Fate mapping analysis reveals a novel murine dermal migratory Langerhans-like cell population
Figures
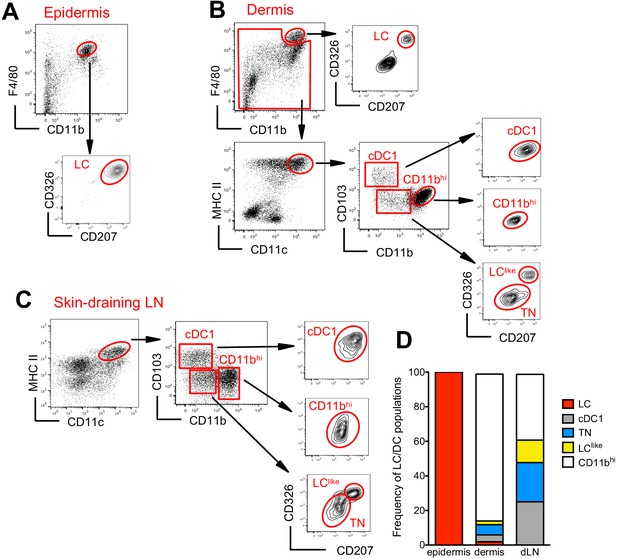
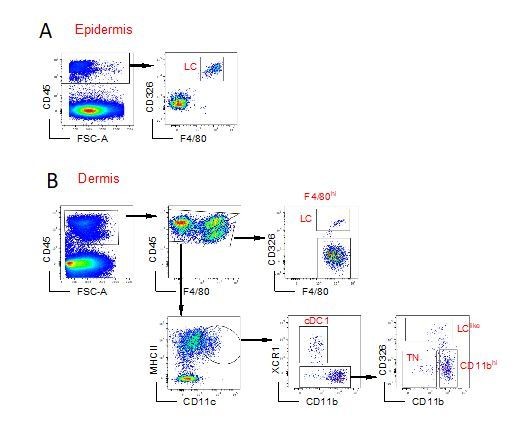
Characterization of cutaneous Langerhans cell (LC) and dendritic cell (DC) subpopulations.
(A) Representative flow cytometry dot plots for LC characterization in the epidermis. Cells from epidermis were first gated for FSC, SSC, and CD45 (not shown). Then, CD45+ cells were analysed for CD11b and F4/80 expression. The CD11bhiF4/80hi cell fraction was further analysed for CD207 and CD326 expression to identify classical bona fide LCs. (B) Representative flow cytometry dot plots for dermal LC and DC subpopulations. Isolated dermis cells were first gated for FSC, SSC, and CD45 (not shown). CD45+ cells were then analysed for CD11b and F4/80 expression. The CD11bhi F4/80hi fraction contained classical CD207+CD326+ LCs. The remaining cells were gated for CD11c+MHC II+ DCs and separated into three subsets based on CD103 and CD11b expression: CD103+CD11b- cells (labelled cDC1), CD103-CD11bhi DCs (labelled CD11bhi), and CD11blow/neg. CD207 and CD326 expression was detectable on cDC1 but not CD11bhi DCs, whereas CD11blow cells were further separated into CD207-CD326- (labelled triple negative [TN]) and CD207+CD326+ (labelled LClike). (C) Representative flow cytometry dot plots for cutaneous DC subpopulations in auricular skin-draining LNs. LN cells were first gated for FSC and SSC to exclude small lymphocytes before F4/80 and CD11b staining. The cell fraction excluding F4/80hi/CD11bhi cells was separated by CD11c and MHCII. CD11chiMHCIIhi migratory DCs were gated and analysed for CD103, CD11b, CD207, and CD326 expression. Four subsets were detected: CD103+CD11b-CD207+CD326+ (cDC1), CD103-CD11blowCD207-CD326- (TN), CD103-CD11blowCD207+CD326+ (LClike), and CD103-CD11bhiCD207-CD326-/+(CD11bhi). (D) Frequency of each DC subpopulation (LC, cDC1, LClike, TN, and CD11bhi) present in epidermis, dermis, and cutaneous lymph node (LN), respectively.
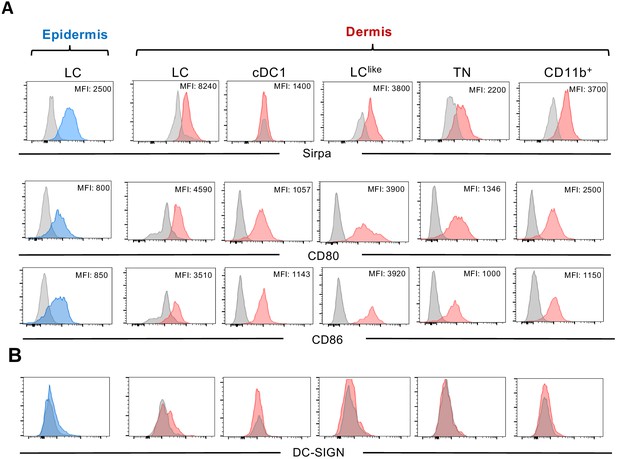
Expression profiles of myeloid marker signal regulatory protein α (Sirpa) and costimulatory molecules (CD80 and CD86) (A) and DC-SIGN (CD209a) (B).
Blue histograms show the relative expression of the markers in Langerhans cells (LCs) in epidermis and red histograms relative expression of the markers in dermal LCs, cDC1, LClike, triple negative (TN), and CD11b+ dendritic cells (DCs) following the gating strategy shown in Figure 1. Grey histogram depicts the staining of isotype control samples. MFI: mean of fluorescence intensity.
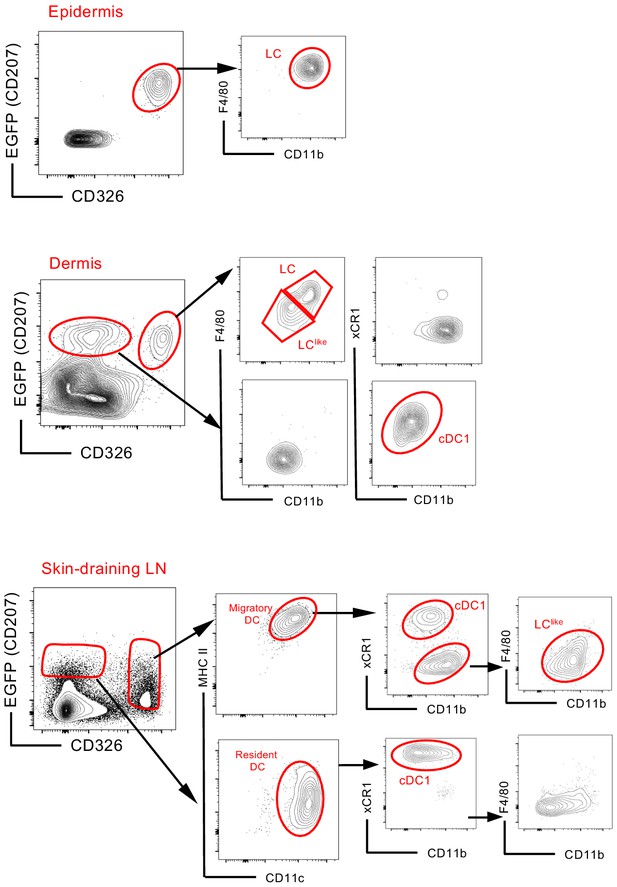
Flow cytometry analysis and characterization of EGFP+ cells in the epidermis (upper panel), dermis (middle panel), and skin-draining lymph nodes (LNs) (lower panel) obtained from the Lang-EGFP mouse.
Pre-gating performed on alive CD45+ cells is not shown.
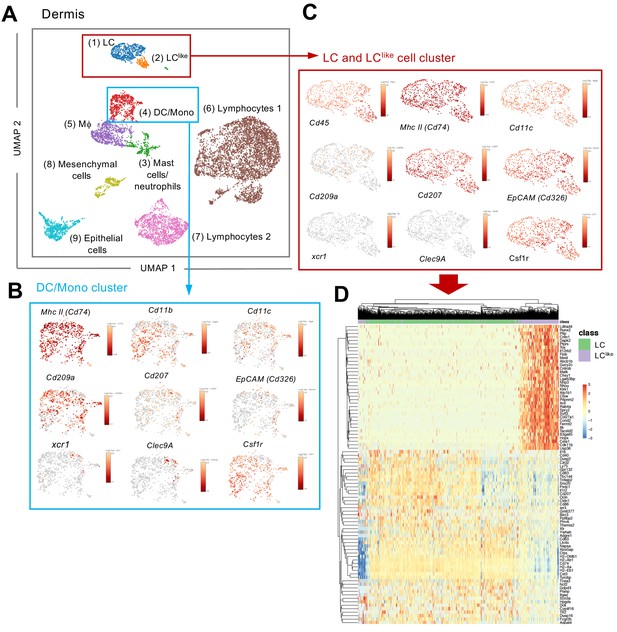
Single-cell RNA-seq analysis reveals Langerhans cell (LC) and LClike cells as two distinct cell populations in the dermis.
9605 cells pooled from the dermis collected from six mice which passed QC were imported for Seurat analysis. (A) Uniform manifold approximation and projection (UMAP) plot is revealing the existence of nine distinct cell clusters (1) LC (blue), (2) LClike(orange), (3) mast cells/neutrophils (green), (4) dendritic cell (DC)/monocytes (red), (5) macrophages (purple), (6) lymphocytes 1 (brown), (7) lymphocytes 2 (pink), (8) mesenchymal cells (light green), and (9) epithelial cells (light blue). (B, C) UMAP maps showing the expression of various LC signature genes in DC/mono (B) and LC/LClike clusters (C). (D) Heat-map of single-cell gene expression data based on the top differentially expressed genes discriminating LC/LClike clusters. Cells (LC in green; LClike in purple) are shown in rows and genes in columns.
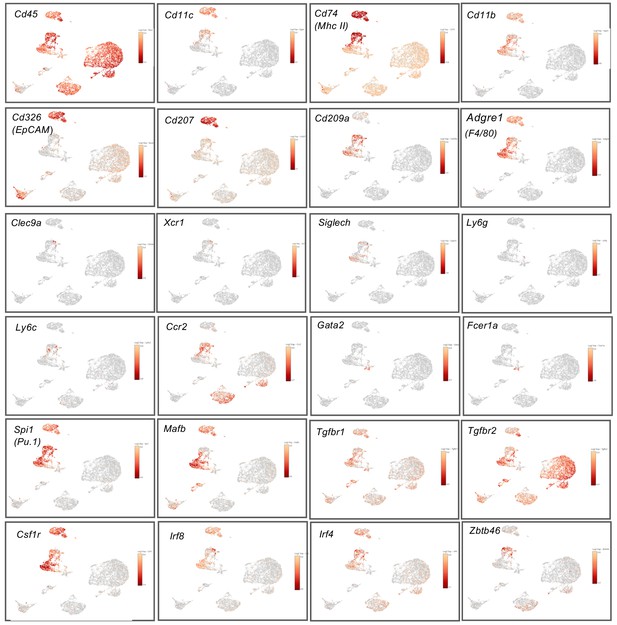
Expression of myeloid receptors and transcription factors among dermal CD45+ cells.
Feature plots showing the expression of Cd45, Cd11c, Cd74, Cd11b, Cd326, Cd207, Cd209a, Adgre1, Clec9a, Xcr1, Siglech, Ly6g, Ly6C, Ccr2, Gata2, Fcer1a, Spi1, Mafb, Tgfbr1, Tgfbr2, Csfr1, Irf8, Irf4 and Zbtb46, respectively.
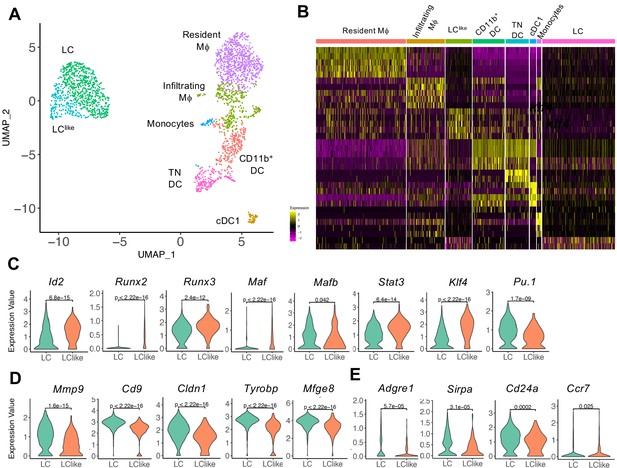
Detailed uniform manifold approximation and projection (UMAP) analysis from clusters 1, 2, 4, and 5 visualizes eight distinct Langerhans cell (LC)/dendritic cell (DC) and macrophage subpopulations.
(A) UMAP plot showing eight distinct LC/DC and macrophage clusters: LC (emerald green), LClike (turquoise), cDC1 (ocher), CD11b+ (red orange), triple negative (TN) DCs (magenta), resident macrophages (purple), monocytes (light blue), and infiltrating macrophages (green). Colours indicate unsupervised clustering by PhenoGraph. (B) Heat-map of single-cell gene expression data based on the top differentially expressed genes between the eight cell clusters. Yellow: upregulated; purple: downregulated. (C) Violin plots comparing transcription factor (TF) expression in LC and LClike cells. (D) Violin plots showing mRNA expression profile of LC signature genes in LC and LClike cells. (E) Violin plots showing Adgre1 (F4/80), Sirpa, Cd24a, and chemokine receptor Ccr7 expression in LC and LClike cells.
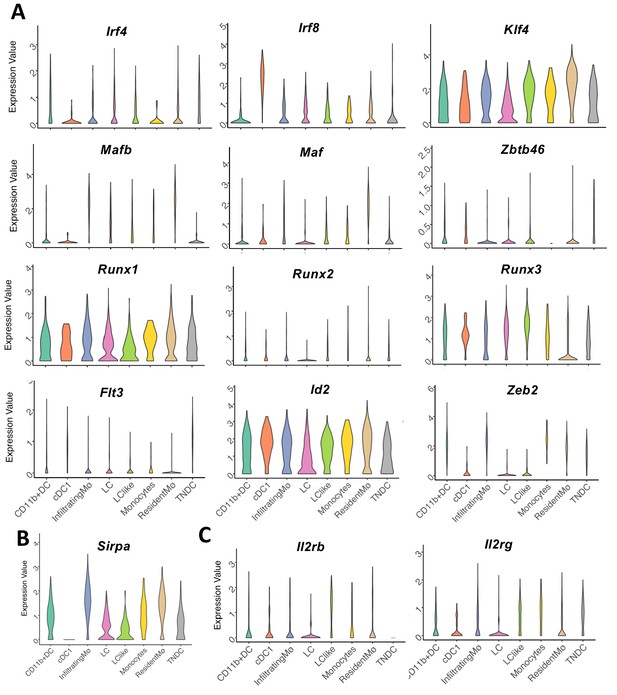
Violin plots showing mRNA expression profile of (A) transcription factor (TF) genes, (B) Sirpa, and (C) IL2 receptor genes (Il2rb, Il2rg) in eight distinct dermal Langerhans cell (LC), dendritic cell (DC), and macrophage subpopulations.
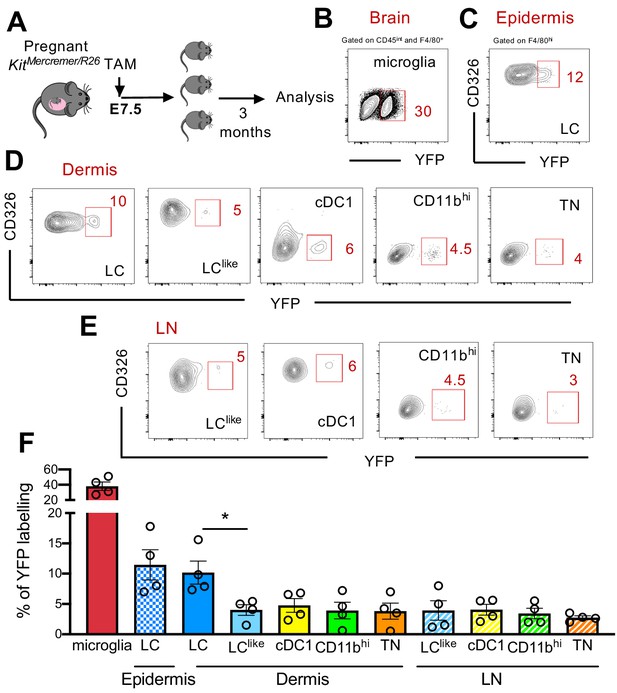
Distinct embryonic origin between Langerhans cell (LC) and LClike cells.
(A) Single pulse of TAM at E7.5 was given to label KitMercreMer/R26 embryos and the percentages of labelled brain microglia (positive control, gated on CD45intF4/80hi), epidermal LCs, and dermal LC/dendritic cell (DC) subpopulations were measured at 3 months of age. (B–D) Flow cytometry analysis of YFP labelling of microglia (B), and each LC and DC subpopulation in the epidermis (C), dermis (D), and lymph node (LN) (E) in KitMerCreMer/R26 fate mapping mice. Representative contour plots are shown. (F) The mean percentage of YFP+ cells of brain microglia, epidermal LC, and dermal DC subpopulations (LC, cDC1, LClike, CD11bhi, and triple negative [TN] cells). The error bars represent the SEM (n = 4 samples of two to three pooled mice for epidermis/dermis and n = 5 mice for LN). Data from two independent experiments. *p<0.05; two-way ANOVA followed by Bonferroni test. For clarity, non-significant values are not shown.
-
Figure 4—source data 1
Percentage of YFP+ cells of brain microglia, epidermal LC,dermal and LN DC subpopulations (LC, cDC1, LClike, CD11bhi,and TN cells).
- https://cdn.elifesciences.org/articles/65412/elife-65412-fig4-data1-v2.xlsx
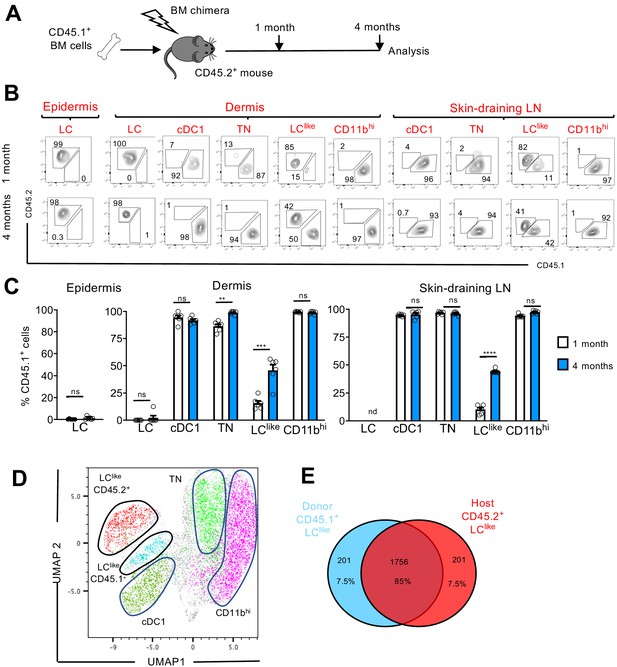
LClike cells derived from embryonic and adult haematopoiesis have a similar transcriptomic signature.
(A) Generation of BM chimeras: CD45.1+ WT BM cells (106) were transferred into lethally irradiated CD45.2+ recipient mice. The epidermis, dermis, and draining lymph nodes (LNs) obtained from the reconstituted chimeras were analysed 1 and 4 months later by flow cytometry. (B) Flow cytometry analysis of donor (CD45.1+) and host (CD45.2+) chimerism in different epidermal, dermal, and skin-draining LN LC and dendritic cell (DC) subpopulations, 1 and 4 months after reconstitution. LC, cDC1, triple negative (TN), LClike, and CD11bhi subsets were gated and analysed for CD45.1 (x-axis) and CD45.2 (y-axis) expression. (C) The percentage of CD45.1 donor cells detected in the epidermis, dermis, and skin-draining LNs of chimeras, 1 or 4 months after reconstitution. Data are represented as mean ± SEM; n = 6 single mice; **p<0.01; ***p<0.001; ****p<0.0001; ns, non-significant; two-tailed Student’s t-test. (D) Uniform manifold approximation and projection (UMAP) analysis of distinct LN DC subpopulations obtained from chimeras 4 months after reconstitution, based on the expression of different markers (CD11c, MHCII, CD103, CD11b, CD326, CD207, CD45.1, CD45.2). (E) Transcriptome analysis of LN CD45.1+ LClike cells (n = 3) and LN CD45.2+ LClike (n = 3) cells collected from 10 mice. The Venn diagram shows the percentage of overlapping genes expressed by CD45.1+ and CD45.2+ LClike cells.
-
Figure 5—source data 1
Percentage of CD45.1+ donor cells detected in the epidermis, dermis and skin-draining LNs of mouse chimeras, 1 or4monthsafter reconstitution.
- https://cdn.elifesciences.org/articles/65412/elife-65412-fig5-data1-v2.xlsx
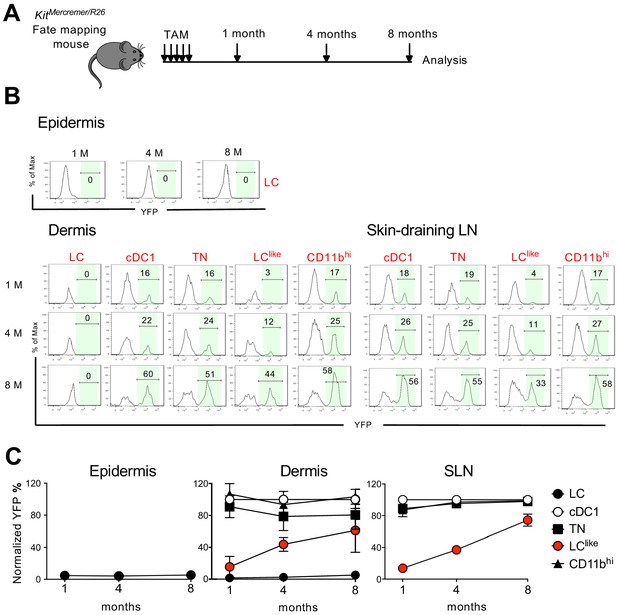
Slow turnover kinetics for dermal and lymph node (LN) LClike cells.
(A) KitMerCreMer/R26 mice aged 6 weeks old were injected with tamoxifen five times and groups of six animals were sacrificed 1, 4, and 8 months later for fate mapping analysis. (B) Flow cytometry analysis of YFP labelling of each LC and dendritic cell (DC) subpopulation in the epidermis, dermis, and skin-draining LNs in KitMerCreMer/R26 fate mapping mice. Representative histograms are shown. (C) The mean percentage of YFP+ cells after normalization to cDC1. Epidermis (left), dermis (middle), and skin-draining LNs (right) were analysed. The error bars represent the SEM (n = 6 mice).
-
Figure 6—source data 1
Percentage of YFP+ epidermal, dermal and LN cells analysed at 1, 4 and 8 months after tamoxifen injection.
Values have been normalized to cDC1 YFP labelling.
- https://cdn.elifesciences.org/articles/65412/elife-65412-fig6-data1-v2.xlsx
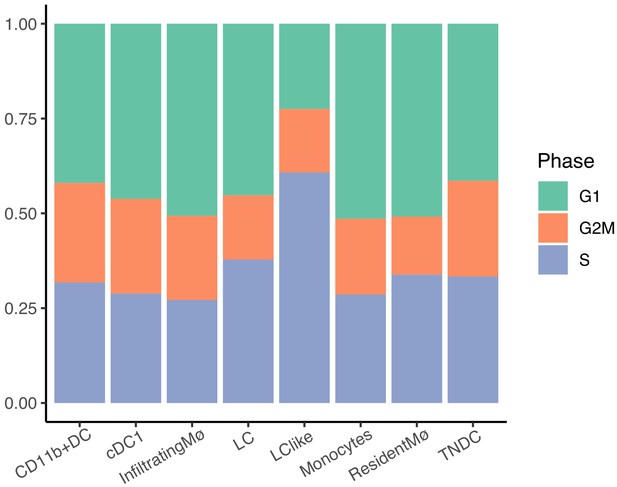
Bar plots showing distinct dermal myeloid cell subset distributions across cell cycle phases.
Blocks represent individual cell cycle phases. Green: G1; orange: G2M; and Blue: S.
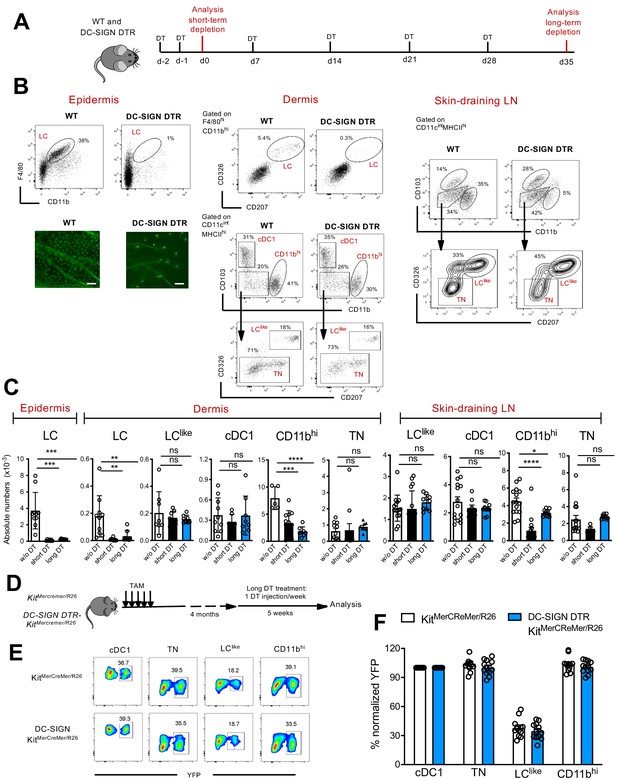
Classical Langerhans cells (LCs), but not LClike cells, are ablated in vivo in DC-SIGN DTR mice.
(A) The short-term and long-term depletion protocol in DC-SIGN-DTR mice. (B) Representative flow cytometry dot plots of single-cell suspensions from the epidermis (left), dermis (middle), and skin-draining lymph nodes (LNs) (right) obtained from diphteria toxin (DT)-injected WT and DC-SIGN-DTR mice. All mice were injected (i.p.) with 10 ng/g DT on days −2 and −1 and analysed on day 0. The gating strategy shown in Figure 1 was followed. Epidermal sheets obtained from WT and DC-SIGN mice were stained for MHC class II (green fluorescence) and analysed by immunofluorescence microscopy (lower left panel). (C) The absolute numbers of each gated myeloid cell subset (LC, cDC1, triple negative [TN], LClike, and CD11bhi cells) obtained from the epidermis, dermis, and skin-draining LNs of DT-injected WT and DC-SIGN DTR mice. Data are represented as mean ± SEM; n = 8–10 single mice. ****p<0.0001; ***p<0.001; **p<0.01; ns, non-significant; two-way ANOVA statistical test Bonferroni test. (D) Fate mapping analysis in DC-SIGN DTR-KitMerCreMer/R26 mice. Mice aged 6 weeks old were orally gavaged with TAM. After 4 months, DT was injected i.p. weekly for 5 weeks to ensure long-term LC depletion. (E) Representative contour plots showing the YFP labelling of distinct LN dendritic cell (DC) subpopulations in DT-treated KitMerCreMer/R26 and DC-SIGN DTR-KitMerCreMer/R26 mice. (F) The percentage of normalized YFP labelling detected in DC subpopulations (LC, cDC1, TN, LClike, and CD11bhi cells) of the skin-draining LNs. Normalization was performed as described in Figure 6; data are represented as mean ± SEM; n = 12 single mice.
-
Figure 7—source data 1
Absolute numbers of epidermal, dermal and LN DC subpopulations after short and long term depletion in DC-SIGN-DTR mice.
- https://cdn.elifesciences.org/articles/65412/elife-65412-fig7-data1-v2.xlsx
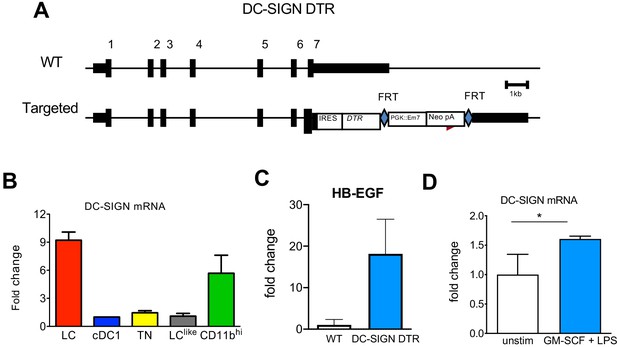
Epidermal LCs isolated from DC-SIGN DTR mice express human HB-EGF.
(A) The experimental strategy to generate DC-SIGN-DTR mice. (B) Quantitative PCR (qPCR) analysis of the mRNA Cd209a expression in epidermal Langerhans cells (LCs) and distinct lymph node (LN) dendritic cell (DC) subpopulations. (C) qPCR analysis of HBEGF expression in LCs obtained from WT and DC-SIG DTR mice. (D) qPCR analysis of Cd209a expression in unstimulated purified LCs and in vitro stimulated purified LCs (GM-CSF + LPS 16 hr).
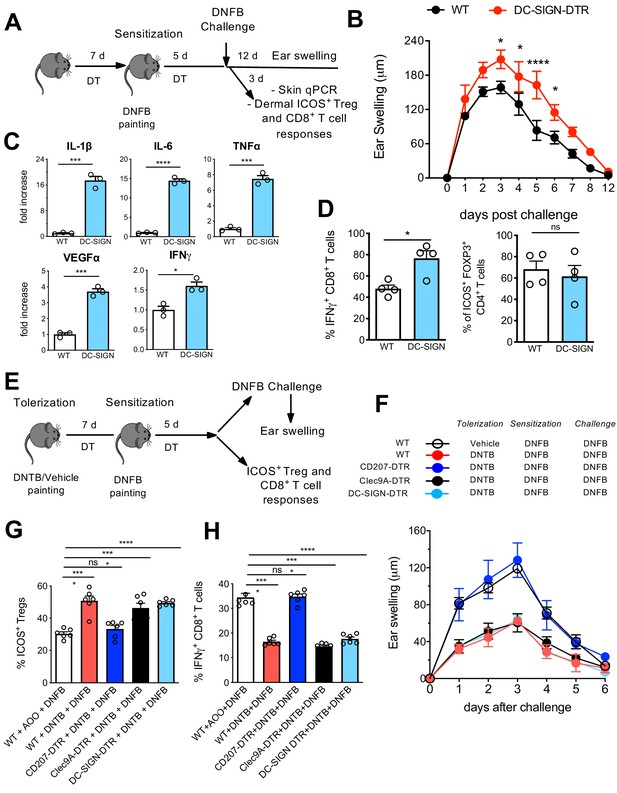
Differential contribution of Langerhans cell (LC) and LClike in skin immune responses.
(A) Diphteria toxin (DT)-treated WT and DC-SIGN DTR mice were sensitized with 0.5% 2,4-dinitrofluorobenzene (DNFB) (applied to the shaved back skin) and ear-challenged 5 days later with 0.1% DNFB. (B) Ear swelling response of challenged WT and DC-SIGN DTR mice was determined over a 12-day period post challenge. (C) Quantitative PCR analysis of distinct inflammatory cytokines and growth factors in ears collected at day 3 post challenge. The error bars represent the SEM (n = 3 mice). *p<0.05; ***p<0.001; ****p<0.0001; two-tailed Student’s t-test. (D) Percentages of dermal IFN-γ-producing CD8+ T-cells and dermal CD4+Foxp3+ICOS+ regulatory T-cells at day 3 post challenge. Single-cell suspensions were generated from the dermis and the cells were re-stimulated (5 hr) with PMA/Ionomycin to detect IFN-γ production or directly stained for determination of activated Tregs. The error bars represent the SEM (n = 4 mice). *p<0.05; ns, non-significant; two-tailed Student’s t-test. Gating strategy is shown in Figure 8—figure supplement 1. (E) DT-treated WT, CD207-DTR, Cleac9A-DTR, and DC-SIGN DTR mice were tolerized with 1% 2,4-dinitrothiocyanobenzene hapten (DNTB) (applied to the shaved abdomen skin). After 7 days, the mice were sensitized with 0.5% DNFB (applied to the shaved back skin) and ear-challenged 5 days later with 0.1% DNFB. (F) The ear swelling response of painted mice was determined over a 6-day period post challenge. (G) Percentage of activated CD4+Foxp3+ICOS+ regulatory T-cells in the draining lymph nodes (LNs) of mice, 5 days after vehicle or DNTB painting. The data were pre-gated on singlet, live CD3+CD4+Foxp3+ Treg cells. The error bars represent the SEM (n = 6 mice). ****p<0.0001; ns, non-significant; two-way ANOVA. (H) Percentages of IFN-γ-producing CD8+ T-cells in the draining LNs of vehicle or DNTB-painted mice followed by DNFB sensitization. Single-cell suspensions were generated from the LNs and the cells were re-stimulated (5 hr) with PMA/Ionomycin to detect IFN-γ production. The data were pre-gated on singlet, live CD3+ CD8+ T-cells. The error bars represent the SEM (n = 6 mice). ****p<0.0001; ns, non-significant; two-way ANOVA.
-
Figure 8—source data 1
Percentages of IFNgamma+ CD8+ T cells and ICOS+ FOXP3+ T regs measured in distinct skin immune repsonses.
- https://cdn.elifesciences.org/articles/65412/elife-65412-fig8-data1-v2.xlsx
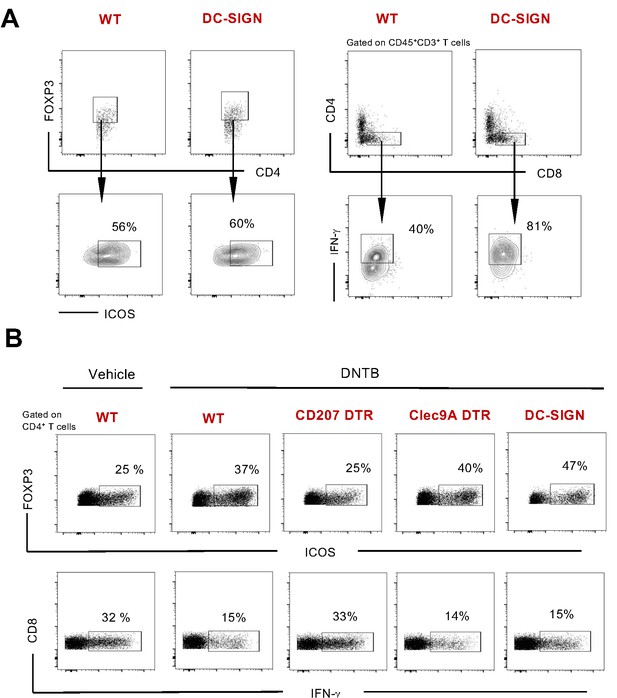
Gating strategy with representative flow cytometry samples.
(A) Contact hypersensitivity (CHS) reactivity in WT and DC-SIGN-DTR mice. Representative dot and contour plots for dermal IFN-γ-producing CD8+ T-cells and CD4+Foxp3+ICOS+ regulatory T-cells at day 3 post challenge. (B) 2,4-Dinitrothiocyanobenzene hapten (DNTB)-mediated tolerance assay in WT, CD207 DTR, Clec9A-DTR, and DC-SIGN DTR mice. Representative dot plots for LN IFN-γ-producing CD8+ T-cells and CD4+Foxp3+ICOS+ regulatory T-cells at day 5.
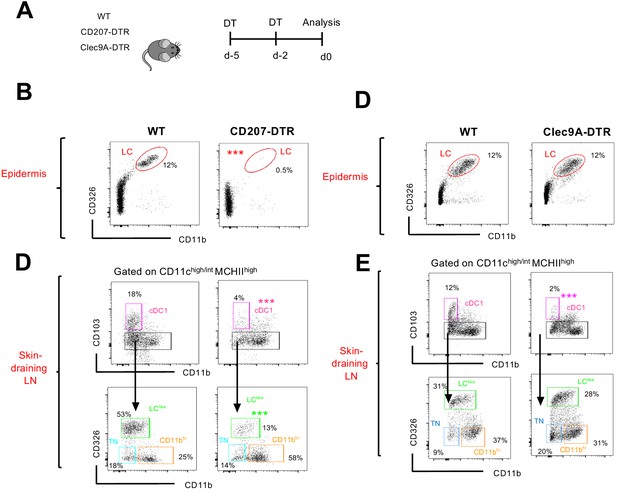
Transgenic DTR mouse strains used for in vivo abaltion of CD207+ and Clec9A+ cells.
(A) The experimental strategy to delete Langerhans cell (LC), cDC1, and LClike cells dendritic cell (DC) in CD207 DTR and Clec9A DTR mice. (B) Representative flow cytometry CD11b/CD326 dot plots for LC characterization in the epidermis of diphteria toxin (DT)-injected WT and CD207-DTR mice. (C) Representative flow cytometry CD103/CD11b and CD11b/CD326 dot plots for migratory DC subset characterization in the skin-draining LNs of WT and CD207-DTR mice following the gating strategy described in Figure 1. (D) Representative flow cytometry CD11b/CD326 dot plots for LC characterization in the epidermis of DT-injected WT and Clec9A DTR mice. (E) Representative flow cytometry CD103/CD11b and CD11b/CD326 dot plots for migratory DC subset characterization in the skin-draining LNs of WT and Clec9A DTR mice.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | Rat anti-mouse CD45 (30-F11) | Biolegend | Cat#: 103108; RRID: AB_312972 | FACS (1:600; 100 μl per test) |
| Antibody | Rat anti-mouse CD45 (30-F11) | Biolegend | Cat#: 103114; RRID: AB_312978 | FACS (1:600; 100 μl per test) |
| Antibody | Rat anti-mouse CD11b (M1/70) | Becton Dickinson- BD | Cat#: 565976; RRID:AB_2721166 | FACS (1:600; 100 μl per test) |
| Antibody | Rat anti-mouse F4/80 (BM8) | Biolegend | Cat#: 123114; RRID: AB_893490 | FACS (1:600; 100 μl per test) |
| Antibody | Rat anti-mouse Ly6c (HK1.4) | Biolegend | Cat#: 128036; RRID: AB_2562352 | FACS (1:600; 100 μl per test) |
| Antibody | Rat anti-mouse I-A/I-E antibody (M5/114.15.2) | Biolegend | Cat#: 107632; RRID: AB_10900075 | FACS (1:600; 100 μl per test) |
| Antibody | Hamster anti-mouse CD11c (N418) | Biolegend | Cat#: 117324; RRID: AB_830646 | FACS (1:600; 100 μl per test) |
| Antibody | Hamster anti-mouse CD103 (2E7) | Biolegend | Cat#: 121416; RRID: AB_1574957 | FACS (1:600; 100 μl per test) |
| Antibody | Mouse anti-mouse CD209 (clone: MMD3) | Thermo Fisher Scientific | Cat#: 50-2094-82; RRID:AB_11219065 | FACS (1:600; 100 μl per test) |
| Antibody | Rat anti-mouse CD326 (G8.8) | Biolegend | Cat#: 118231; RRID: AB_2632774 | FACS (1:600; 100 μl per test) |
| Antibody | Mouse anti-mouse CD207 (clone: 4C7) | Biolegend | Cat#: 144204; RRID: AB_2561498 | FACS (1:600; 100 μl per test) |
| Antibody | Mouse anti-mouse CD45.1 (A20) | Biolegend | Cat#: 110726; RRID: AB_893347 | FACS (1:600; 100 μl per test) |
| Antibody | Mouse anti-mouse CD45.2 (104) | Biolegend | Cat#: 109830; RRID: AB_1186103 | FACS (1:600; 100 μl per test) |
| Antibody | Rat anti-mouse CD3 (17A2) | Biolegend | Cat#: 100306; RRID: AB_312670 | FACS (1:500; 100 μl per test) |
| Antibody | Rat anti-mouse CD4 (GK1.5) | Biolegend | Cat#: 100414; RRID: AB_312699 | FACS (1:600; 100 μl per test) |
| Antibody | Rat anti-mouse CD8 (53–6.7) | Biolegend | Cat#: 100722; RRID: AB_312761 | FACS (1:600; 100 μl per test) |
| Antibody | Rat anti-mouse FOXP3 (MF-14) | Biolegend | Cat#: 126407; RRID: AB_1089116 | FACS (1:600; 100 μl per test) |
| Antibody | Hamster anti-mouse ICOS (15F9) | Biolegend | Cat#: 107705; RRID: AB_313334 | FACS (1:600; 100 μl per test) |
| Antibody | Rat anti-IFN-gamma (XMG1.2) | Biolegend | Cat#: 505810; RRID:AB_315404 | FACS (1:600; 100 μl per test) |
| Antibody | Fc-R block (2.4G2) | Self-made | N/A | Blocking step (1:100; 1000 ml per sample) |
| Chemical compound, drug | Brefeldin A | Sigma-Aldrich | Cat#: B7651 | 10 μg/ml |
| Chemical compound, drug | Phorbol 12-myristate 13-acetate | Sigma-Aldrich | Cat#: 79346 | 10 μg/ml |
| Chemical compound, drug | Ionomycin | Sigma-Aldrich | Cat#: I0634 | 10 μg/ml |
| Chemical compound, drug | Collagenase D | Roche | Cat#: 11088882001 | 1 mg/ml |
| Chemical compound, drug | Dispase II | Gibco | Cat#: 17105041 | 1 U/ml |
| Chemical compound, drug | Ficoll-Paque | GE Healthcare | Cat#: 17144003 | |
| Chemical compound, drug | Percoll | Merck | Cat#: P4937-500ML | |
| Chemical compound, drug | Diphtheria toxin | Sigma-Aldrich | Cat#: D0564 | 20 ng DT/g body weight, i.p. |
| Chemical compound, drug | Tamoxifen | Sigma-Aldrich | Cat#: T5648 | 4 mg TAM for 5 consecutive days by oral gavage for adult labelling. Pregnant mice (E7.5) were injected once with 16 mg TAM for embryo labelling. |
| Chemical compound, drug | IMDM | Thermo Fisher | Cat#: 12440046 | |
| Chemical compound, drug | Ammonium thiocyanate | Sigma-Aldrich | Cat#: 221988 | |
| Chemical compound, drug | 5,5'-Dithio-bis- 2-nitrobenzoic acid (DNTB) | Sigma-Aldrich (Lancaster Synthesis) | Cat#: D8130 | |
| Chemical compound, drug | 1-Fluoro-2,4- dinitrobenzene (DNFB) | Sigma-Aldrich | Cat#: D1529 | |
| Chemical compound, drug | Acetone | Sigma-Aldrich | Cat#: 650501 | |
| Chemical compound, drug | Saponin | Sigma-Aldrich | Cat#: S7900 | |
| Chemical compound, drug | TRIzol reagent | Thermo Fisher Scientific | Cat#: 15596026 | |
| Commercial assay or kit | RNAsimple Total RNA kit | Tiangen Biotech Ltd | Cat#: DP419 | |
| Commercial assay or kit | Foxp3 staining buffer | eBioscience | Cat#: 00-5521-00 | |
| Commercial assay or kit | Cytofix/cytoperm | eBioscience | Cat#: 51-2090KZ | |
| Commercial assay or kit | Ovation Universal RNA-seq system | NuGEN Technologies | Cat#: 0343–32 | |
| Commercial assay or kit | DNA High Sensitivity Reagent Kit | Agilent, Santa Clara, CA, USA | Cat#: 5067–4626 | |
| Commercial assay or kit | 10× Chromium Controller | 10X Genomics | Cat #: 120263 | |
| Commercial assay or kit | Chromium Single Cell v3 reagent kit | 10X Genomics | Cat #: PN-100009 | |
| Software, algorithm | FlowJo | TreeStar | FlowJo 10.6 RRID:SCR_008520 | |
| Software, algorithm | GraphPad Prism | GraphPad Software | GraphPad 9.0 RRID:SCR_002798 | |
| Strain, strain background (mouse) | C57BL/6J | The Jackson Laboratory | Stock Nr. 000664 RRID:IMSR_JAX:000664 | |
| Strain, strain background (mouse) | B6.SJL-Ptprca Pepcb/BoyJ | The Jackson Laboratory | Stock Nr. 002014 RRID:IMSR_JAX:002014 | |
| Strain, strain background (mouse) | KitMerCreMer/Rosa26-LSL-eYFP (called KitMerCreMer/R26) | Nanyang Technological University, Singapore Sheng et al., 2015 | ||
| Strain, strain background (mouse) | Clec9A-DTR | Nanyang Technological University, Singapore Piva et al., 2012 | ||
| Strain, strain background (mouse) | CD207-DTR | SIgN, A*Star, Singapore Kissenpfennig et al., 2005 | ||
| Strain, strain background (mouse) | DC-SIGN-DTR | Nanyang Technological University, Singapore | Sheng et al., this paper | |
| Strain, strain background (mouse) | DC-SIGN-DTR-KitMerCreMer/R26 | Nanyang Technological University, Singapore | Sheng et al., this paper | |
| Strain, strain background (mouse) | B6.129S2-Cd207tm2Mal/J (Lang-EGFP) | The Jackson Laboratory | Stock Nr. 016939 RRID:IMSR_JAX:016939 | |
| Sequenced-based reagent | Ifng_F | This paper | PCR primers | GACAATCAGGCCATCAGCAAC |
| Sequenced-based reagent | Ifng_R | This paper | PCR primers | ACTCCTTTTCCGCTTCCTGAG |
| Sequenced-based reagent | Il6_F | This paper | PCR primers | TGATGGATGCTACCAAACTGG |
| Sequenced-based reagent | Il6_R | This paper | PCR primers | CCAGGTAGCTATGGTACTCCAGA |
| Sequenced-based reagent | Tnfa_F | This paper | PCR primers | AATTCGAGTGACAAGCCTGTAG |
| Sequenced-based reagent | Tnfa_R | This paper | PCR primers | TTGAGATCCATGCCGTTGG |
| Sequenced-based reagent | Il1b_F | This paper | PCR primers | GGGCCTCAAAGGAAAGAATC |
| Sequenced-based reagent | Il1b_R | This paper | PCR primers | TTCTTCTTTGGGTATTGCTTGG |
| Sequenced-based reagent | Vegfa_F | This paper | PCR primers | GCAGCTTGAGTTAAACGAACG |
| Sequenced-based reagent | Vegfa_R | This paper | PCR primers | GGTTCCCGAAACCCTGAG |
| Sequenced-based reagent | HBEGF_F | This paper | PCR primers | ATGACCACACAACCATCCTG |
| Sequenced-based reagent | HBEGF_R | This paper | PCR primers | CCAGCAGACAGACAGATGACA |
| Sequenced-based reagent | cd209a_F | This paper | PCR primers | CCAAGAACTGACCCAGTTGAA |
| Sequenced-based reagent | cd209a_R | This paper | PCR primers | CTTCTGGGCCACAGAGAAGA |
| Sequenced-based reagent | Actb_F | This paper | PCR primers | AAGGCCAACCGTGAAAAGAT |
| Sequenced-based reagent | Actb_R | This paper | PCR primers | CCTGTGGTACGACCAGAGGCATACA |