BUB-1 targets PP2A:B56 to regulate chromosome congression during meiosis I in C. elegans oocytes
Figures
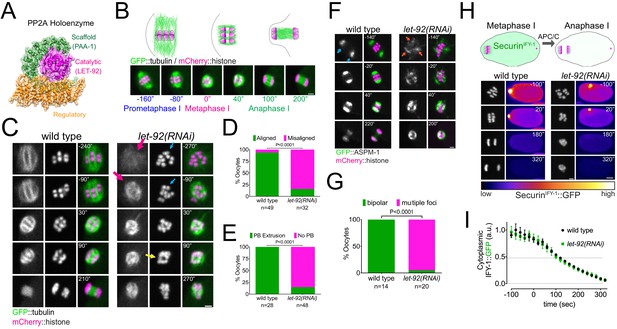
Protein Phosphatase 2A (PP2A) is essential for meiosis I in C. elegans oocytes.
(A) Schematic of the PP2A heterotrimer highlighting the single catalytic and scaffold subunits in C. elegans (LET-92 and PAA-1, respectively). The schematic was generated from the PDB structure 2npp (Xu et al., 2006). (B) Schematic of the timescales used throughout the paper. Metaphase I was defined as time zero and chosen as the frame prior to the one where chromosome separation was detected. (C) Microtubule and chromosome dynamics were followed in wild-type and let-92(RNAi) oocytes expressing GFP::tubulin and mCherry::histone. Magenta arrows point to defective spindle structure; the cyan arrow points to misaligned chromosomes; the yellow arrow shows an apparent separation between chromosome masses. Inset numbers represent the time relative to metaphase I in seconds. Scale bar, 2 µm. See Figure 1—video 1. (D) The number of oocytes with misaligned chromosomes in metaphase I was compared between wild-type and let-92(RNAi) oocytes and the percentage is represented (p<0.0001, Fisher’s exact test). The number of oocytes analysed (n) is shown. (E) The number of oocytes with an extruded polar body (PB) after meiosis I in wild-type and let-92(RNAi) oocytes was analysed and the percentage is represented (p<0.0001, Fisher’s exact test). The number of oocytes analysed (n) is shown. (F) ASPM-1 and chromosome dynamics were followed in wild-type and let-92 (RNAi) oocytes expressing GFP::ASPM-1 and mCherry::histone. Inset numbers represent the time relative to metaphase I in seconds. Cyan arrows point to spindle poles, whereas orange arrows highlight the unfocused ASPM-1 cumuli. Scale bar, 2 µm. See Figure 1—video 2. (G) The number of oocytes with defective spindle at prometaphase I in wild-type and let-92(RNAi) oocytes was analysed, and the percentage is represented (p<0.0001, Fisher’s exact test). The number of oocytes analysed (n) is shown. (H) SecurinIFY-1 degradation was used as a proxy for anaphase progression and followed in wild-type and let-92(RNAi) oocytes. Greyscale images of the chromosomes are shown as well as whole oocyte images using the ‘fire’ LUT. The intensity scale is shown in the bottom. Inset numbers represent the time relative to metaphase I in seconds. Scale bar, 2 µm. See Figure 1—video 3. (I) Cytoplasmic SecurinIFY-1 levels were measured throughout meiosis I, and the mean ± s.e.m is shown in the graph.
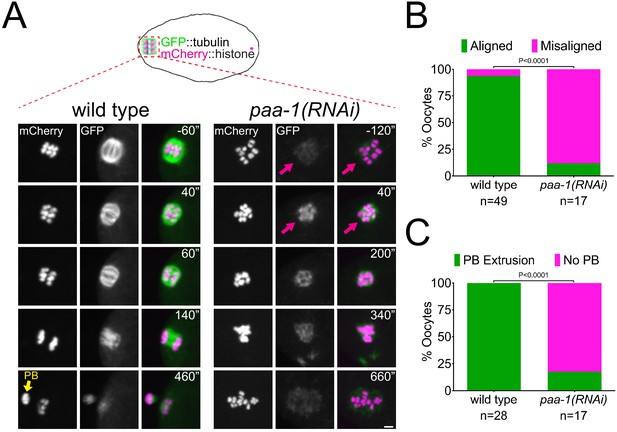
PP2A is essential for Meiosis I in C. elegans oocytes.
(A) Microtubule and chromosome dynamics were followed in wild-type and paa-1(RNAi) oocytes expressing GFP::tubulin and mCherry::histone. Yellow arrow shows polar body (PB) and magenta arrows point to the lack of a bipolar spindle in paa-1(RNAi) oocytes. Inset numbers represent the time relative to metaphase I in seconds. Scale bar, 2 µm. (B) The number of oocytes with misaligned chromosomes at metaphase I in wild-type and paa-1(RNAi) oocytes was analysed and the percentage is represented (p<0.0001, Fisher’s exact test). (C) The number of oocytes with an extruded polar body (PB) after meiosis I in wild-type and paa-1(RNAi) oocytes was analysed and the percentage is represented (p<0.0001, Fisher’s exact test).
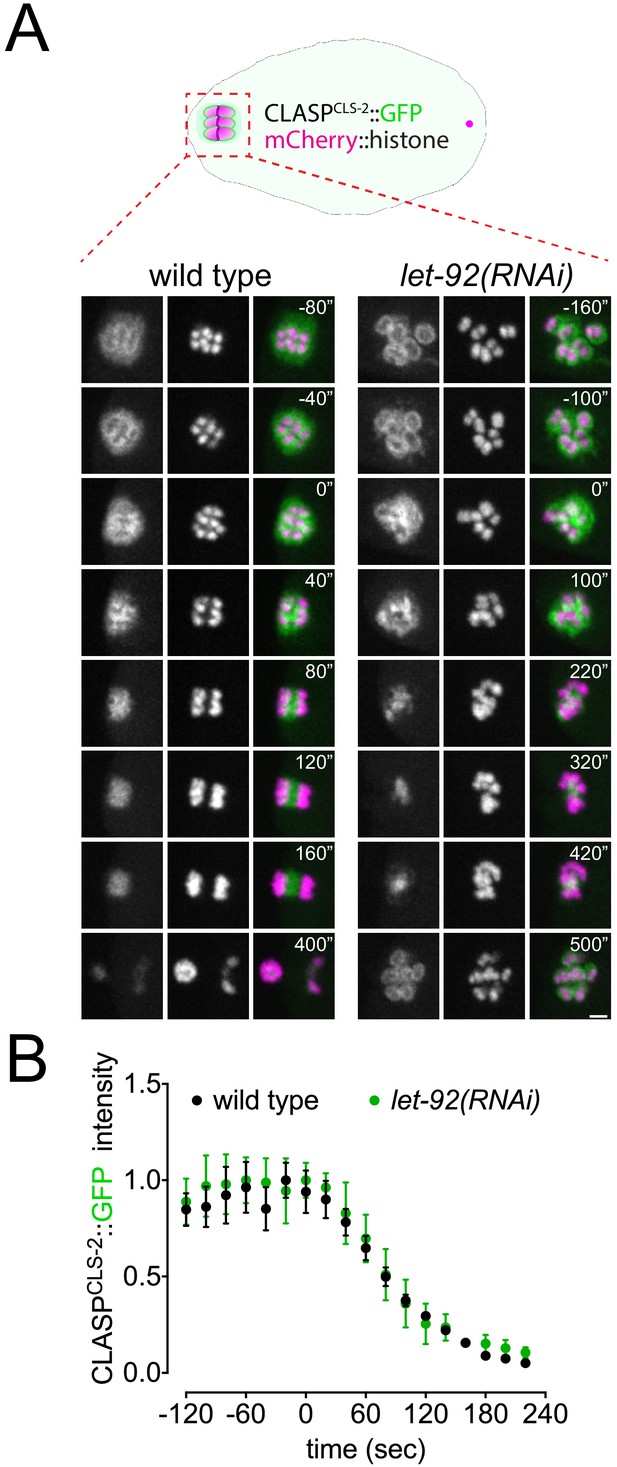
CLASPCLS-2 intensity and localisation is not affected after let-92 depletion.
(A) CLASPCLS-2 and chromosome dynamics were followed in wild-type and let-92(RNAi) oocytes expressing CLASPCLS-2::GFP and mCherry::histone. Inset numbers represent the time relative to metaphase I in seconds. Scale bar, 2 µm. See Figure 1—video 4. (B) CLASPCLS-2::GFP levels were measured throughout meiosis I in wild-type and let-92(RNAi), and the mean ± s.e.m is shown in the graph.
Wild type and let-92(RNAi) oocytes expressing GFP::tubulin and mCherry::histone were dissected and recorded.
Labels indicate the channels, time (relative to metaphase I), and scale bar. First slice indicates additionally indicates the intensity scale, the dimensiones of each channel image, and any additional filtering applied to the images.
Wild type and let-92(RNAi) oocytes expressing GFP::ASPM-1 and mCherry::histone were dissected and recorded.
Labels indicate the channels, time (relative to metaphase I), and scale bar. First slice indicates additionally indicates the intensity scale, the dimensiones of each channel image, and any additional filtering applied to the images.
Wild type and let-92(RNAi) oocytes expressing SecurinIFY-1::GFP and mCherry::histone were dissected and recorded.
Labels indicate the channels, time (relative to metaphase I), and scale bar. First slice indicates additionally indicates the intensity scale, the dimensiones of each channel image, and any additional filtering applied to the images.
Wild type and let-92(RNAi) oocytes expressing CLASPCLS-2::GFP and mCherry::histone were dissected and recorded.
Labels indicate the channels, time (relative to metaphase I), and scale bar. First slice indicates additionally indicates the intensity scale, the dimensiones of each channel image, and any additional filtering applied to the images.
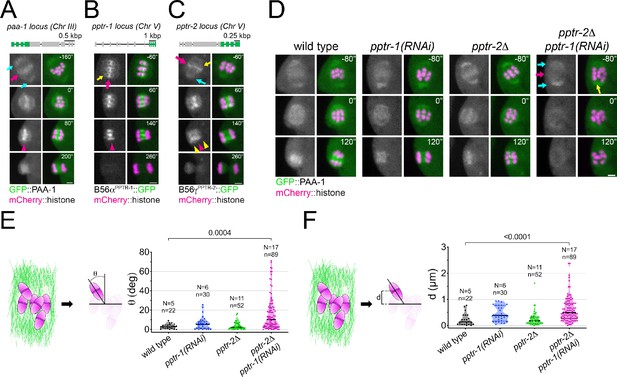
Caenorhabditis elegans B56 regulatory subunits PPTR-1 and PPTR-2 are required for normal meiosis I.
(A) Top: Schematic of the paa-1 gene structure and its tagging with gfp. Bottom: The PP2A scaffold subunit GFP::PAA-1 was followed throughout meiosis I in a dissected oocyte. Magenta arrows point to the midbivalent; cyan arrows point to spindle poles; and magenta arrowhead highlights the central-spindle localisation. Inset numbers represent the time relative to metaphase I in seconds. Scale bar, 2 µm. See Figure 2—video 1. (B) Top: Schematic of the pptr-1 gene structure and its tagging with gfp. Bottom: PPTR-1 and chromosome dynamics were followed in oocytes expressing PPTR-1::GFP and mCherry::histone. The magenta arrow points to the midbivalent and the yellow arrow points to the kinetochore. Magenta arrowhead highlights the central-spindle localisation. Inset numbers represent the time relative to metaphase I in seconds. Scale bar, 2 µm. See Figure 2—video 2. (C) Top: Schematic of the pptr-2 gene structure and its tagging with gfp. Bottom: PPTR-2 and chromosome dynamics were followed in oocytes expressing PPTR-2::GFP and mCherry::histone. The magenta arrow points to the midbivalent, the yellow arrow points to the kinetochore, and the cyan arrow points to the spindle pole. Inset numbers represent the time relative to metaphase I in seconds. Scale bar, 2 µm. Magenta arrowhead highlights the central-spindle localisation, and yellow arrowhead highlights the chromosome-associated signal. See Figure 2 —video 3. (D) PAA-1 and chromosome dynamics were followed in wild-type, pptr-1(RNAi), pptr-2Δ, and pptr-2Δ+pptr-1(RNAi) oocytes expressing GFP::PAA-1 and mCherry::histone. Magenta arrows point to the absence of GFP signal on prometaphase chromosomes, and the cyan arrows point to spindle poles. The yellow arrows highlight one misaligned bivalent. Inset numbers represent the time relative to metaphase I in seconds. Scale bar, 2 µm. See Figure 2 —video 4. (E) On the left, the schematic depicts how angles were measured relative to the spindle pole-to-pole axis. On the right, the angle of bivalents at 80 s before metaphase I in wild-type, pptr-1(RNAi), pptr-2Δ, and pptr-2Δ+pptr-1(RNAi) oocytes. Violin plot includes each data point (chromosome), the median (straight black line), and the interquartile range (dashed black lines). N represents number of oocytes and n number of bivalents measured. p values shown in the figure were obtained using a Kruskal–Wallis test. (F) On the left, the schematic depicts how distances were measured relative to the centre of the spindle. On the right, the distance of bivalents 80 s before metaphase I was measured in wild-type, pptr-1(RNAi), pptr-2Δ, and pptr-2Δ+pptr-1(RNAi) oocytes. Violin plot includes each data point (chromosome), the median (straight black line), and the interquartile range (dashed black lines). N represents number of oocytes and n number of bivalents measured. p values shown in the figure were obtained using a Kruskal–Wallis test.
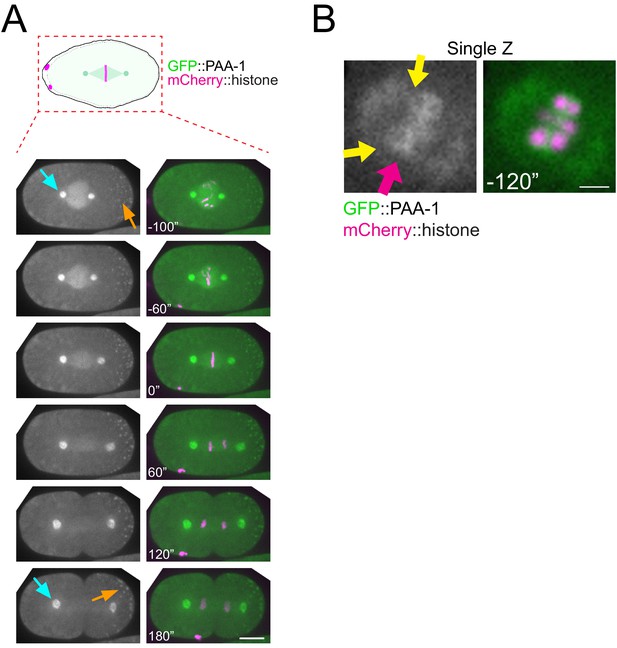
Localisation of GFP::PAA-1 during mitosis.
(A) PAA-1 localisation was followed during the first mitotic division using endogenously tagged GFP::PAA-1. Cyan arrows point to centrosomes, and orange arrows point to P-bodies. Inset numbers represent the time relative to metaphase in seconds. Scale bar, 10 µm. (B) A single Z-plane from one time point (−120’) from the same video as in Figure 2A is shown to highlight that the chromosomal PAA-1 signal is a combination of midbivalent (magenta arrow) and kinetochore (yellow arrows).
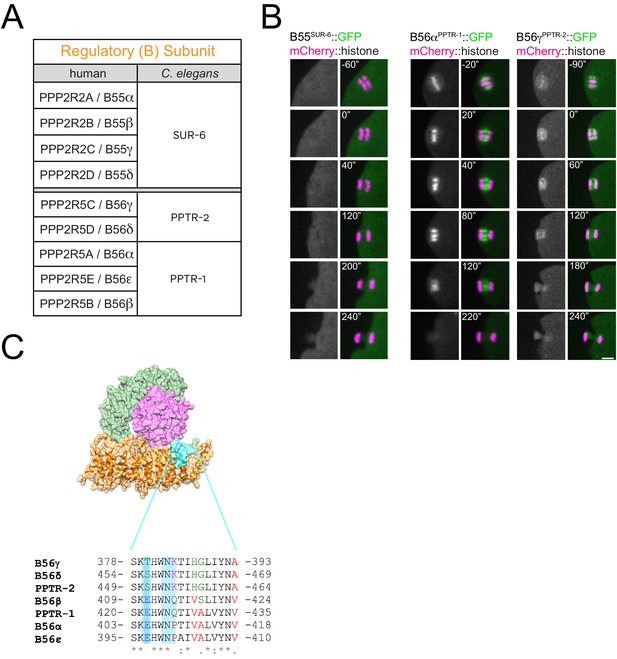
Localisation of C.
elegans B55 and B56 subunits. (A) Chart showing the C. elegans orthologues of the human B55 and B56 regulatory B subunits. The chart is based on the alignment provided in Table 1 and Supplementary file 1. (B) The respective B subunit was endogenously tagged with GFP and their localisation was followed by imaging of dissected oocytes. Scale bar, 2 µm. (C) Alignment of the human and C. elegans B56 subunits in the C-terminal stretch shown to be important for kinetochore vs centromere localisation of B56 in human cells during mitosis (Vallardi et al., 2019).
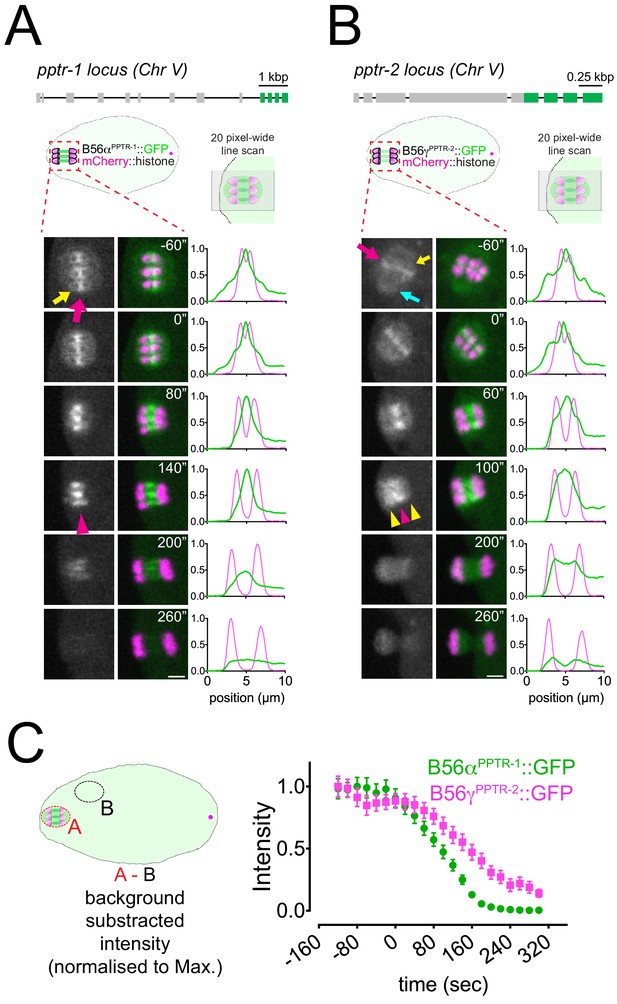
Localisation of C.
elegans B55 and B56 subunits. (A) Top, Schematic of the pptr-1 gene structure and its tagging with gfp. Bottom, PPTR-1 and chromosome dynamics were followed in oocytes expressing PPTR-1::GFP and mCherry::histone. The magenta arrow points to the midbivalent, and the yellow arrow points to the kinetochore. Magenta arrowhead highlights the central-spindle localisation. Inset numbers represent the time relative to metaphase I in seconds. Scale bar, 2 µm. Representative, spindle-wide (20 pixels) line profiles are shown on the right of each time point. See Figure 2—video 2. (B) Top, Schematic of the pptr-2 gene structure and its tagging with gfp. Bottom, PPTR-2 and chromosome dynamics were followed in oocytes expressing PPTR-2::GFP and mCherry::histone. The magenta arrow points to the midbivalent, the yellow arrow points to the kinetochore, and the cyan arrow points to the spindle pole. Inset numbers represent the time relative to metaphase I in seconds. Scale bar, 2 µm. Magenta arrowhead highlights the central-spindle localisation, and yellow arrowhead highlights the chromosome-associated signal. Representative, spindle-wide (20 pixels) line profiles are shown on the right of each time point. See Figure 2—video 3. (C) PPTR-1::GFP and PPTR-2::GFP levels were measured throughout meiosis I, and the mean ± s.e.m is shown in the graph.
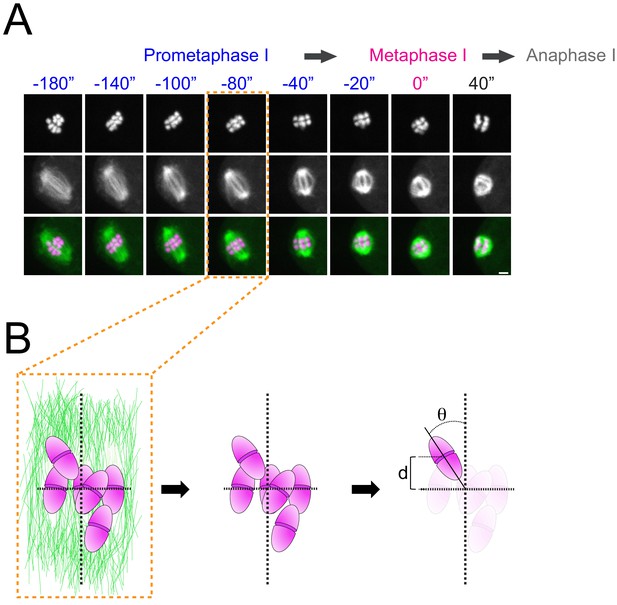
Schematics of how congression and alignment were anotated in this study.
(A) Microtubules and chromosomes of a wild-type oocyte are shown in the panel. For the congression/alignment analysis during prometaphase I, we chose to analyse 80 s before metaphase I, as chromosome are readily aligned in most wild-type oocytes. (B) Schematic of a prometaphase I spindle indicating how the angle and the distance of each bivalent was measured.
An oocyte expressing GFP::PAA-1 and mCherry::histone was dissected and recorded.
Labels indicate the channels, time (relative to metaphase I), and scale bar. First slice indicates additionally indicates the intensity scale, the dimensiones of each channel image, and any additional filtering applied to the images.
An oocyte expressing PPTR-1::GFP and mCherry::histone was dissected and recorded.
Labels indicate the channels, time (relative to metaphase I), and scale bar. First slice indicates additionally indicates the intensity scale, the dimensiones of each channel image, and any additional filtering applied to the images.
An oocyte expressing PPTR-2::GFP and mCherry::histone was dissected and recorded.
Labels indicate the channels, time (relative to metaphase I), and scale bar. First slice indicates additionally indicates the intensity scale, the dimensiones of each channel image, and any additional filtering applied to the images.
Wild type, pptr-1(RNAi), pptr-2Δ, and pptr-2Δ+pptr-1(RNAi) oocytes expressing GFP::PAA-1 and mCherry::histone were dissected and recorded.
Labels indicate the channels, time (relative to metaphase I), and scale bar. First slice indicates additionally indicates the intensity scale, the dimensiones of each channel image, and any additional filtering applied to the images.
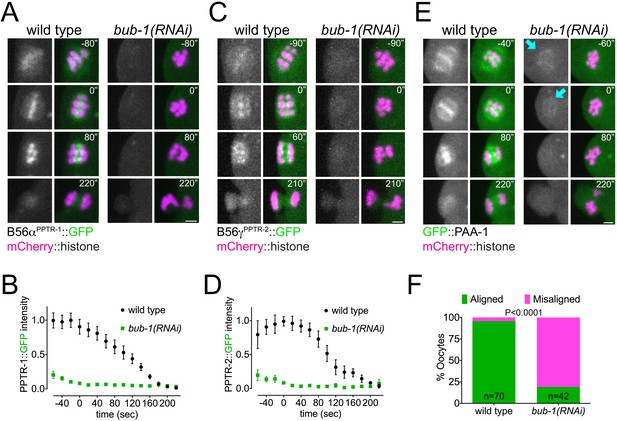
BUB-1 recruits B56αPPTR-1 and B56γPPTR-2 during oocyte meiosis.
(A) B56αPPTR-1 and chromosome dynamics were followed in wild-type and bub-1(RNAi) oocytes expressing PPTR-1::GFP and mCherry::histone. Scale bar, 2 µm. See Figure 3—video 1. (B) PPTR-1::GFP levels were measured in wild-type and bub-1(RNAi) oocytes throughout meiosis I and the mean ± s.e.m is shown in the graph. (C) B56γPPTR-2 and chromosome dynamics were followed in wild-type and bub-1(RNAi) oocytes expressing PPTR-2::GFP and mCherry::histone. Scale bar, 2 µm. See Figure 3—video 2. (D) PPTR-2::GFP levels were measured in wild-type and bub-1(RNAi) oocytes throughout meiosis I and the mean ± s.e.m is shown in the graph. (E) Scaffold subunit PAA-1 and chromosome dynamics were followed in wild-type and bub-1(RNAi) oocytes expressing GFP::PAA-1 and mCherry::histone. Cyan arrows highlight the GFP::PAA-1 remaining in bub-1(RNAi) oocytes. Scale bar, 2 µm. See Figure 3—video 3. (F) The number of oocytes with misaligned chromosomes at metaphase I in wild-type and bub-1(RNAi) oocytes was analysed, and the percentage is represented (p<0.0001, Fisher’s exact test).
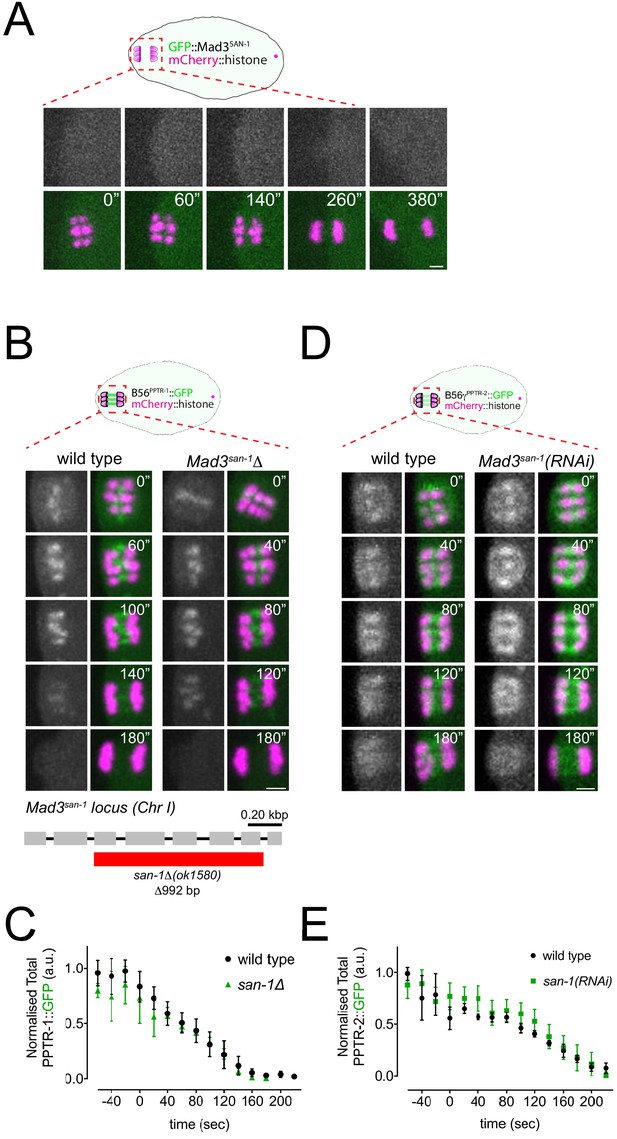
Mad3SAN-1 does not play a major role in B56αPPTR-1 and B56γPPTR-2 targeting.
(A) FP::Mad3SAN-1 was imaged during oocyte meiosis along with mCherry::histone to follow chromosomes. Time insets are relative to metaphase I (‘t=0’). Scale bar, 2 µm. (B) B56αPPTR-1 and chromosome dynamics were followed in wild type and in Mad3SAN-1Δoocytes expressing PPTR-1::GFP and mCherry::histone. Scale bar, 2 µm. The bottom panel shows a schematic of the san-1Δ allele. (C) PPTR-1::GFP levels were measured in wild type and Mad3SAN-1Δoocytes throughout meiosis I and the mean ± s.e.m is shown in the graph. (D) B56γPPTR-2 and chromosome dynamics were followed in wild type and in Mad3san-1(RNAi) oocytes expressing PPTR-2::GFP and mCherry::histone. Scale bar, 2 µm. (E) PPTR-2::GFP levels were measured in wild type and Mad3san-1(RNAi)oocytes throughout meiosis I and the mean ± s.e.m is shown in the graph.
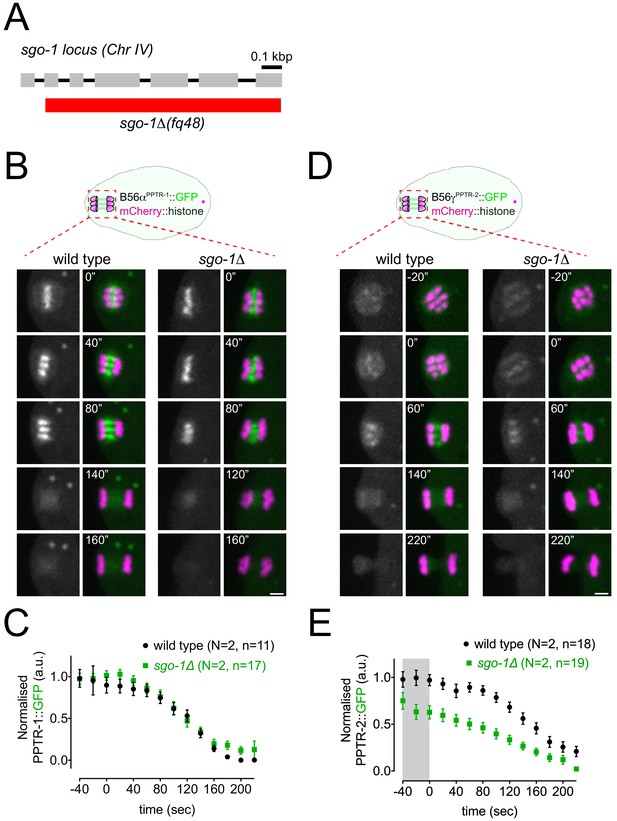
ShugoshinSGO-1 does not play a major role in B56αPPTR-1 and B56γPPTR-2 targeting.
(A) Schematic of the sgo-1Δ allele. (B) B56αPPTR-1 and chromosome dynamics were followed in wild-type and in Shugoshinsgo-1 oocytes expressing PPTR-1::GFP and mCherry::histone. Scale bar, 2 µm. (C) PPTR-1::GFP levels were measured in wild-type and in Shugoshinsgo-1 oocytes throughout meiosis I and the mean ± s.e.m is shown in the graph. N represents number of experiments, and n represents number of oocytes quantified. (D) B56γPPTR-2 and chromosome dynamics were followed in wild-type and Shugoshinsgo-1 oocytes expressing PPTR-2::GFP and mCherry::histone. Scale bar, 2 µm. (E) PPTR-1::GFP levels were measured in wild-type and in Shugoshinsgo-1 oocytes throughout meiosis I, and the mean ± s.e.m is shown in the graph. N represents number of experiments, and n represents number of oocytes quantified.
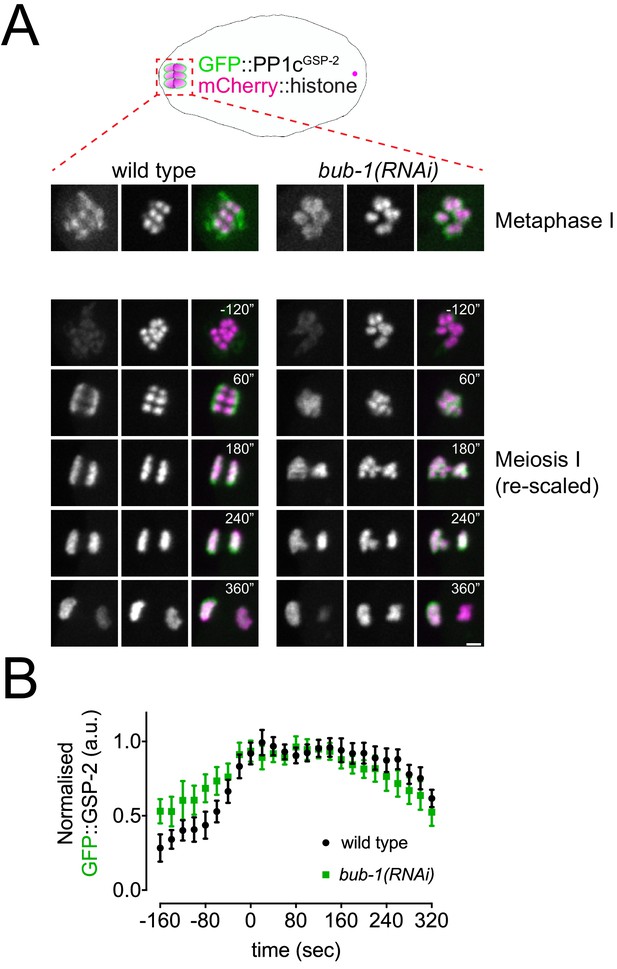
PP1 catalytic subunit localisation is not regulated by BUB-1.
(A) GFP::PP1cGSP-2 and chromosome dynamics were followed in wild-type and bub-1(RNAi) oocytes. Since the GFP signal increases during anaphase, one scale was chosen to show kinetochore localisation during metaphase I (top) and another for the composite panel (bottom). Scale bar, 2 µm. See Figure 3—video 4. (B) GFP::PP1cGSP-2 levels were measured in wild-type and bub-1(RNAi) oocytes throughout meiosis I, and the mean ± s.e.m is shown in the graph.
Wild type and bub-1(RNAi) oocytes expressing PPTR-1::GFP and mCherry::histone were dissected and recorded.
Labels indicate the channels, time (relative to metaphase I), and scale bar. First slice indicates additionally indicates the intensity scale, the dimensiones of each channel image, and any additional filtering applied to the images.
Wild type and bub-1(RNAi) oocytes expressing PPTR-2::GFP and mCherry::histone were dissected and recorded.
Labels indicate the channels, time (relative to metaphase I), and scale bar. First slice indicates additionally indicates the intensity scale, the dimensiones of each channel image, and any additional filtering applied to the images.
Wild type and bub-1(RNAi) oocytes expressing GFP::PAA-1 and mCherry::histone were dissected and recorded.
Labels indicate the channels, time (relative to metaphase I), and scale bar. First slice indicates additionally indicates the intensity scale, the dimensiones of each channel image, and any additional filtering applied to the images.
Wild type and bub-1(RNAi) oocytes expressing GFP::PP1cGSP-2 and mCherry::histone were dissected and recorded.
Labels indicate the channels, time (relative to metaphase I), and scale bar. First slice indicates additionally indicates the intensity scale, the dimensiones of each channel image, and any additional filtering applied to the images.
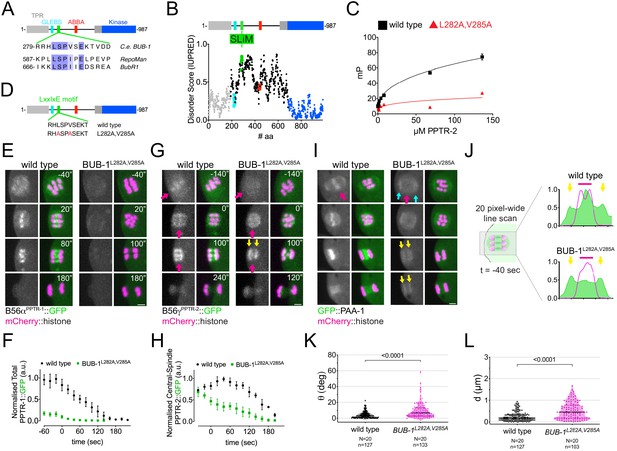
B56αPPTR-1 and B56γPPTR-2 are targeted through a LxxIxE motif in BUB-1.
(A) C. elegans BUB-1 LxxIxE SLiM is shown aligned with BubR1 and Repoman short linear motifs (SLiMs). The only change within the LxxIxE sequence itself is the presence of a valine instead of isoleucine, which still fits within the consensus (Wang et al., 2016). The alignment was performed using Clustal Omega and visualised with Jalview (Waterhouse et al., 2009). (B) Disorder prediction of full-length BUB-1 was done using IUPRED2A (Erdős and Dosztányi, 2020). All prevuously characterised BUB-1 domains fall within ordered regions < 0.5. The putative SLiM is in a disordered region (IUPRED score ~0.8). (C) PPTR-2 interaction with a LxxIxE motif-containing synthetic peptide was assessed using fluorescence polarisation. Increasing amounts of purified recombinant PPTR-2 were incubated with FITC-labelled wild-type or L282A,V285A mutant peptide. The graph was taken from a representative experiment and shows the mean ± s.d. of technical triplicates. (D) Schematic showing the LxxIxE motif in BUB-1 and the BUB-1L282A,V285A mutant. (E) B56αPPTR-1 and chromosome dynamics were followed in wild-type and in BUB-1L282A,V285A oocytes expressing PPTR-1::GFP and mCherry::histone. Scale bar, 2 µm. Inset numbers represent the time relative to metaphase I in seconds. See Figure 4—video 1. (F) PPTR-1::GFP levels were measured in wild-type and BUB-1L282A,V285A oocytes throughout meiosis I and the mean ± s.e.m is shown in the graph. (G) B56γPPTR-2 and chromosome dynamics were followed in wild-type and in BUB-1L282A,V285A oocytes expressing PPTR-2::GFP and mCherry::histone. Magenta arrows point towards prometaphase chromosomes and anaphase central spindle, whereas yellow arrows point towards the chromosome-associated anaphase signal. Scale bar, 2 µm. Inset numbers represent the time relative to metaphase I in seconds. See Figure 4—video 2. (H) Midbivalent/central spindle PPTR-2::GFP levels were measured in wild-type and BUB-1L282A,V285A oocytes throughout meiosis I, and the mean ± s.e.m is shown in the graph. (I) GFP::PAA-1 is present on chromosomes in the BUB-1L282A,V285A mutant. PAA-1 and chromosome dynamics were followed in wild-type and BUB-1L282A,V285A oocytes expressing GFP::PAA-1 and mCherry::histone. Magenta arrows point towards prometaphase chromosomes and anaphase central spindle, whereas yellow arrows point towards the chromosome-associated anaphase signal. Cyan arrows point towards spindle poles. Scale bar, 2 µm. Inset numbers represent the time relative to metaphase I in seconds. See Figure 4—video 3. (J) Representative, spindle-wide (20 pixels) line profiles are shown for wild-type (top) and BUB-1L282A,V285A mutant (bottom) measured in prometaphase I (40 s before metaphase I). Green arrows point to the PAA-1 pole signal and blue line to the chromosome associated population. (K) The angle of bivalents 80 s before segregation in meiosis I relative to the average angle of the spindle was measured in wild-type and in BUB-1L282A,V285A oocytes. Violin plot includes each data point (chromosome), the median (straight black line), and the interquartile range (dashed black lines). N represents number of oocytes and n number of bivalents measured. p value shown in the figure was obtained using a Mann–Whitney test. (L) Distance of bivalents 80 s before segregation in meiosis I relative to the centre of the spindle was measured in wild-type and BUB-1L282A,V285A oocytes. Violin plot includes each data point (chromosome), the median (straight black line), and the interquartile range (dashed black lines). N represents number of oocytes and n number of bivalents measured. p value shown in the figure was obtained using a Mann–Whitney test.
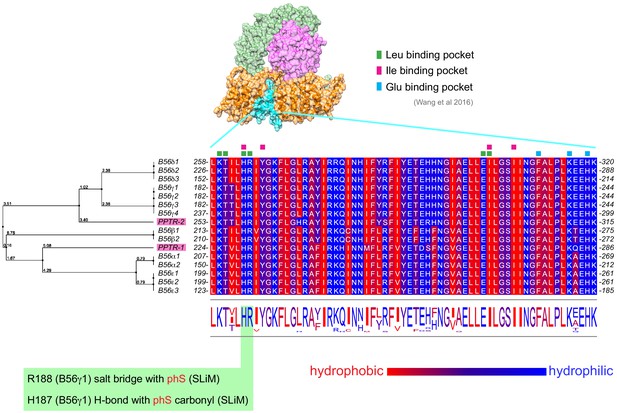
Alignment of the B56 subunits LxxIxE motif binding pocket.
C. elegans and human B56 subunits LxxIxE motif binding pocket were aligned using Clustal Omega and Jalview. The scale from blue to red represents increasing hydrophobicity. Key residues as reported in Wang et al., 2016 are highlighted. Additionally, residues making contact with the phospho serine are highlighted with green background. The tree on the left was calculated from the distance matrix generated from sequence pairwise scores and confirms the relationship between C. elegans and human B56s.
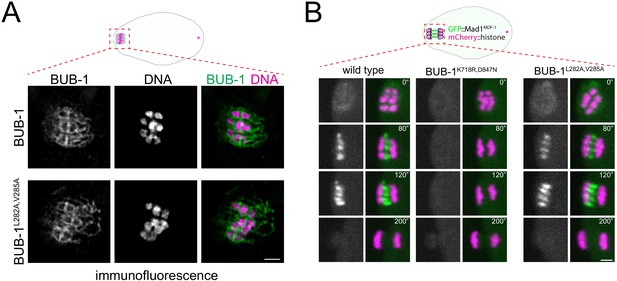
Mad1MDF-1 localisation is not affected by a LxxIxE motif mutation in BUB-1.
(A) BUB-1 localisation was analysed in fixed wild-type and BUB-1L282A,V285A mutant oocytes by immunofluorescence using BUB-1 specific antibodies. Scale bar, 2 µm. (B) GFP::Mad1MDF-1 was imaged during oocyte meiosis along with mCherry::histone to follow chromosomes. Time insets are relative to metaphase I (t = 0’). The BUB-1K718R,D847N mutation completely abolishes Mad1MDF-1 localisation. On the other hand, GFP::Mad1MDF-1 remained unaltered in the the BUB-1L282A,V285A mutant. Scale bar, 2 µm.
Wild type and BUB-1L282A,V285A oocytes expressing PPTR-1::GFP and mCherry::histone were dissected and recorded.
Labels indicate the channels, time (relative to metaphase I), and scale bar. First slice indicates additionally indicates the intensity scale, the dimensiones of each channel image, and any additional filtering applied to the images.
Wild type and BUB-1L282A,V285A oocytes expressing PPTR-2::GFP and mCherry::histone were dissected and recorded.
Labels indicate the channels, time (relative to metaphase I), and scale bar. First slice indicates additionally indicates the intensity scale, the dimensiones of each channel image, and any additional filtering applied to the images.
Wild type and BUB-1L282A,V285A oocytes expressing GFP::PAA-1 and mCherry::histone were dissected and recorded.
Labels indicate the channels, time (relative to metaphase I), and scale bar. First slice indicates additionally indicates the intensity scale, the dimensiones of each channel image, and any additional filtering applied to the images.
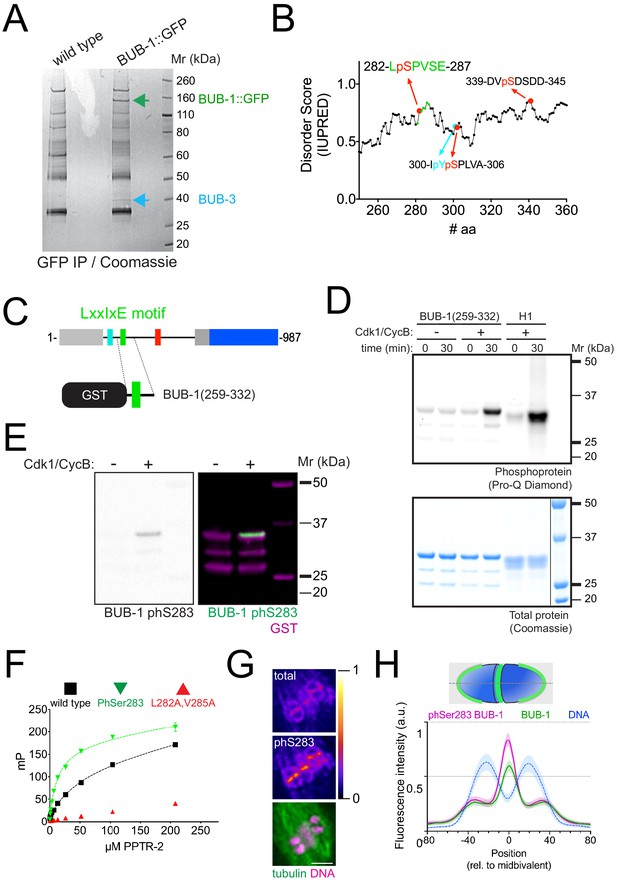
Phosphorylation of Ser 283 within the LxxIxE motif regulates B56 subunit binding.
(A) BUB-1::GFP was immunoprecipitated and the eluted proteins were run on sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) followed by Coomassie staining. The position of BUB-1::GFP and BUB-3 is highlighted with green and blue arrows, respectively. The band corresponding to BUB-1::GFP was further analysed by mass spectrometry to look for phospho-modified peptides. (B) Serine 283 is phosphorylated in vivo. In addition, three other phosphorylation sites were identified within this disordered region. (C) A fragment of BUB-1 (259–332) containing the LxxIxE motif (282–287) was expressed in bacteria fused to a GST tag. (D) GST-BUB-1 (259–332) was phosphorylated in vitro using Cdk1/CyclinB (‘Cdk1/CycB’). Histone H1 was used as a positive control. Reactions were run on 4–12% SDS–PAGE and subject to phosphoprotein staining (top) followed by total protein staining (bottom). (E) GST-BUB-1 (259–332) was phosphorylated in vitro using Cdk1/CyclinB (‘Cdk1/CycB’) and subject to western blotting using a specific antibody against phosphorylated serine 283. Anti-GST served as a loading control for the substrate. (F) PPTR-2 interaction with a LxxIxE motif-containing synthetic peptide was assessed using fluorescence polarisation. Increasing amounts of purified recombinant PPTR-2 were incubated with FITC-labelled wild-type, serine 283 phosphorylated, or L282A,V285A mutant peptide. The graph was taken from a representative experiment and shows the mean ± s.d. of technical triplicates. (G) Fixed oocytes were subject to immunofluorescence using labelled BUB-1 (‘total’), phospho Ser 283-BUB-1 (‘phS283’), and tubulin. Single-channel images for BUB-1 and phospho Ser 283-BUB-1 are shown in ‘fire’ LUT, and the bottom panel shows tubulin (green) and DNA (magenta). (H) Line profile analysis was performed in samples co-stained with labelled BUB-1 (Alexa488) and phospho Ser 283-BUB-1 (Alexa647) as described in the Materials and methods section. The lines represent the mean, and the shaded area represents the s.d. of the indicated number of bivalents.
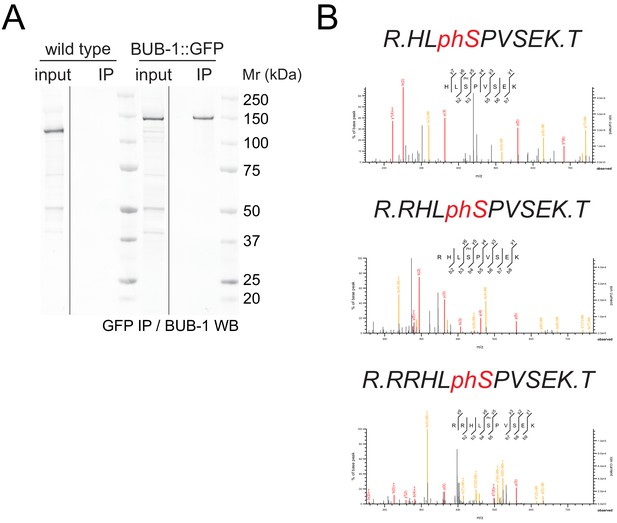
BUB-1::GFP immunoprecipitation and selected MS spectra of peptides containing phospho serine 283.
(A) Embryo extracts from wild-type (N2) and BUB-1::GFP worms were immunoprecipitated using a GFP nanobody coupled to magnetic beads. The inputs and immunoprecipitates were resolved on SDS–PAGE and subject to BUB-1 western blot. BUB-1::GFP is readily pulled down form the extracts, whereas untagged BUB-1 is not. Note the size difference due to the GFP tag. MW marker is shown on the right side. (B) Seleted MS spectra without (top), with one (middle), or with two (bottom) trypsin miscleavages are shown.
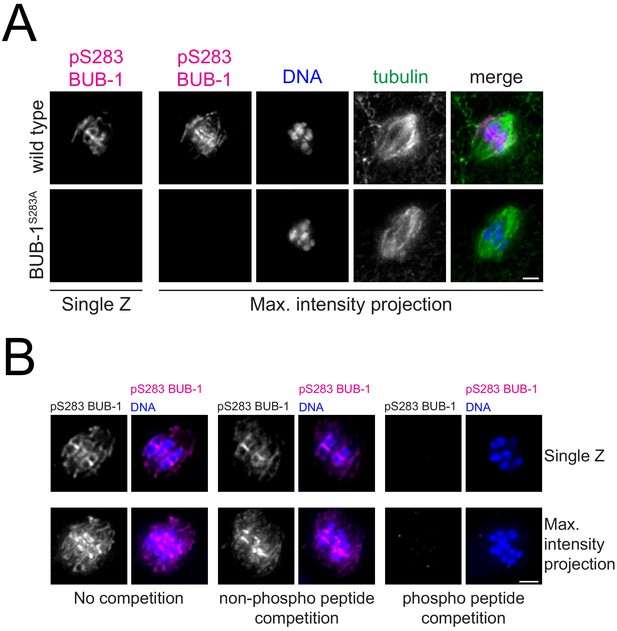
Validation of anti-phospho Ser 283 BUB-1 specific antibody.
(A) Fixed wild-type and S283A oocytes were stained with the anti-phospho Ser 283 BUB-1 antibody (‘pS283 BUB-1’). Scale bar, 2 µm. (B) Fixed oocytes were stained with the pS283 BUB-1 antibody in the absence of competing peptide, competed with a non-phosphorylated peptide, or competed with the same concentration of the phosphorylated peptide.
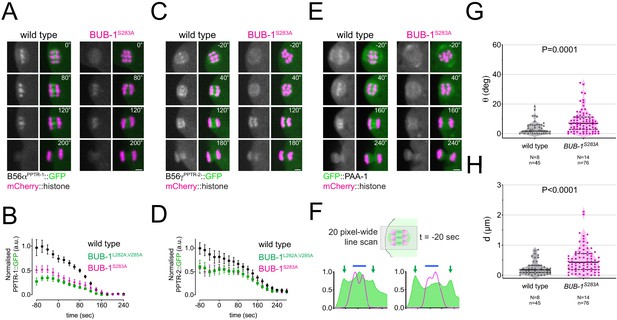
LxxIxE motif phosphorylation regulates the recruitment of B56s subunits in vivo.
(A) B56αPPTR-1 and chromosome dynamics were followed in wild-type and in BUB-1S283A oocytes expressing PPTR-1::GFP and mCherry::histone. Scale bar, 2 µm. Inset numbers represent the time relative to metaphase I in seconds. See Figure 6—video 1. (B) PPTR-1::GFP levels were measured in wild-type, BUB-1L282A,V285A and in BUB-1S283A oocytes throughout meiosis I and the mean ± s.e.m. is shown in the graph. (C) B56γPPTR-2 and chromosome dynamics were followed in wild-type and in BUB-1S283A oocytes expressing PPTR-2::GFP and mCherry::histone. Scale bar, 2 µm. Inset numbers represent the time relative to metaphase I in seconds. See Figure 6—video 2. (D) PPTR-2::GFP levels were measured in wild-type, BUB-1L282A,V285A, and BUB-1S283A oocytes throughout meiosis I, and the mean ± s.e.m is shown in the graph. (E) GFP::PAA-1 is present on chromosomes in the BUB-1S283A mutant. PAA-1 and chromosome dynamics were followed in wild-type and in BUB-1S283A oocytes expressing GFP::PAA-1 and mCherry::histone. Yellow dotted line shows that chromosome associated PAA-1 is lost in the BUB-1S283A mutant. Scale bar, 2 µm. Inset numbers represent the time relative to metaphase I in seconds. See Figure 6—video 3. (F) Representative, spindle-wide (20 pixels) line profiles are shown for wild-type (left) and BUB-1S283A mutant (right) measured in prometaphase I (20 s before metaphase I). Green arrows point to the PAA-1 pole signal and blue line to the chromosome associated population. (G) Angle of bivalents 80 s before segregation in meiosis I relative to the average angle of the spindle was measured in wild-type and in BUB-1S283A oocytes. Violin plot includes each data point (chromosome), the median (straight black line), and the interquartile range (dashed black lines). N represents number of oocytes and n number of bivalents measured. p value shown in the figure was obtained using a Mann–Whitney test. (H) Distance of bivalents 80 s before segregation in meiosis I relative to the centre of the spindle was measured in wild-type and in BUB-1S283A oocytes. Violin plot includes each data point (chromosome), the median (straight black line), and the interquartile range (dashed black lines). N represents number of oocytes and n number of bivalents measured. p value shown in the figure was obtained using a Mann–Whitney test.
Wild type and BUB-1S283A oocytes expressing PPTR-1::GFP and mCherry::histone were dissected and recorded.
Labels indicate the channels, time (relative to metaphase I), and scale bar. First slice indicates additionally indicates the intensity scale, the dimensiones of each channel image, and any additional filtering applied to the images.
Wild type and BUB-1S283A oocytes expressing PPTR-2::GFP and mCherry::histone were dissected and recorded.
Labels indicate the channels, time (relative to metaphase I), and scale bar. First slice indicates additionally indicates the intensity scale, the dimensiones of each channel image, and any additional filtering applied to the images.
Wild type and BUB-1S283A oocytes expressing GFP::PAA-1 and mCherry::histone were dissected and recorded.
Labels indicate the channels, time (relative to metaphase I), and scale bar. First slice indicates additionally indicates the intensity scale, the dimensiones of each channel image, and any additional filtering applied to the images.
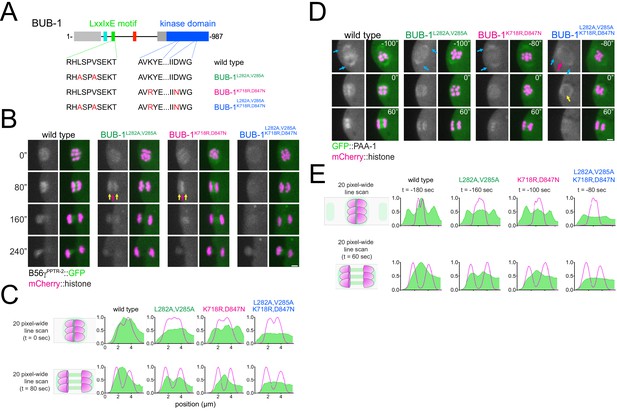
Role of BUB-1 kinase domain in B56γPPTR-2 chromosomal targeting.
(A) Schematic showing the LxxIxE motif and the kinase domain in BUB-1 of the wild-type, the BUB-1L282A,V285A, BUB-1K718R,D847N, and BUB-1L282A,V285A, K718R,D847N mutant. (B) B56γPPTR-2 and chromosome dynamics were followed in wild-type, the BUB-1L282A,V285A, BUB-1K718R,D847N, and BUB-1L282A,V285A, K718R,D847N oocytes expressing PPTR-2::GFP and mCherry::histone. Magenta arrows point towards the central spindle and yellow arrows towards chromosomes. Scale bar, 2 µm. Inset numbers represent the time relative to metaphase I in seconds. See also Figure 7—video 2. (C) Representative, spindle-wide (20 pixels) line profiles are shown for wild-type, the BUB-1L282A,V285A, BUB-1K718R,D847N, and BUB-1L282A,V285A, K718R,D847N. On top profiles of metaphase plate just in anaphase onset, on the bottom profiles 80 s after segregation to show central spindle levels. (D) Scaffolding subunit PAA-1 and chromosome dynamics were followed in wild-type, the BUB-1L282A,V285A, BUB-1K718R,D847N, and BUB-1L282A,V285A, K718R,D847N oocytes expressing GFP::PAA-1 and mCherry::histone. Cyan arrows point to pole population of PAA-1, magenta arrow signals chromosomes, and yellow arrow highlight the levels of PAA-1 in anaphase onset. Scale bar, 2 µm. Inset numbers represent the time relative to metaphase I in seconds. See also Figure 7—video 3. (E) Representative, spindle-wide (20 pixels) line profiles are shown for wild-type, the BUB-1L282A,V285A, BUB-1K718R,D847N, and BUB-1L282A,V285A, K718R,D847N. On top profiles before segregation starts (different time points matching the first panel on D) to highlight the PAA-1 in the poles and on the bottom profiles 60 s after segregation to show central spindle levels.
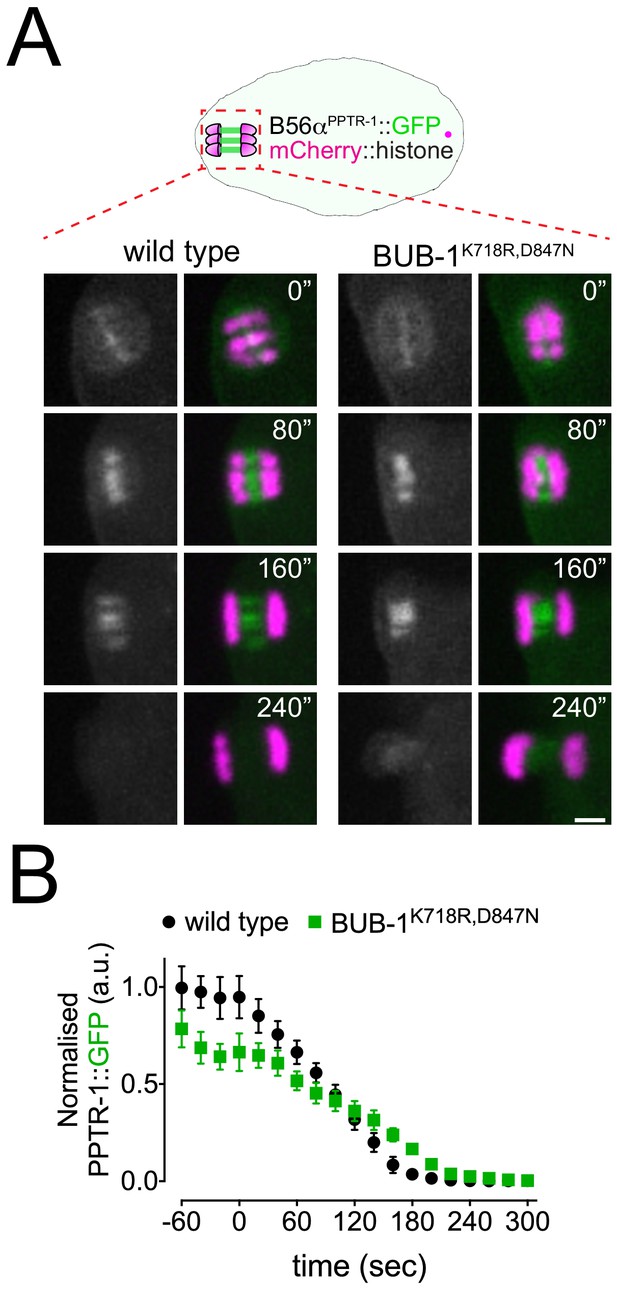
Kinase domain in BUB-1 does not regulate targeting of B56αPPTR-1 regulatory subunit in meiosis I.
(A) PPTR-1 and chromosome dynamics were followed in wild-type and BUB-1K718R,D847N oocytes expressing PPTR-1::GFP and mCherry::histone. Inset numbers represent the time relative to metaphase I in seconds. Scale bar, 2 µm. See Figure 7—video 1. (B) PPTR-1::GFP levels were measured throughout meiosis I in wild-type and BUB-1K718R,D847N oocytes and the mean ± s.e.m is shown in the graph.
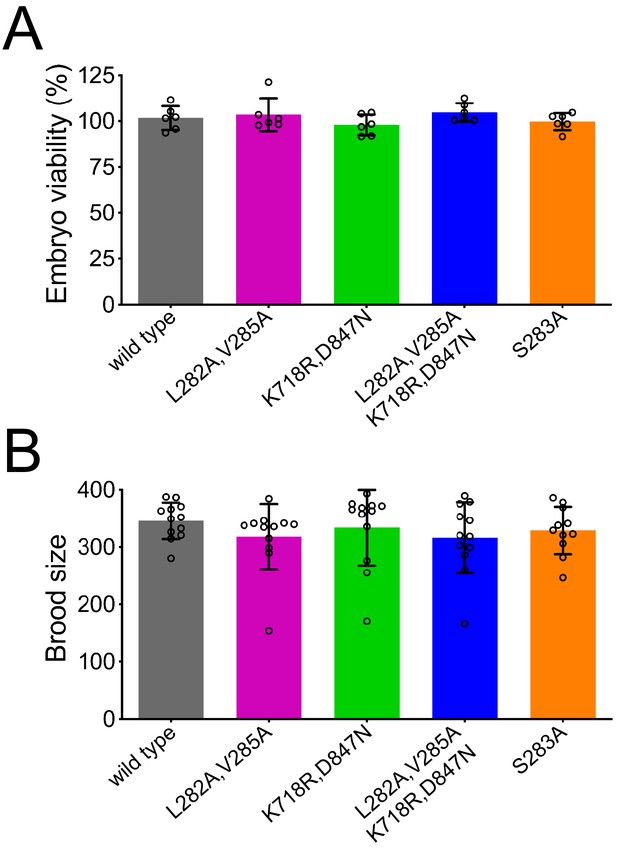
Embryo viability and brood size analysis of BUB-1 mutants.
(A) For the embryo viability assay, worms were synchronised by bleaching and 24 hr after L4, three worms were allowed to lay eggs during 3 hr in the same plate. Three plates per strain, for a total of 9 worms, were analysed per experiment. After 3 hr, adults were removed and eggs were counted. After 48 hr, viable progeny was quantified. Viability was scored as percentage of larvae/embryo per plate. This experiment was done twice. (B) For analysis of the brood size, the same strains as in the viability assay were used. Worms were synchronized by bleaching and 3 days post-L4, six adult worms per strain were transferred into new plates. Twenty-four hours after transferring the adult hermaphrodites into a new plate, progeny was scored in each plate. Total accumulated progeny graphs were obtained by merging two experiments.
Wild type and BUB-1K718R,D847N oocytes expressing PPTR-1::GFP and mCherry::histone were dissected and recorded.
Labels indicate the channels, time (relative to metaphase I), and scale bar. First slice indicates additionally indicates the intensity scale, the dimensiones of each channel image, and any additional filtering applied to the images.
Wild type and BUB-1L282A,V285A, BUB-1K718R,D847N, BUB-1 L282A,V285A;K718R,D847N oocytes expressing PPTR-2::GFP and mCherry::histone were dissected and recorded.
Labels indicate the channels, time (relative to metaphase I), and scale bar. First slice indicates additionally indicates the intensity scale, the dimensiones of each channel image, and any additional filtering applied to the images.
Wild type and BUB-1L282A,V285A, BUB-1K718R,D847N, BUB-1 L282A,V285A;K718R,D847N oocytes expressing GFP::PAA-1 and mCherry::histone were dissected and recorded.
Labels indicate the channels, time (relative to metaphase I), and scale bar. First slice indicates additionally indicates the intensity scale, the dimensiones of each channel image, and any additional filtering applied to the images.
Tables
Sequence identity between full-length mammalian B56 isoforms and C. elegans orthologues.
| PPTR-1 | B56β1 | B56ε3 | B56α2 | PPTR-2 | B56δ3 | B56γ1 | |
|---|---|---|---|---|---|---|---|
| PPTR-1 | 60.12 | 71.87 | 68.30 | 52.26 | 60.17 | 64.48 | |
| PPTR-2 | 52.26 | 56.36 | 63.94 | 60.84 | 66.19 | 68.97 |
-
Table was created using Clustal Omega version 2.1. See Supplementary file 1.
Additional files
-
Supplementary file 1
Percent Identity (%) Matrix of the full length sequence alignment of mammalian B56 isoforms and C. elegans orthologues PPTR-1 and PPTR-2.
Created with Clustal Omega version 2.1.
- https://cdn.elifesciences.org/articles/65307/elife-65307-supp1-v3.docx
-
Supplementary file 2
List of C. elegans strains used in this study.
- https://cdn.elifesciences.org/articles/65307/elife-65307-supp2-v3.xlsx
-
Supplementary file 3
List of primers used for genotyping.
- https://cdn.elifesciences.org/articles/65307/elife-65307-supp3-v3.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/65307/elife-65307-transrepform-v3.docx





