND, not determined.
NMA, no measurable activity.
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Homo sapiens) | SYNJ1 | NCBI | Gene ID: 8867 | |
| Strain, strain background (Escherichia coli) | BL21(DE3) pLysS | Weiner et al., 1994 | Genotype: F-hsdSB (rB-mB-) gal dcm (DE3) pLysS (CmR) | Chemically (CaCl2) competent |
| Strain, strain background (E. coli) | WK6 (Su-) | Zell and Fritz, 1987 (PMID:3038536) | Genotype: Δ(lac-proAB) galE strA/F’ [lacIq lacZΔM15 proA+B+] | Chemically (CaCl2) competent |
| Strain, strain background (M13 helper phage) | Kanamycin-resistant VCSM13 | Stratagene | 200251 | |
| Recombinant DNA reagent | pET28a (plasmid) | Novagen | 69864 | |
| Recombinant DNA reagent | pGEX-4T1 (plasmid) | GE Healthcare | GE28-9545-49 | |
| Recombinant DNA reagent | pMESy4 (plasmid) | Pardon et al., 2014 (DOI: 10.1038/nprot.2014.039) | GenBank KF415192 | |
| Chemical compound, drug | IP6 (D-myo-inositol 1,2,3,4,5,6-hexakis phosphate) | Merck Millipore | 407125 | |
| Chemical compound, drug | IP3 (D-myo-inositol 1,4,5-trisphosphate) | Merck Millipore | 407137 | |
| Chemical compound, drug | diC8-PI(5)P | Echelon Biosciences | P-5008 | |
| Chemical compound, drug | diC8-PI(4,5)P2 | Echelon Biosciences | P-4508 | |
| Chemical compound, drug | diC8-PI(3,5)P2 | Echelon Biosciences | P-3508 | |
| Chemical compound, drug | diC8-PI(3,4,5)P3 | Echelon Biosciences | P-3908 | |
| Chemical compound, drug | disodium-4- nitrophenyl phosphate (DNPP) | Sigma | N-4645 | |
| Sequence-based reagent | Y793C_F | This paper | PCR primers | CGACTGTGACACCA GTGAAAAGTGCCG |
| Sequenced-based reagent | Y793C_R | This paper | PCR primers | CTGGTGTCACAG TCGTCAGAAAACAAG |
| Sequence-based reagent | R800C_F | This paper | PCR primers | GTGCTGCACCCCTG CCTGGACAGAC |
| Sequenced-based reagent | R800C_R | This paper | PCR primers | GGTGCAGCACTTTT CACTGGTGTC |
| Sequence-based reagent | Y849C_F | This paper | PCR primers | CACTGTGGAAGAG CTGAGCTGAAG |
| Sequenced-based reagent | Y849C_R | This paper | PCR primers | CTTCCACAGTGCAGC AAAGTGCCTGG |
| Peptide, recombinant protein | CaptureSelect Biotin anti-C-tag conjugate | Thermo Fisher Scientific | 7103252100 | |
| Peptide, recombinant protein | Streptavidin Alkaline Phosphatase | Promega | V5591 | |
| Commercial assay or kit | Malachite Green Phosphate Assay kit | Gentaur | POMG-25H | |
| Software, algorithm | autoPROC | Vonrhein et al., 2011 (DOI: 10.1107/S0907444911007773) | RRID:SCR_015748 https://www.globalphasing.comautoproc/ | |
| Software, algorithm | STARANISO | Tickle et al., 2018 | RRID:SCR_018362 http://staraniso.globalphasing.org/cgi-bin/staraniso.cgi | |
| Software, algorithm | Phaser | McCoy et al., 2007 (DOI:10.1107/S0021889807021206) | RRID:SCR_014219 https://www.phenix-online.org/documentation/reference/phaser.html | |
| Software, algorithm | Phenix.Ligand Fit | Terwilliger et al., 2006 (DOI:10.1107/S0907444906017161) | https://www.phenix-online.org/documentation/reference/ligandfit.html | |
| Software, algorithm | Phenix.Refine | Afonine et al., 2012 (DOI: 10.1107/S0907444912001308) | RRID:SCR_016736 https://www.phenix-online.org/documentation/reference/refinement.html | |
| Software, algorithm | Coot | Emsley et al., 2010 (DOI: 10.1107/S0907444910007493) | RRID:SCR_014222 https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ | |
| Software, algorithm | MolProbity | Chen et al., 2010 (DOI: 10.1107/S0907444909042073) | RRID:SCR_014226 http://molprobity.biochem.duke.edu | |
| Software, algorithm | PDB-REDO server | Joosten et al., 2014 (DOI: 10.1107/S2052252514009324) | RRID:SCR_018936 https://pdb-redo.eu/ | |
| Software, algorithm | PyMOL (version 2.0) | Schrödinger | RRID:SCR_000305 https://pymol.org/2/ | |
| Software, algorithm | GraphPad Prism (version 8) | Graphpad Software | RRID:SCR_002798 | |
| Software, algorithm | CCP4 suite | Winn et al., 2011 (DOI: 10.1107/S0907444910045749) | RRID:SCR_007255 http://www.ccp4.ac.uk/ | |
| Software, algorithm | Clustal Omega | Madeira et al., 2019 (DOI: 10.1093/nar/gkz268) | RRID:SCR_001591 http://www.ebi.ac.uk/Tools/msa/clustalo/ | |
| Software, algorithm | ESPript | Robert and Gouet, 2014 (DOI: 10.1093/nar/gku316) | RRID:SCR_006587 http://espript.ibcp.fr/ESPript/ESPript/ | |
| Software, algorithm | ACD/ChemSketch (version 2019.2.1) | Advanced Chemistry Development | http://www.acdlabs.com |
| diC8‐PI(3,4,5)P3‐bound Synj1528‐873 | |||
|---|---|---|---|
| STARANISO 2.73 Å | Without STARANISO 2.73 Å | Without STARANISO 3.00 Å | |
| PDB number | 7A17 | ||
| Data collection | |||
| Resolution range (Å)a | 87.39‐2.73 (3.02‐2.73) | 87.39‐2.73 (2.83‐2.73) | 87.39‐3.00 (3.16‐3.00) |
| Spherical completeness (%)a | 76.2 (22.8) | 99.8 (100) | 99.8 (99.9) |
| Ellipsoidal completeness (%)a | 91.5 (57.3) | / | / |
| Unique reflections | 32149 | 42013 | 31720 |
| Mean (I)/SD(I)a | 5.3 (1.4) | 4.1 (0.7) | 5.2 (1.4) |
| CC(1/2)a | 0.964 (0.474) | 0.953 (0.195) | 0.962 (0.448) |
| Multiplicitya | 3.5 (3.6) | 3.5 (3.6) | 3.5 (3.4) |
| Rmeas (%)a | 28.2 (123.7) | 37.1 (259.9) | 28.1 (114.4) |
R, Rfree and R‐Rfree statistics for structures in the resolution range 2.50‐2.90 Å as given by phenix.r_factor_statistics.
| Histogram of Rwork for models in PDB at resolution 2.50‐2.90 Å : 0.119 ‐ 0.171 : 281 structures 0.171 ‐ 0.223 : 5171 structures 0.223 ‐ 0.274 : 3263 structures 0.274 ‐ 0.326 : 122 structures 0.326 ‐ 0.378 : 4 structures |
| Histogram of Rfree for models in PDB at resolution 2.50‐2.90 Å: 0.159 ‐ 0.220 : 469 structures 0.220 ‐ 0.281 : 6011 structures 0.281 ‐ 0.343 : 2315 structures 0.343 ‐ 0.404 : 45 structures 0.404 ‐ 0.465 : 1 structures |
| Histogram of Rfree‐Rwork for all model in PDB at resolution 2.50‐2.90 Å: 0.001 ‐ 0.021 : 408 structures 0.021 ‐ 0.041 : 2370 structures 0.041 ‐ 0.060 : 3841 structures 0.060 ‐ 0.080 : 1809 structures 0.080 ‐ 0.100 : 413 structures |
| Number of structures considered: 8841 |
Additional files
-
Supplementary file 1
(A) Comparison of the 5PPase domain of human Synj1 (Synj1528–873) with the corresponding 5PPase domain of the other human inositol polyphosphate 5-phosphatases and SPSynj. The root-mean-square deviation (rmsd) after superposition of the structures was determined using CCP4 SUPERPOSE. Furthermore, the sequence identity was determined using Clustal Omega. (B) Residues interacting with the diC8-PI(3,4,5)P3 substrate in Synj1528–873 and the corresponding residues in the other nine human 5PPases and SPSynj. Completely conserved residues are written in white with a red background and similar residues are written in red.
- https://cdn.elifesciences.org/articles/64922/elife-64922-supp1-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/64922/elife-64922-transrepform-v2.docx