eNOS-induced vascular barrier disruption in retinopathy by c-Src activation and tyrosine phosphorylation of VE-cadherin
Figures
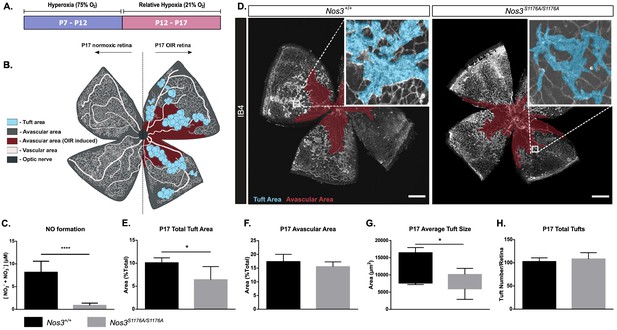
Suppressed tuft formation in the Nos3S1176A/S1176A retina after OIR-challenge.
(A) Outline of OIR-challenge protocol; pups were placed in 75% O2 (hyperoxia) between P7 and P12, followed by return to normal atmosphere (relative hypoxia) until P17. (B) Schematic representation of vascular abnormalities after OIR in P17 retinas. (C) Nitric oxide formation determined using a Griess assay, expressed as the combined concentration of nitrite and nitrate, the end products of NO, reacting with molecules in biological fluids. Mean ± S.E.M. n = 3 mice/genotype. ****p<0.0001; t-test. (D) Representative images of whole mount retinas from Nos3+/+ and Nos3S1176A/S1176A mice, collected at P17 after OIR-challenge, stained with isolectin B4 (IB4). Avascular area is marked in magenta and tufts in blue. Scale bar = 500 µm. (E, F) Tuft area (E) and avascular area (F) expressed as percentage of total vascular area at P17. (G, H) Tuft size in µm2 (G) and total number/FOV in P17 mice (H). For (E–H): mean ± S.E.M. n = 7 (Nos3+/+) and 5 (Nos3S1176A/S1176A) mice. *p<0.05; t-test.
-
Figure 1—source data 1
Excel file containing numerical values collected from NO formation assays and OIR-induced tuft formation experiments in Nos3+/+ and Nos3S1176A/S1176A mice shown in Figure 1, Figure 1—figure supplements 1–4.
- https://cdn.elifesciences.org/articles/64944/elife-64944-fig1-data1-v2.xlsx
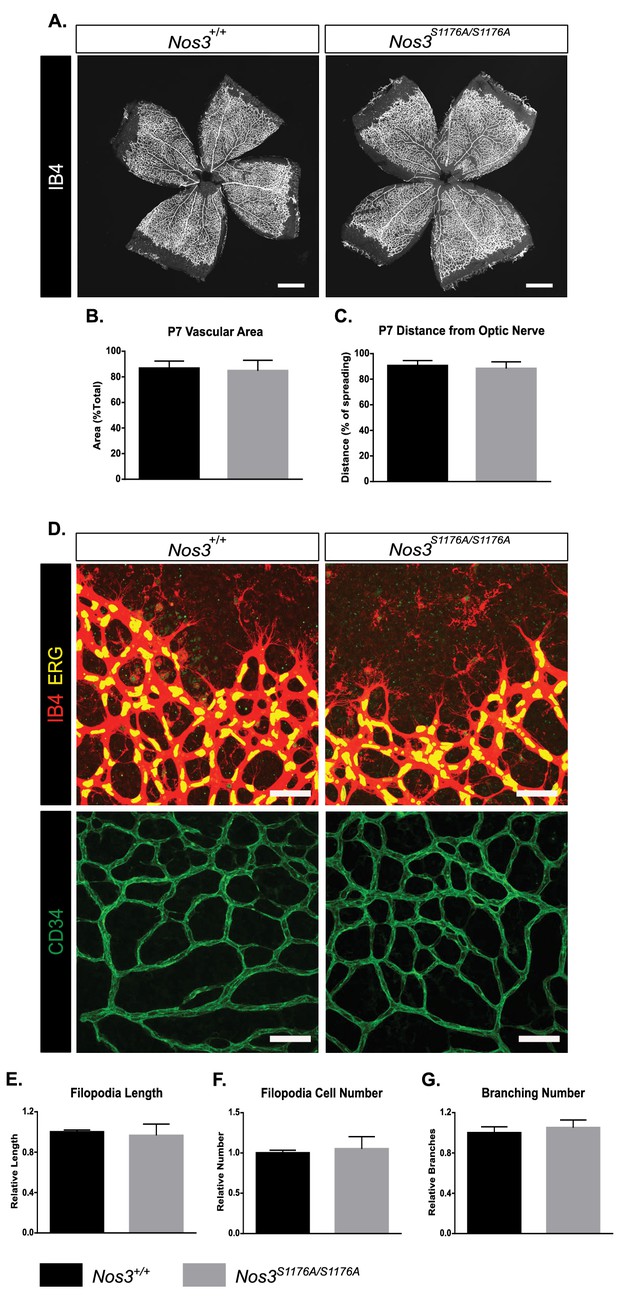
Postnatal development of Nos3+/+ and Nos3S1176A/S1176A retinal vasculature.
(A) Representative images of Nos3+/+ and Nos3S1176A/S1176A retinas collected at P7, stained with isolectin B4 (IB4). (B, C) Quantification of vascular area (B) and outgrowth from the optic nerve (C) at P7 in Nos3+/+ and Nos3S1176A/S1176A pups. (D) Representative images of the vessel front from whole mount retinas collected at P7 stained with Isolectin-B4 (IB4, red, upper), ERG (yellow, upper), and CD34 (green, lower), to visualize vessel outgrowth and tip cells in Nos3+/+ and Nos3S1176A/S1176A retinal vasculature. Scale bar = 50 µm. (E–G) Filopodia length (E), tip cell number (F), branch points (G) in Nos3+/+ and Nos3S1176A/S1176A retinas at P7. Mean ± S.E.M. n = 3 (Nos3+/+) and 4 (Nos3S1176A/S1176A) mice, five to six images per mouse; t-test.
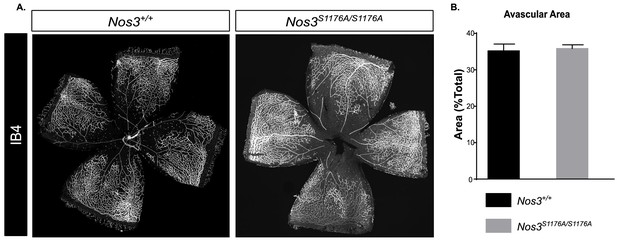
Retina characteristics in Nos3+/+ and Nos3S1176A/S1176A P12 pups.
(A) Representative images of whole mount Nos3+/+ and Nos3S1176A/S1176A retinas collected at P12 after the vessel destruction phase of OIR and before vessel regrowth, stained with isolectin B4 (IB4). Scale bar = 500 µm. (B) Avascular area in Nos3+/+ and Nos3S1176A/S1176A retinas at P12 after OIR. n = 4 (Nos3+/+) and 5 (Nos3S1176A/S1176A) mice, three independent experiments; t-test.
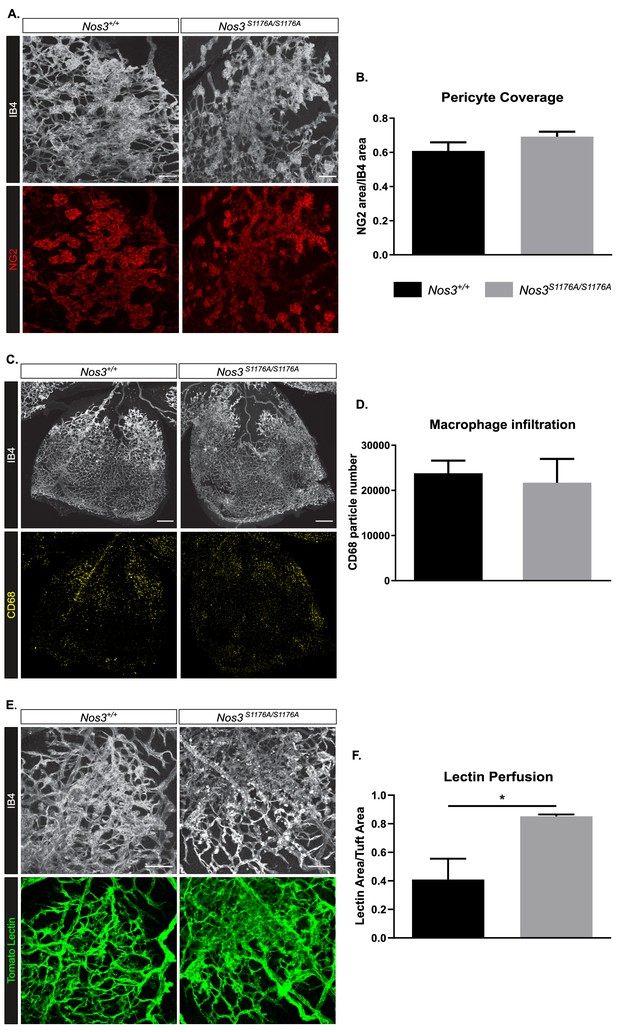
Pericytes, macrophage influx, and vascular perfusion in OIR-challenged Nos3+/+ and Nos3S1176A/S1176A retinas.
(A) Representative images of tufts visualized by IB4 binding and co-stained for NG2 (red) to mark pericytes in Nos3+/+ and Nos3S1176A/S1176A retinas collected at P17. Scale bar = 100 μm. (B) Pericyte coverage expressed as the NG2-positive area normalized to the IB4 area. Mean ± S.E.M. n = 4 (Nos3+/+) and 4 (Nos3S1176A/S1176A) mice. t-test. (C) Representative images of one leaf of the retina (IB4) co-stained for CD68 (yellow) to mark circulatory/tissue macrophages in Nos3+/+ and Nos3S1176A/S1176A retinas collected at P17. Scale bar = 200 μm. (D) The number of CD68-positive macrophages in the entire retina represented by particle number, normalized to the retina area. Mean ± S.E.M. n = 4 (Nos3+/+) and 4 (Nos3S1176A/S1176A) mice. t-test. (E) Representative images of tufts with circulating tomato lectin (green), co-stained with IB4 in Nos3+/+ and Nos3S1176A/S1176A retinas collected at P17. Scale bar = 100 μm. (F) Tuft perfusion, expressed as the area of tomato lectin normalized to the IB4 area. Mean ± S.E.M. n = 4 (Nos3+/+) and 4 (Nos3S1176A/S1176A) mice. *p<0.05; t-test.
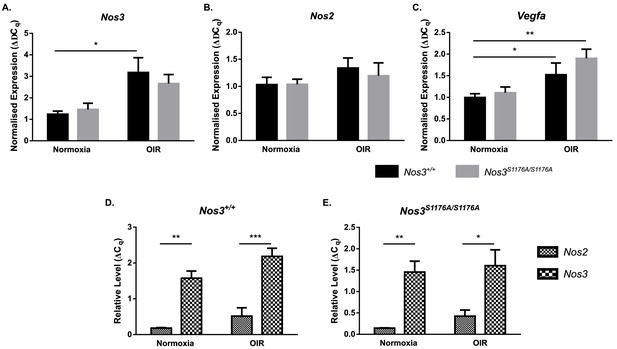
Expression of Nos2, Nos3, and Vegfa in Nos3+/+ and Nos3S1176A/S1176A retinas.
(A–C) qPCR of Nos3 (A), Nos2 (B), and Vegfa (C) expression in P17 normoxic and OIR-challenged Nos3+/+ and Nos3S1176A/S1176A mouse retinas. (D, E) Relative levels of Nos2 and Nos3 compared against standard curves of TBP and UBC in Nos3+/+ (D) and Nos3S1176A/S1176A (E) retinas. Mean ± S.E.M. n = 5 (Nos3+/+) and 5 (Nos3S1176A/S1176A) mice. Three independent experiments. *, **, ***p<0.05, 0.01, 0.001; two-way ANOVA, Sidak’s multiple comparison test.
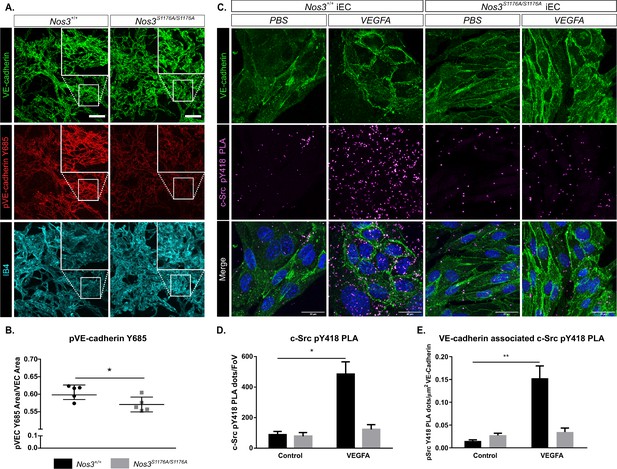
Suppressed c-Src Y418 and VE-cadherin Y685 phosphorylation in Nos3S1176A/S1176A retinas and isolated endothelial cells.
(A) Representative maximum intensity projections of similar sized tufts from Nos3+/+ and Nos3S1176A/S1176A retinas showing VE-cadherin (green), pY685 VE-cadherin (red), and isolectin B4 (IB4; cyan). Scale bar = 50 μm. (B) Ratio of pY685 positive area/total VE-cadherin positive area. Mean ± S.E.M n = 3–6 images per group from 5 (Nos3+/+) and 5 (Nos3S1176A/S1176A) mice, three independent experiments, *p<0.05; t-test. (C) Representative images of VE-cadherin staining (green) and proximity ligation assay (PLA) to detect c-Src pY418 (magenta) in isolated mouse lung endothelial cells (iEC) from Nos3+/+ and Nos3S1176A/S1176A mice. Scale bar = 20 µM. (D) c-Src pY418 PLA dots detected in PBS and VEGFA (50 ng/mL)-treated iECs from Nos3+/+ and Nos3S1176A/S1176A mouse lungs. Data expressed as the number of dots per field of view. (E) c-Src pY418 PLA dots co-localized with VE-cadherin (green), normalized against total VE-cadherin area in the field of view. Mean ± S.E.M. Cells isolated from n = 4 (Nos3+/+) and 4 (Nos3S1176A/S1176A) mice, from three separate experiments. *, **p<0.05, 0.01; two-way ANOVA, Sidak’s multiple comparisons test.
-
Figure 2—source data 1
Excel file containing numerical values collected from biochemical analyses shown in Figure 2, Figure 2—figure supplements 1–3.
- https://cdn.elifesciences.org/articles/64944/elife-64944-fig2-data1-v2.xlsx
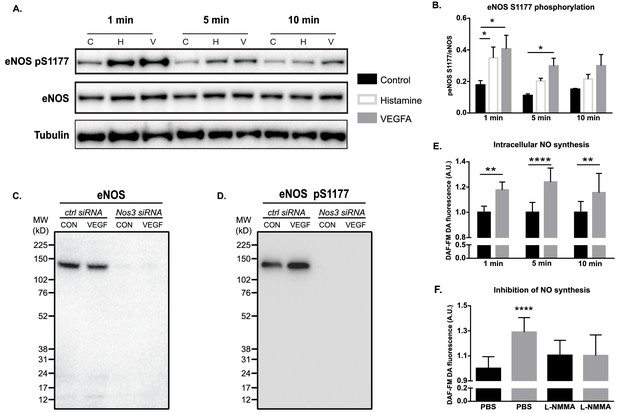
VEGFA induced eNOS phosphorylation and activity in vitro.
(A) Effect of VEGFA (V; 50 ng/mL; 1, 5, 10 min), histamine (H; 10 µM, 1, 5, 10 min), or medium (C, control) on eNOS phosphorylation at S1177 in cultured Human Retinal Microvascular Endothelial Cells (HRMEC). (B) Quantification of eNOS pS1177/total eNOS normalized to tubulin. Mean ± S.E.M. n = 3 independent experiments. *p<0.05; two-way ANOVA, Sidak’s multiple comparison test. (C) Antibody validation by immunoblotting for eNOS on HRMECs transfected with a control siRNA or Nos3-specific siRNA followed by treatment with VEGFA (50 ng/mL; 5 min). (D) Antibody validation by immunoblotting for eNOS pS1177 on HRMECs transfected with a control siRNA or Nos3-specific siRNA followed by treatment with VEGFA (50 ng/mL; 5 min). (E) Quantification of NO production in HRMECs treated with PBS or VEGFA (50 ng/mL, for 1, 5, or 10 min) using the cell-permeable fluorescent probe DAF-FM DA. (F) Quantification of NO production in HRMECs pre-treated with PBS or L-NMMA (1 mM) before VEGFA stimulation (50 ng/mL, 5 min). Mean ± S.E.M. n = 12, three independent experiments. *, **, ****p<0.05, 0.01, 0.0001; two-way ANOVA, Sidak’s multiple comparison test.
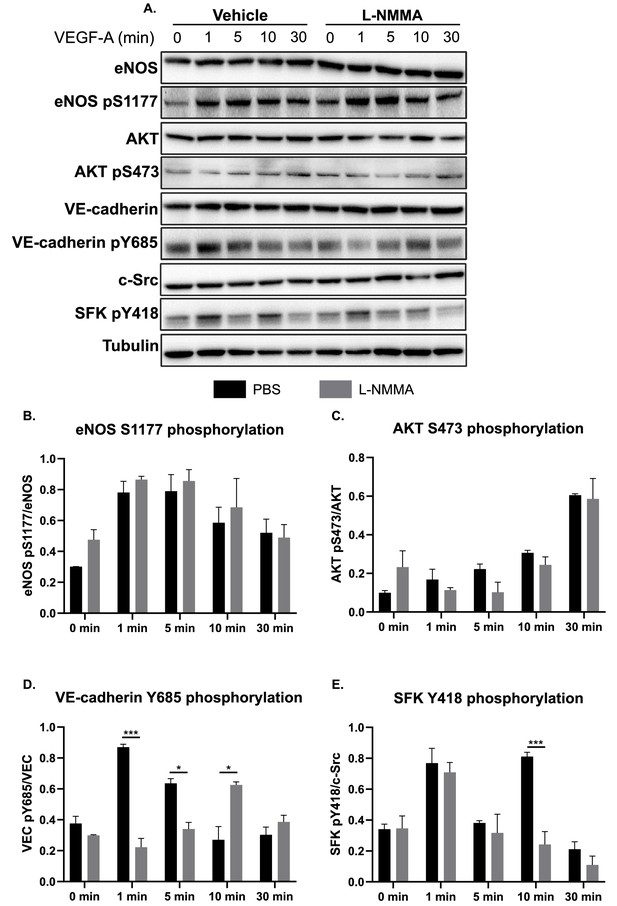
VEGFA induced phosphorylation of eNOS, AKT, VE-cadherin, and c-Src in vitro.
(A) Representative immunoblots and the effect of VEGFA (50 ng/mL; 1, 5, 10, 30 min) on eNOS, eNOS pS1177, AKT, AKT pS473, VE-cadherin, VE-cadherin pY685, c-Src, SFK pY418, and tubulin in cultured Human Retinal Microvascular Endothelial Cells (HRMEC) treated with a PBS vehicle or 1 mM L-NMMA for 1 hr. (B) Quantification of eNOS pS1177/total eNOS (normalized to tubulin). (C) Quantification of AKT pS473/total AKT (normalized to tubulin). (D) Quantification of VE-cadherin pY685/total VE-cadherin (normalized to tubulin). (E) Quantification of SFK pY418/total c-Src (normalized to tubulin) Mean ± S.E.M. n = 3 independent experiments. *, ***p<0.05, 0.001 (indicates significance between vehicle and L-NMMA-treated samples, stimulated with VEGFA as indicated). Two-way ANOVA, Sidak’s multiple comparison test.
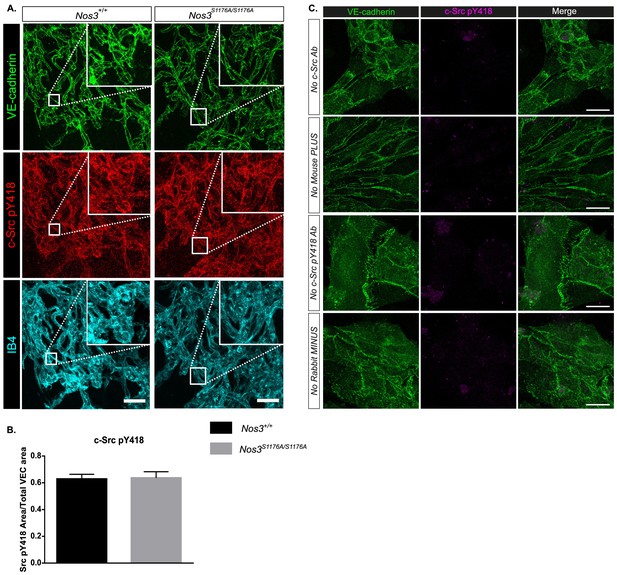
c-Src pY418 immunostaining and PLA controls.
(A) Representative maximum intensity projections of tufts from Nos3+/+ and Nos3S1176A/S1176A retinas immunostained for VE-cadherin (green), c-Src pY418 (red), and IB4 (cyan). (B) Quantification of pY418-positive area/total VE-cadherin area. n = 3 images per group from 3 (Nos3+/+) and 4 (Nos3S1176A/S1176A) mice from two separate experiments; t-test. (C) Representative images of VE-cadherin staining (green) and negative controls for proximity ligation in isolated mouse lung endothelial cells (iEC) from Nos3+/+ mice. Controls were performed by incubation with only one of the necessary antibodies or, one of the PLUS or MINUS PLA probes (mouse and rabbit secondary antibodies), which should inhibit rolling circle DNA synthesis. This allows for the detection of non-specific ligation or uncontrolled rolling circle DNA synthesis. Scale bar = 20 µm.
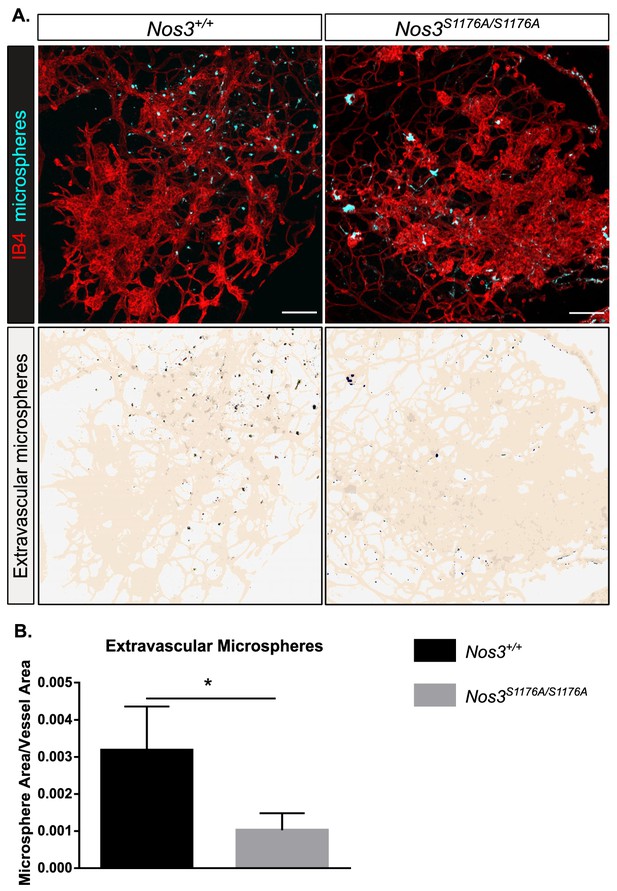
Suppressed microsphere leakage in Nos3S1176A/S1176A retinas.
(A) Representative images of similar sized tufts from Nos3+/+ and Nos3S1176A/S1176A mice showing isolectin B4 (IB4; red), and leakage of tail-vein injected FITC-conjugated 25 nm microspheres (cyan) around the tufts. Scale bar = 100 µM. Lower panels show leakage maps. Dots that do not overlap with vessels (beige) are considered extravascular. (B) Quantification of the average area of extravascular microspheres normalized to IB4 area. Mean ± S.E.M. n = 4 (Nos3+/+) and 4 (Nos3S1176A/S1176A) mice, 6–15 images per mouse. *p<0.05, t-test.
-
Figure 3—source data 1
Excel file containing numerical values collected from microsphere leakage experiments from OIR retinas in Nos3+/+ and Nos3S1176A/S1176A mice shown in Figure 3.
- https://cdn.elifesciences.org/articles/64944/elife-64944-fig3-data1-v2.xlsx
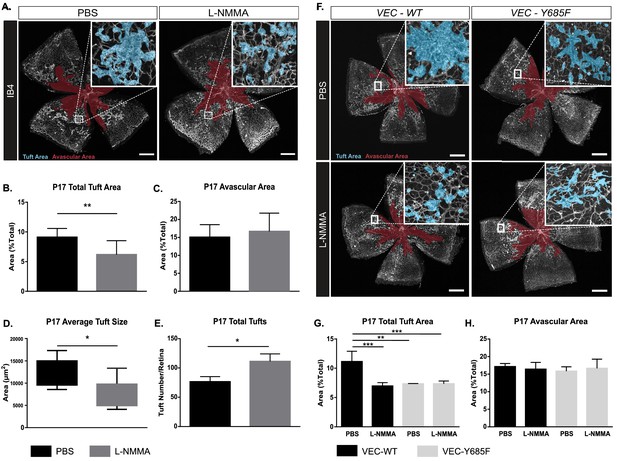
OIR-challenged mice treated with NO inhibitor L-NMMA.
(A) Representative images of whole mount retinas from PBS- and L-NMMA-treated (P12–P16) wild-type C57Bl/6 mice, collected on P17 after OIR-challenge, and stained with isolectin B4 (IB4). Avascular area as a result of OIR is marked in magenta and tufts in blue. Scale bar = 500 µm. (B, C) Tuft area and avascular area expressed as percentage of total vascular area at P17. (D, E) Tuft size in µm2 and total number of tufts/field of vision at P17. Mean ± S.E.M. n = 8 (PBS) and 9 (L-NMMA) treated mice. *, **p<0.05, 0.01; t-test. (F) Representative images of whole mount IB4-stained P17 retinas from OIR-challenged VEC-WT and VEC-Y685F mice injected with PBS or L-NMMA during P12-P16. Avascular area is marked in magenta and tufts in blue. Scale bar = 500 µm. (G) Tuft area normalized to total vascular area in PBS or L-NMMA-treated VEC-WT and VEC-Y685F retinas. (H) Avascular area normalized to total vascular area. Mean ± S.E.M. n = 8 (VEC-WT) and 8 (VEC-Y685F) mice. **, ***p<0.01, 0.001; two-way ANOVA, Sidak’s multiple comparison test.
-
Figure 4—source data 1
Excel file containing numerical values collected from OIR-induced tuft formation experiments in PBS- and L-NMMA-treated WT retinas shown in Figure 4 and Figure 4—figure supplement 1.
- https://cdn.elifesciences.org/articles/64944/elife-64944-fig4-data1-v2.xlsx
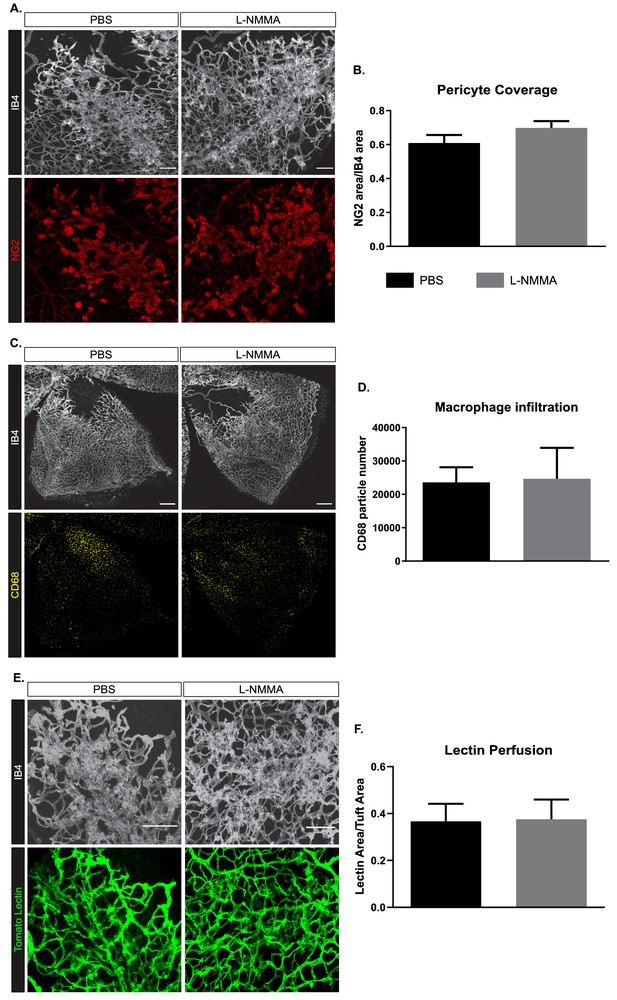
Pericytes, macrophage influx, and vascular perfusion in OIR-challenged WT retinas treated with PBS or L-NMMA.
(A) Representative images of tufts visualized by IB4 binding and co-stained for NG2 (red) to mark pericytes in PBS and L-NMMA-treated retinas collected at P17. Scale bar = 100 μm. (B) Pericyte coverage expressed as the NG2-positive area normalized to the IB4 area. Mean ± S.E.M. n = 4 (PBS) and 4 (L-NMMA) mice. t-test. (C) Representative images of one leaf of the retina (IB4) co-stained for CD68 (yellow) to mark circulatory/tissue macrophages in PBS and L-NMMA treated retinas collected at P17. Scale bar = 200 μm. (D) The number of CD68-positive macrophages in the entire retina represented by particle number, normalized to the retina area. Mean ± S.E.M. n = 4 (Nos3+/+) and 4 (Nos3S1176A/S1176A) mice. t-test. (E) Representative images tufts with circulating tomato lectin (green) co-stained with IB4 in PBS- and L-NMMA-treated retinas collected at P17. Scale bar = 100 μm. (F) Tuft perfusion, expressed as the tomato lectin area normalized to the IB4 area Mean ± S.E.M. n = 4 (PBS) and 4 (L-NMMA) mice. t-test.
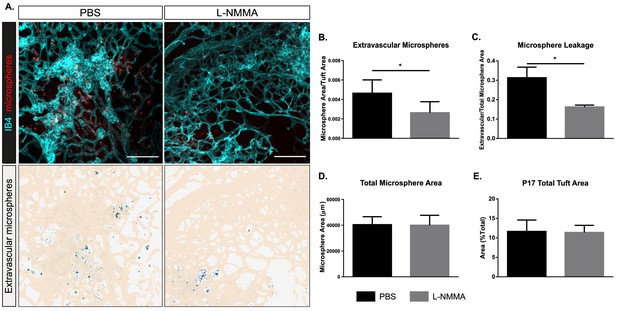
Decreased leakage from retinal vascular tufts after single-dose L-NMMA treatment.
(A) Representative images of tufts from Nos3+/+ (wild type) mice treated with PBS or L-NMMA (60 µg/g body weight) 24 hr before tail-vein injection of 25 nm microspheres. Retinas were stained using isolectin B4 (IB4; cyan), microspheres (red) appear in and around the tufts. Scale bar = 100 µM. Lower panels show leakage maps. Dots that do not overlap with vessels (beige) are considered extravascular. (B) Quantification of the average area of extravascular microspheres normalized to IB4 area. (C) Quantification of the average area of extravascular microspheres normalized to total microsphere area. (D) Quantification of average total microsphere area in PBS- or L-NMMA-treated wild-type mouse retinas. (E) Quantification of tuft area normalized to total vascular area. Mean ± S.E.M. n = 5 (PBS) and 5 (L-NMMA) treated mice. *p<0.05; t-test.
-
Figure 5—source data 1
Excel file containing numerical values collected from microsphere leakage analyses from OIR retinas in single-treated PBS and L-NMMA WT mice shown in Figure 5, Figure 5—figure supplement 1.
- https://cdn.elifesciences.org/articles/64944/elife-64944-fig5-data1-v2.xlsx
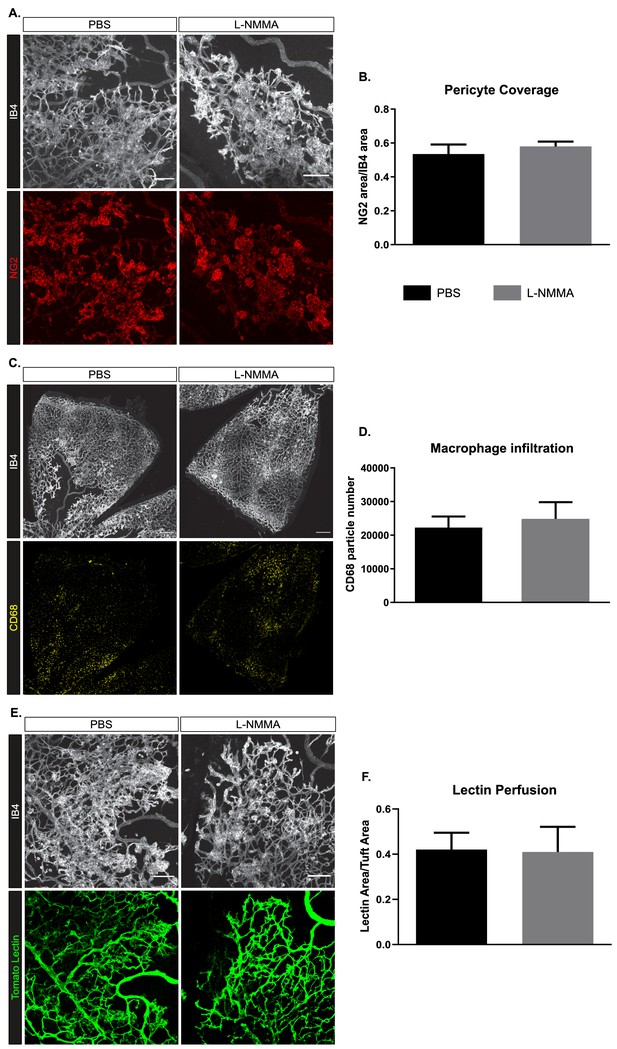
Pericytes, macrophage influx, and vascular perfusion in OIR-challenged WT retinas treated with a single injection of PBS or L-NMMA.
(A) Representative images of tufts visualized by IB4 binding and co-stained for NG2 (red) to mark pericytes in single-dose-treated PBS and L-NMMA retinas collected at P17. Scale bar = 100 μm. (B) Pericyte coverage expressed as the NG2-positive area normalized to the IB4 area. Mean ± S.E.M. n = 4 (PBS) and 4 (L-NMMA) mice. t-test. (C) Representative images of one leaf of the retina (IB4) co-stained for CD68 (yellow) to mark circulatory/tissue macrophages in single-dose-treated PBS and L-NMMA retinas collected at P17. Scale bar = 200 μm. (D) The number of CD68-positive macrophages in the entire retina represented by particle number, normalized to the retina area. Mean ± S.E.M. n = 4 (Nos3+/+) and 4 (Nos3S1176A/S1176A) mice. t-test. (E) Representative images tufts with circulating tomato lectin (green) co-stained with IB4 in single-dose-treated PBS and L-NMMA retinas collected at P17. Scale bar = 100 μm. (F) Tuft perfusion, expressed as the area of tomato lectin normalized to the IB4 area. Mean ± S.E.M. n = 4 (PBS) and 4 (L-NMMA) mice. t-test.
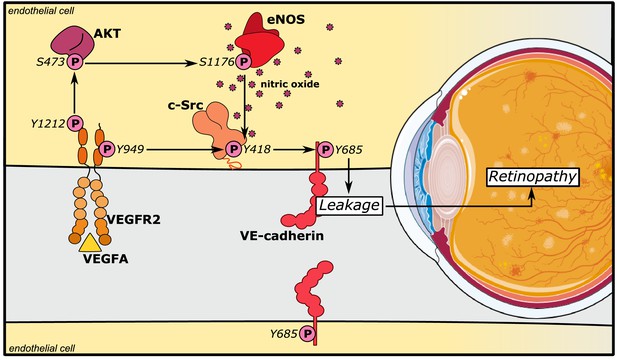
eNOS/NO modulates VE-cadherin Y685 phosphorylation via c-Src in a VEGFA/VEGFR2-dependent manner.
VEGFA through VEGFR2 and its phosphosite Y1212 induces a chain of consecutive reactions in endothelial cells: phosphorylation of AKT at S473 and eNOS at S1176. The VEGFR2 phosphosite Y949 mediates phosphorylation of c-Src at Y418 and of VE-cadherin at Y685. Combined, these activating phosphorylation reactions disrupt the vascular barrier by dissociating VE-cadherin’s homophilic interactions, resulting in macromolecular leakage. eNOS/NO regulates activation of c-Src to enhance VE-cadherin Y685 phosphorylation and internalization.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain; strain background (Mus musculus) | Nos3+/+ (C57BL/6J) | DOI:10.1016/j.bbrc.2012.12.110 | ||
| Strain; strain background (Mus musculus) | Nos3S1176A/S1176A (C57BL/6J) | DOI:10.1016/j.bbrc.2012.12.110 | ||
| Strain; strain background (Mus musculus) | Cdh5-WT (C57BL/6J) | DOI: 10.1038/ni.2824 | Referred to as VEC-WT throughout | |
| Strain; strain background (Mus musculus) | Cdh5-Y685F (C57BL/6J) | DOI: 10.1038/ni.2824 | Referred to as VEC-Y685F throughout | |
| Cell line (Homo-sapiens) | Human retinal microvascular endothelial | Cell Systems | Cat# ACBRI 181 | Primary cells HRMEC |
| Antibody | Anti-VE-cadherin pY685 (rabbit polyclonal) | DOI:10.1038/ncomms2199 | IF (1:50), WB (1:1000) | |
| Antibody | Anti-VE-cadherin (goat polyclonal) | R and D systems | Cat# AF1002 RRID:AB_2077789 | IF (1:200), WB (1:1000) |
| Antibody | Anti-eNOS (mouse monoclonal) | Abcam | Cat# ab76198 RRID:AB_1310183 | WB (1:1000) |
| Antibody | Anti-Src GD11 clone (mouse monoclonal) | Merck Millipore | Cat# 05–184 RRID:AB_2302631 | IF (1:200), WB (1:1000) |
| Antibody | Anti-c-Src pY418 (rabbit polyclonal) | Invitrogen | Cat# 44–660G RRID:AB_1500523 | IF (1:100), WB (1:1000) |
| Antibody | Anti-α-tubulin (mouse monoclonal) | Sigma-Aldrich | Cat# T9026 RRID:AB_477593 | WB (1:1000) |
| Antibody | Anti-eNOS pS1177 (mouse monoclonal) | BD Biosciences | Cat# 612392 RRID:AB_399750 | WB (1:1000) |
| Antibody | Anti-Akt (rabbit polyclonal) | Cell Signaling | Cat# 9272S RRID:AB_329827 | WB (1:1000) |
| Antibody | Anti-Akt pS473 (rabbit monoclonal) | Cell Signaling | Cat# 4058S RRID:AB_331168 | WB (1:1000) |
| Antibody | Anti-CD31 (rat monoclonal) | BD Biosciences | Cat# 553370 RRID:AB_394816 | IF (1:200) |
| Antibody | Anti-CD68 FA-11 clone (rat monoclonal) | BIO-RAD | Cat# MCA1957 RRID:AB_322219 | IF (1:300) |
| Antibody | Anti-NG2 (rabbit polyclonal) | Merck Millipore | Cat# AB5320 RRID:AB_91789 | IF (1:300) |
| Antibody | Anti-ERG (rabbit monoclonal) | Abcam | Cat# Ab92513 RRID:AB_2630401 | IF (1:200) |
| Antibody | Donkey anti-Rabbit IgG | ThermoFisher Scientific | Cat# A-31572 RRID:AB_162543 | IF (1:500) |
| Antibody | Donkey anti-Rat IgG | ThermoFisher Scientific | Cat# A-21208 RRID:AB_141709 | IF (1:500) |
| Antibody | Donkey anti-Goat IgG | ThermoFisher Scientific | Cat# A-11055 RRID:AB_2534102 | IF (1:500) |
| Antibody | Donkey anti-Mouse IgG, (H + L) HRP | ThermoFisher Scientific | Cat# A-16011 RRID:AB_2534685 | WB (1:10000) |
| Antibody | Donkey anti-Rabbit IgG, (H + L) HRP | ThermoFisher Scientific | Cat# A-16023 RRID:AB_2534697 | WB (1:10000) |
| Antibody | Donkey anti-Goat IgG, (H + L) HRP | ThermoFisher Scientific | Cat# A-15999 RRID:AB_2534673 | WB (1:10000) |
| Commercial assay, kit | Griess assay (nitrate/nitrite colorimetric assay kit) | Cayman Chemical | Cat# 780001 | |
| Commercial assay, kit | CD31 microbeads, mouse | Miltenyi Biotec | Cat# 130-097-418 | |
| Commercial assay, kit | RNeasy Mini Kit | QIAGEN | Cat# 74104 | |
| Chemical compound, drug | Nω-Methyl-L-arginine acetate salt (L-NMMA) | Sigma-Aldrich | Cat# M7033 | |
| Commercial assay, kit | Amersham ECL Prime Western Blotting Detection | GE Healthcare | Cat# RPN2232 | |
| Sequence-based reagent | Nos3 forward | ThermoFisher Scientific | PCR primers | AAGGTGATGAGGACTCTGTGGC |
| Sequence-based reagent | Nos3 reverse | ThermoFisher Scientific | PCR primers | GATATCTCGGGCAGCAGCTT |
| Sequence-based reagent | Nos2 forward | ThermoFisher Scientific | PCR primers | GGTGAAGGGACTGAGCTGTTA |
| Sequence-based reagent | Nos2 reverse | ThermoFisher Scientific | PCR primers | TGAGAACAGCACAAGGGGTTT |
| Sequence-based reagent | Vegfa forward | ThermoFisher Scientific | PCR primers | GCACATAGAGAGAATGAGCTTCC |
| Sequence-based reagent | Vegfa reverse | ThermoFisher Scientific | PCR primers | CTCCGCTCTGAACAAGGCT |
| Sequence-based reagent | Tbp forward | ThermoFisher Scientific | PCR primers | CCTTGTACCCTTCACCAATGAC |
| Sequence-based reagent | Tbp reverse | ThermoFisher Scientific | PCR primers | ACAGCCAAGATTCACGGTAGA |
| Sequence-based reagent | Ubc forward | ThermoFisher Scientific | PCR primers | CCCACACAAAGCCCCTCAAT |
| Sequence-based reagent | Ubc reverse | ThermoFisher Scientific | PCR primers | AAGATCTGCATCGTCTCTCTCAC |
| Peptide, recombinant protein | VEGFA, recombinant, mouse | Peprotech | Cat# 450–32 | |
| Commercial assay, kit | Duolink In Situ PLA Probe anti-Rabbit MINUS | Sigma-Aldrich | Cat# DUO92005 RID:AB_2810942 | |
| Commercial assay, kit | Duolink In Situ PLA Probe anti-Mouse PLUS | Sigma-Aldrich | Cat# DUO92001 RRID:AB_2810939 | |
| Commercial assay, kit | Duolink In Situ Detection Reagent (Orange) | Sigma-Aldrich | Cat# DUO92007 | |
| Other | SuperScript III Reverse Transcriptase | Invitrogen | Cat# 18080093 | |
| Other | DAF-FM diacetate (DA) | Sigma-Aldrich | Cat# D1946-1MG | |
| Other | Lycopersicon Esculentum (Tomato) Lectin (LEL, TL), Fluorescein | Vector Laboratories | Cat# FL-1171–1 | |
| Other | Fluoro-Max Dyed Green Aqueous Fluorescent Particles | ThermoFisher Scientific | Cat# G25 | |
| Other | Hoechst 33342 | ThermoFisher Scientific | Cat# H3570 | IF (1:1000) |
| Other | Alexa Fluor 488-Isolectin B4 | Sigma-Aldrich | Cat# I21411 RRID:AB_2314662 | IF (1:500) |
| Other | Alexa Fluor 647-Isolectin B4 | Sigma-Aldrich | Cat# I32450 RRID:SCR_014365 | IF (1:500) |
| Software | ImageJ | NIH, Bethesda, MD | RRID:SCR_003070 | |
| Software | GraphPad Prism | GraphPad | RRID:SCR_002798 |
Additional files
-
Supplementary file 1
Body weights for OIR experiments.
- https://cdn.elifesciences.org/articles/64944/elife-64944-supp1-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/64944/elife-64944-transrepform-v2.pdf





