DDK regulates replication initiation by controlling the multiplicity of Cdc45-GINS binding to Mcm2-7
Figures
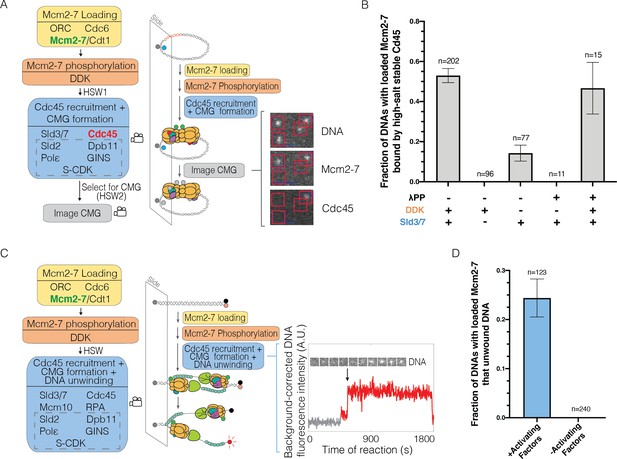
Single-molecule reaction for Cdc45-Mcm2-7-GINS (CMG) formation and DNA unwinding.
(A) Schematic for the single-molecule CMG formation reaction. Alexa-Fluor-488-labeled (blue circle) circular origin DNA molecules were tethered to the slide surface. Purified Mcm2-74SNAP549 (green circle), Cdc45SORT649 (red circle), and other indicated proteins necessary to form the CMG were incubated with slide-coupled DNA in the steps shown. Members of the group of proteins referred to as SDPGC are in the dashed box. A high-salt wash (HSW1) was performed after Mcm2-7 phosphorylation to remove helicase-loading intermediates. Colocalization of fluorescently labeled proteins with fluorescently labeled DNA was monitored (indicated by camera icon) during CMG formation and after a second high-salt wash (HSW2). Example images show a small subregion of the microscope field of view taken at a single time point after the CMG formation reaction recorded in the color channels for DNA488, Mcm2-74SNAP549, and Cdc45SORT649. Red squares are centered on DNA locations. Spots within the red square indicate stable binding of Mcm2-74SNAP549 or Cdc45SORT649 after HSW2. (B) Cdc45 binding depends on Sld3/7 and DDK phosphorylation. The fraction (± SE) of n DNA molecules with bound Mcm2-74SNAP549 that exhibited high-salt-resistant Cdc45SORT649 at the first time point after the HSW2 is plotted for each of the conditions. DNAs are counted if they contain Mcm2-74SNAP549 at the beginning of the CMG formation reaction. Where indicated, lambda phosphatase (λPP) was used to dephosphorylate Mcm2-74SNAP549 prior to the Mcm2-7 loading reaction. The first bar represents the results of three replicates, the second and third bars represent the results of two replicates, and the remaining bars are the result of a single experiment. (C) Schematic of the single-molecule DNA-unwinding reaction. Cy5- and BHQ-2-labeled (red and black circles) linear origin-DNA molecules were coupled to microscope slides. The same stepwise incubations as with the single-molecule CMG-formation assay were used, except Mcm10 and replication protein A (RPA) were added to the 'CMG formation and DNA unwinding step'. The plot displays a representative background-corrected fluorescence-intensity record (see Materials and methods) for DNACy5 (increase in fluorescence indicated by arrow). An objective image-analysis algorithm (Friedman and Gelles, 2015) detected a spot of DNA fluorescence at time points shown in red. (D) DNA unwinding is dependent on activating factors. Helicase-activating factors eliminated were S-CDK, Sld2, Dpb11, Pol ε, GINS, Sld3/7, Cdc45, Mcm10, and RPA. The fraction (± SE) of n DNA molecules that were unwound is plotted for both conditions.
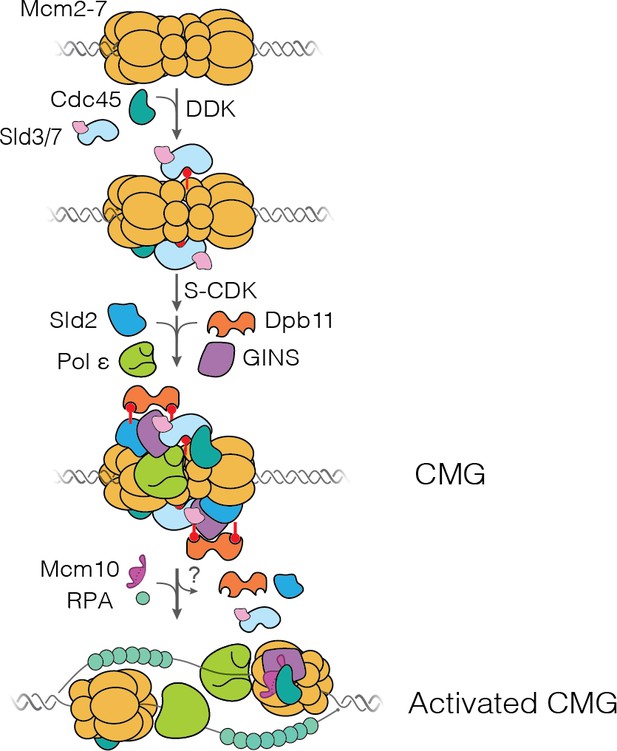
Schematic representation of DNA replication initiation.
The scheme illustrates the first time that each factor is proposed to be required.
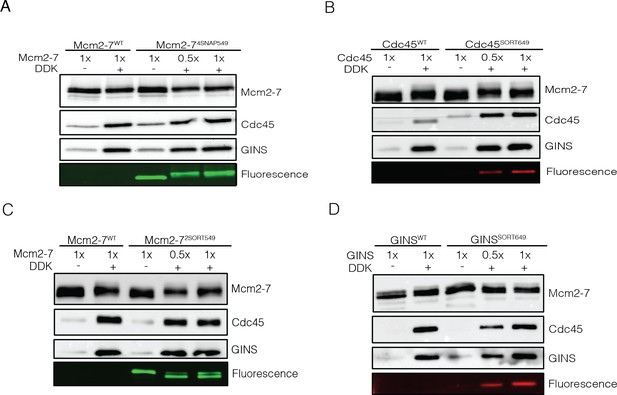
Fluorescently labeled proteins function at wild-type levels in ensemble Cdc45-Mcm2-7-GINS (CMG)-formation assays.
Mcm2-7 was loaded onto bead-coupled 3.8 kb plasmids and subsequently phosphorylated with Dbf4-dependent kinase (DDK). The phosphorylation reaction mix was removed without washing the beads prior to addition of helicase-activation proteins. Beads were washed with high-salt buffer, and Mcm2-7 proteins, Cdc45, and GINS were detected by immunoblot. Omission of DDK was used as a control for non-specific DNA binding of Cdc45 and GINS. (A) Mcm2-74SNAP549 does not interfere with Mcm2-7 loading or Cdc45/GINS recruitment. (B) Cdc45SORT649 does not interfere with Cdc45 or GINS recruitment to CMG. (C) Mcm2-72SORT549 does not interfere with Mcm2-7 loading or Cdc45/GINS recruitment. (D) GINSSORT649 does not hinder Cdc45 or GINS recruitment to CMG.
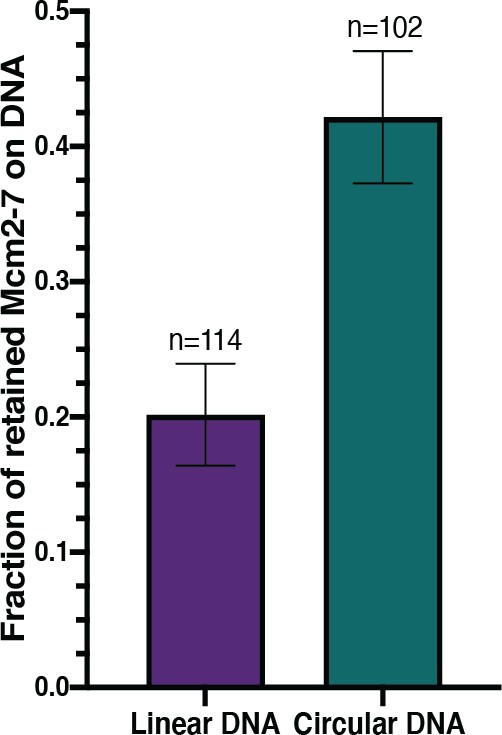
Circular DNA results in a higher number of retained Mcm2-7 during helicase loading.
Bar graphs indicate the fraction (± SE) of n DNA molecules with retained Mcm2-7 during helicase loading. Origin-containing linear (purple) and circular (green) DNA are plotted.
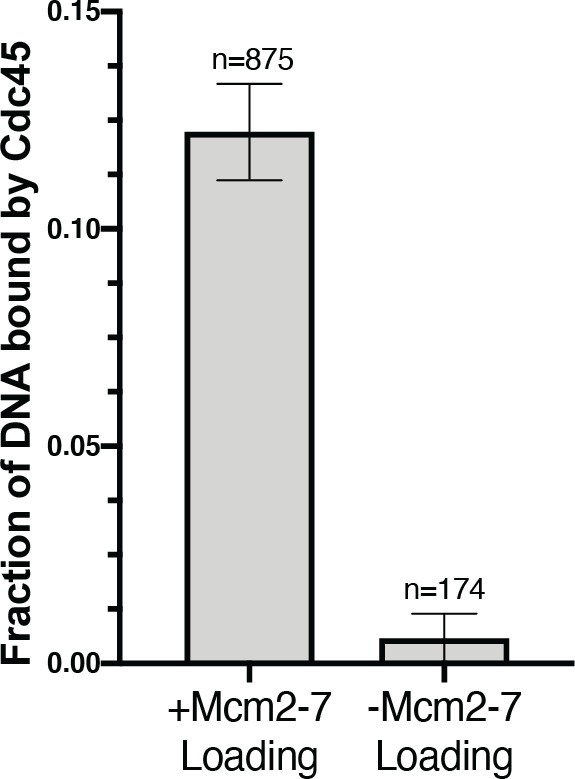
Cdc45 binds DNA in a Mcm2-7-loading-dependent manner.
The fraction (± SE) of n DNA molecules with bound Cdc45SORT649 after the HSW2 high-salt wash is plotted. Cdc6 was omitted from the −Mcm2-7 loading reaction (right bar).
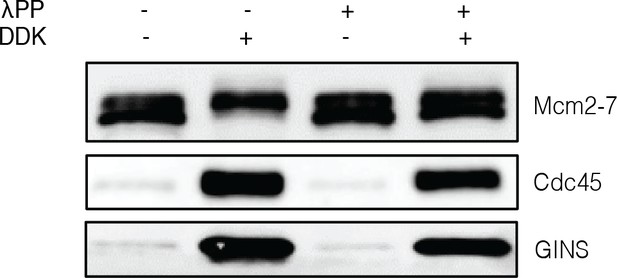
Phosphatase-treated Mcm2-7 remains functional after treatment with phosphatase.
Ensemble Cdc45-Mcm2-7-GINS-formation assays were performed as described in Materials and methods. Immunoblots for Mcm2-7, Cdc45, and GINS after HSW2 are shown. As indicated, Mcm2-7 was treated with lambda phosphatase and/or Dbf4-dependent kinase as described in Materials and methods.
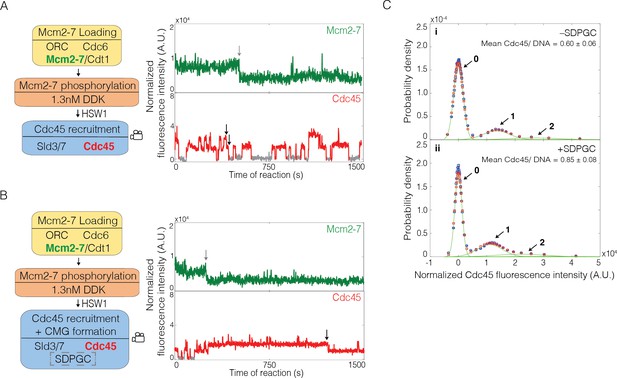
Sld2, Dpb11, Pol ε, GINS, and S-CDK (SDPGC) increases the frequency of Cdc45 binding events to the Mcm2-7 double hexamer.
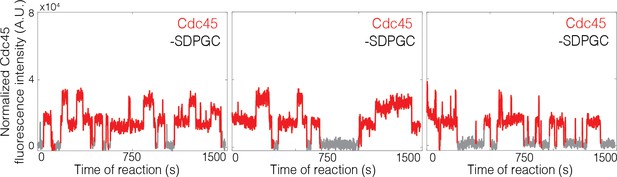
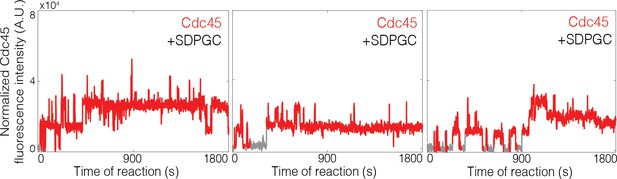
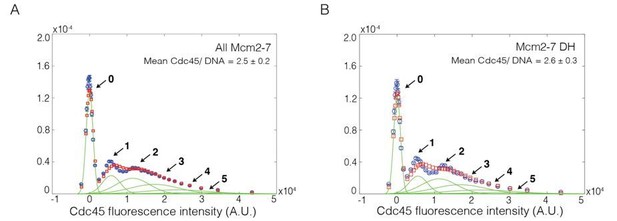
(A) Experiment protocol and representative fluorescence-intensity record for Mcm2-74SNAP549 and Cdc45SORT649 at an origin-DNA location with minimal set of proteins required to recruit Cdc45. An objective image-analysis algorithm (Friedman and Gelles, 2015) detected a spot of protein fluorescence at time points shown in red or green. Photobleaching of Mcm2-74SNAP549 is marked by a gray arrow, and a set of likely dissociations of Cdc45SORT649 molecules are marked by black arrows. (B) Experiment protocol and representative fluorescence-intensity record for Mcm2-74SNAP549 and Cdc45SORT649 at an origin-DNA location with all the factors required for Cdc45-Mcm2-7-GINS (CMG) formation (+ SDPGC = Sld2, Dpb11, Pol ε, GINS, and S-CDK). Photobleaching of Mcm2-74SNAP549 is marked by a gray arrow, and a potential dissociation of Cdc45SORT649 molecules is marked by a black arrow. (C) Multiple Cdc45SORT649 molecules bind to Mcm2–74SNAP549-bound DNA. Cdc45SORT649 fluorescence-intensity histograms (blue) are shown for two conditions: – SDPGC (i) and + SDPGC (ii). The histogram data were fit to a sum-of-Gaussians model (red; see Materials and methods). Fit parameters and calculated area fractions of individual Gaussian components (green) corresponding to the presence of the indicated numbers of Cdc45SORT649 molecules are given in Supplementary file 1a. The mean (±SE) numbers of Cdc45 molecules per DNA are calculated from the fit parameters and the fraction Cdc45 labeled. In all three panels, intensity values were background corrected and normalized as described in Materials and methods.
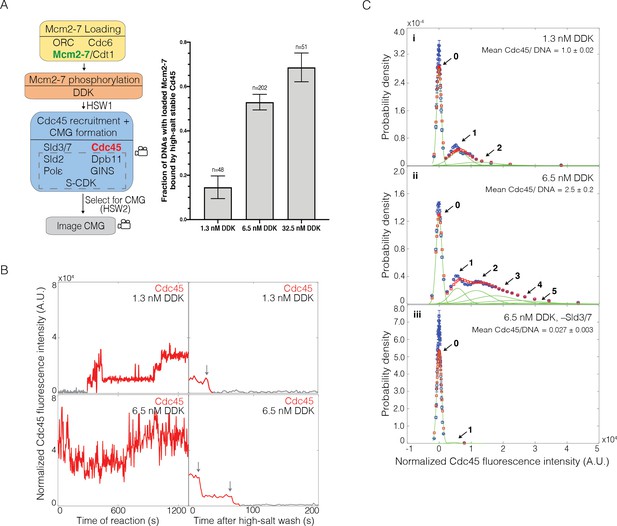
Dbf4-dependent kinase (DDK) modulates the number of Cdc45 binding events and Cdc45-Mcm2-7-GINS (CMG) formation efficiency.
(A) Increasing DDK concentration results in more efficient CMG formation. The fraction (± SE) of n DNA molecules with loaded Mcm2-74SNAP549 that were bound by high-salt-stable Cdc45SORT649 are reported for reactions using 1.3 nM, 6.5 nM, or 32.5 nM DDK. Experimental protocol was as in Figure 1A. The data from 6.5 nM DDK condition is the same as Figure 1B, left bar. (B) Representative fluorescence-intensity records for Cdc45SORT649 under 1.3 nM and 6.5 nM DDK concentrations. Left panel is during CMG formation and right panel is after high-salt wash (HSW2). Points colored as in Figure 2B. Photobleaching of Cdc45SORT649 after the HSW2 is marked by gray arrows. Fluorescence intensities after HSW2 were adjusted to account for the higher excitation laser intensity used during photobleaching. (C) Multiple Cdc45SORT649 molecules bind toMcm2-74SNAP549. Cdc45SORT649 fluorescence-intensity histograms (blue) are shown for three conditions: (i) 1.3 nM DDK; (ii) 6.5 nM DDK; and (iii) 6.5nM DDK in the absence of Sld3/7. The histogram data were fit to a sum-of-Gaussians model (red; see Materials and methods). Fit parameters and calculated area fractions of individual Gaussian components (green) corresponding to the presence of the indicated numbers of Cdc45SORT649 molecules are given in Supplementary file 1a. The mean (± SE) numbers of Cdc45 molecules per DNA are calculated from the fit parameters and the fraction Cdc45 labeled.
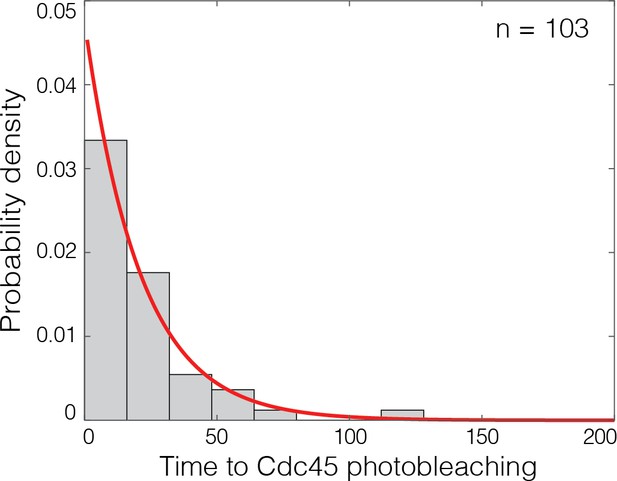
Cdc45SORT649 photobleaching events are temporally resolved.
Distribution of time intervals until photobleaching for Cdc45SORT649 bound to DNA with loaded Mcm2-7 prepared at 6.5 nM Dbf4-dependent kinase, as exemplified by the lower-right record in Figure 3B. For records that showed two separate photobleaching steps, the interval was taken to be the time from the first to the second bleaching step. Exponential fit (line) yielded time constant τ = 21 s. This implies that only a small fraction (less than exp (−3 τ Δt)=7%) of sequential photobleaching steps were mistakenly counted as one rather than two steps based on the conservative assumption that all steps separated by three or more Δt = 2.6 s video frame intervals were resolved.
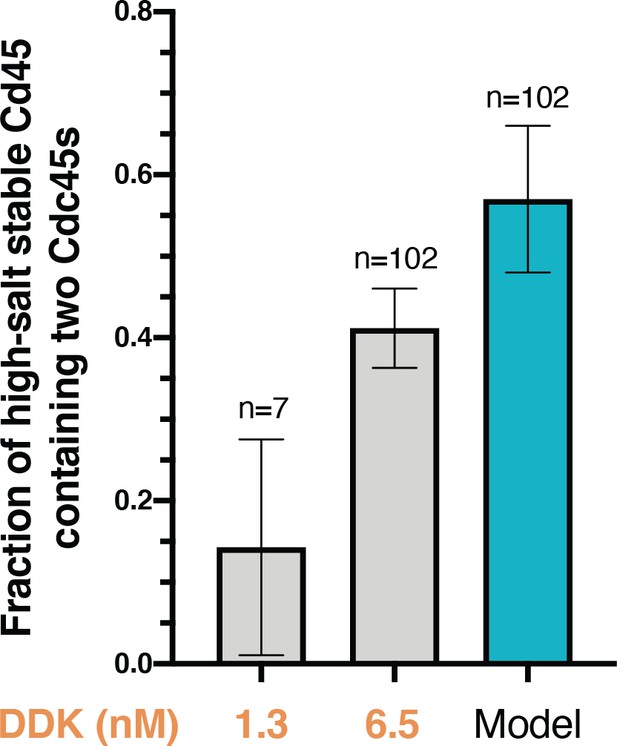
Cdc45-Mcm2-7-GINS (CMG) formation on each Mcm2-7 in the double hexamer can occur independently.
The fraction (± SE) of n high-salt-stable Cdc45 SORT649 molecules that contained two Cdc45SORT649 after the HSW2 high-salt wash is plotted for conditions with 1.3 nM and 6.5 nM Dbf4-dependent kinase (gray bars). A model calculating the amount of bound high-salt-resistant Cdc45SORT649 that should include two labeled Cdc45s if CMG formation always occurred on both Mcm2-7 molecules in a double hexamer is shown (teal bar).
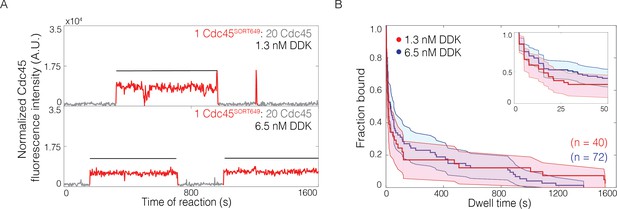
The lifetimes of individual bound Cdc45 molecules are not significantly changed by Dbf4-dependent kinase (DDK) levels.
(A) Representative fluorescence-intensity records for Cdc45SORT649 under conditions in which only a small fraction (0.036 ± 0.004) of Cdc45 molecules were fluorescently labeled at 1.3 nM (top) and 6.5 nM (bottom) DDK. Points colored as in Figure 2B. Black lines indicates single Cdc45SORT649 binding. (B) Survival function for Cdc45SORT649 dwell times on Mcm2-7-bound DNA. The vertical axis represents the fraction of Cdc45SORT649 molecules that remain bound after the dwell interval indicated on the horizontal axis. Shaded areas represent the 95% confidence intervals for each curve. Inset: magnified view.
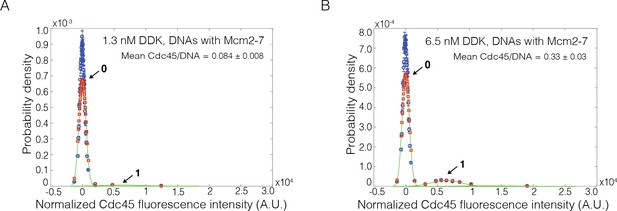
Reducing fraction of labeled Cdc45 results in the detection of individual binding events.
(A, B) Cdc45-Mcm2-7-GINS formation assays were performed with 3.6% of Cdc45 molecules being fluorescently labeled. Cdc45SORT649 fluorescence-intensity histograms (blue) are shown. The histogram data were fit to a sum-of-Gaussians model (red; see Materials and methods). Fit parameters and calculated area fractions of individual Gaussian components (green) corresponding to the presence of the indicated numbers of Cdc45SORT649 molecules are given in Supplementary file 1a. The mean (± SE) numbers of Cdc45 molecules per DNA are calculated from the fit parameters and the fraction Cdc45 labeled. (A) Reactions with 1.3 nM Dbf4-dependent kinase (DDK) on Mcm2-7-bound DNA molecules. (B) Reactions with 6.5 nM DDK on Mcm2-7-bound DNA molecules.
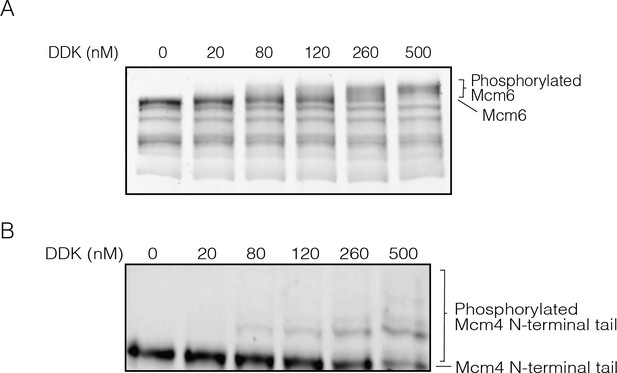
Higher Dbf4-dependent kinase (DDK) concentrations result in higher numbers of phosphorylation events on Mcm6 and Mcm4.
(A) Phosphorylation of loaded wild-type Mcm2-7 with increasing concentrations of DDK. After loading onto DNA beads, Mcm2-7 complexes were phosphorylated with the indicated concentrations of DDK. Phosphorylated Mcm2-7 was separated on a 10% SuperSep Phos-tag gel and stained with Krypton Protein Stain to separate Mcm2-7 subunits with different numbers of phosphorylation sites. (B) The Mcm4 N-terminal tail fused to SNAP-tag protein was phosphorylated with increasing concentrations of DDK. The phosphorylated Mcm4 N-terminal tail was separated on a 10% SuperSep Phos-tag gel and stained with Krypton Protein Stain.
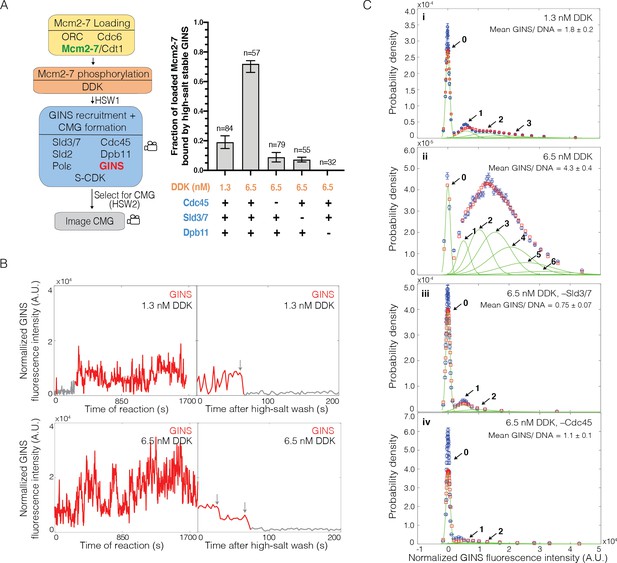
Multiple GINS bind to Mcm2-7.
(A) Increasing Dbf4-dependent kinase (DDK) concentration results in more efficient Cdc45-Mcm2-7-GINS (CMG) formation that is dependent on the presence of Cdc45, Sld3/7, and Dpb11. Left: schematic representation of the experiment. Right: graph showing the fraction of n Mcm2-72SORT549-bound DNA molecules that were bound by high-salt-stable GINSSORT649 under the indicated conditions. (B) Representative GINSSORT649 fluorescence-intensity records at two DNA molecules in the presence of 1.3 nM or 6.5 nM DDK. Left panel is during CMG formation and right panel is the same molecule after the second high-salt wash (HSW2). Points colored as in Figure 2B. Photobleaching of GINSSORT649 after HSW2 is marked by gray arrows. (C) GINSSORT649 fluorescence-intensity histograms (blue) are shown for four conditions: (i) 1.3 nM DDK; (ii) 6.5 nM DDK; (iii) 6.5 nM DDK in the absence of Sld3/7; and (iv) 6.5 nM DDK in the absence of Cdc45. The histogram data were fit to a sum-of-Gaussians model (red; see Materials and methods). Fit parameters and calculated area fractions of individual Gaussian components (green) corresponding to the presence of the indicated numbers of GINSSORT649 molecules are given in Supplementary file 1b. The mean (± SE) numbers of GINS molecules per DNA are calculated from the fit parameters and the fraction GINS labeled.
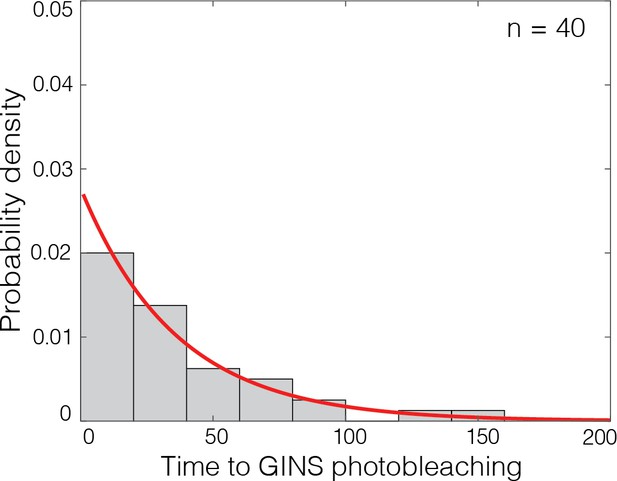
Photobleaching GINSSORT649 events are temporally resolved.
Distribution of time intervals until photobleaching for GINSSORT649 bound to DNA with loaded Mcm2-7 prepared at 6.5 nM Dbf4-dependent kinase, as exemplified by the lower-right record in Figure 5B. For records that showed two separate photobleaching steps, the interval was taken to be the time from the first to the second bleaching step. Exponential fit (line) yielded time constant τ = 36 s. This implies that only a small fraction (less than exp (−τ/(3Δt))=1%) of sequential photobleaching steps were mistakenly counted as one rather than two steps, based on the conservative assumption that all steps separated by three or more Δt = 2.6 s video frame intervals were resolved.
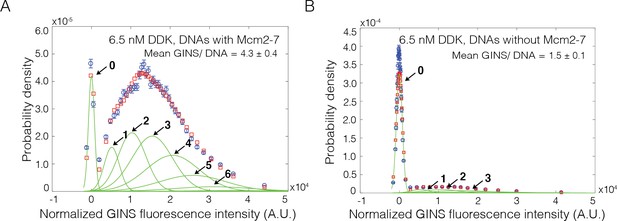
Mcm2-72SORT549 dependence of GINSSORT649 binding.
GINSSORT649 fluorescence-intensity histograms (blue) from the same recording on (A) Mcm2-72SORT549-bound DNA (same data as Figure 5Cii) and (B) DNA molecules without bound Mcm2-72SORT549. The histogram data were fit to a sum-of-Gaussians model (red; see Materials and methods). Fit parameters and calculated area fractions of individual Gaussian components (green) corresponding to the presence of the indicated numbers of GINSSORT649 molecules are given in Supplementary file 1b. The mean (± SE) numbers of GINS molecules per DNA calculated from the fit parameters and the fraction GINS labeled are indicated. Data in (B) may overestimate the amount of Mcm2-7-independent GINS binding because of the possible presence of a subpopulation of unlabeled Mcm2-7.
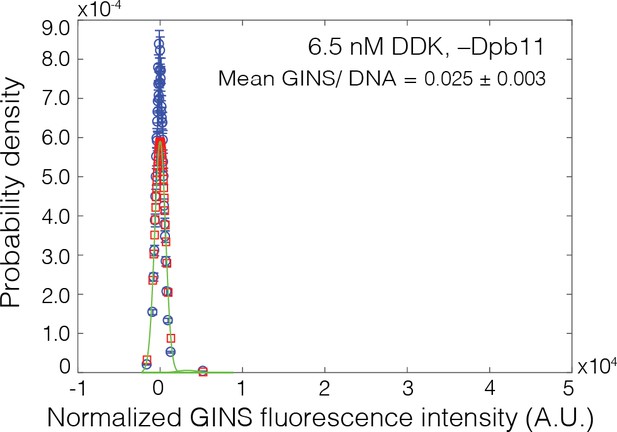
Dpb11 dependence of GINSSORT649 binding.
GINSSORT649 fluorescence-intensity histograms (blue) on Mcm2-72SORT549-bound DNA in the absence of Dpb11. The histogram data were fit to a sum-of-Gaussians model (red; see Materials and methods). Fit parameters and calculated area fractions of individual Gaussian components (green) corresponding to the presence of the indicated numbers of GINSSORT649 molecules are given in Supplementary file 1b. The mean (± SE) number of GINS molecules per DNA are calculated from the fit parameters and the fraction GINS labeled.
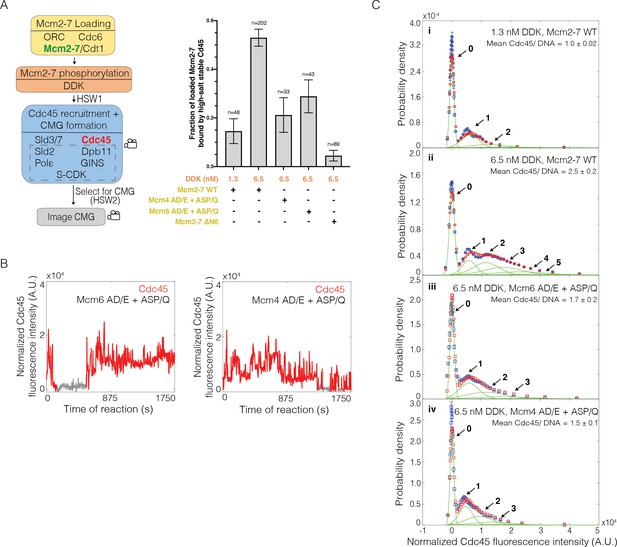
Loss of the Mcm6 N-terminal tail reduces Cdc45 binding events and Cdc45-Mcm2-7-GINS (CMG) formation.
(A) Reduction of Dbf4-dependent kinase (DDK)-dependent phosphorylation sites on either Mcm6 or Mcm4 and deletion of the Mcm6 N-terminal tail reduced high-salt-stable Cdc45 binding to Mcm2-7-bound DNAs, indicative of CMG formation. The data from 1.3 nM and 6.5 nM DDK (first and second bars) are the same as Figure 3A. (B) Representative fluorescence-intensity records for Cdc45SORT649 binding to Mcm2–74SNAP549–6AD/E+ASP/Q- and Mcm2-72SORT549–4AD/E+ASP/Q-bound DNA during CMG formation at 6.5 nM DDK. Points colored as in Figure 2B. (C) Binding of Cdc45SORT649 to Mcm2–74SNAP549–6AD/E+ASP/Q- and Mcm2-72SORT549–4AD/E+ASP/Q-bound DNA. Cdc45SORT649 fluorescence-intensity histograms (blue) are shown for four conditions: (i) 1.3 nM DDK with wild-type (WT) Mcm2-74SNAP549, (ii) 6.5 nM DDK with WT Mcm2-74SNAP549, (iii) 6.5 nM DDK with Mcm2-74SNAP549–6AD/E+ASP/Q, and (iv) 6.5 nM DDK with Mcm2-72SORT549–4AD/E+ASP/Q. The data from 1.3 nM (i) and 6.5 nM (ii) DDK are the same as Figure 3C. Fit parameters and calculated area fractions of individual Gaussian components (green) corresponding to the presence of the indicated numbers of Cdc45SORT649 molecules are given in Supplementary file 1a. The mean (± SE) number of Cdc45 molecules per DNA are calculated from the fit parameters and the fraction Cdc45 labeled.
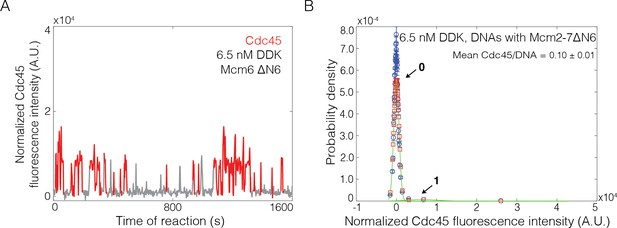
Loss of the Mcm6 N-terminal tail reduces Cdc45 binding events.
(A) Representative fluorescence-intensity record for Cdc45SORT649 binding to Mcm2-74SNAP549-6ΔN-bound DNA during Cdc45-Mcm2-7-GINS formation at 6.5 nM Dbf4-dependent kinase. Points colored as in Figure 2B. (B) Binding of Cdc45SORT649 to Mcm2-74SNAP5496ΔN-bound DNA. Fit parameters and calculated area fractions of individual Gaussian components (green) corresponding to the presence of the indicated numbers of Cdc45SORT649 molecules are given in Supplementary file 1a. The mean (± SE) number of Cdc45 molecules per DNA are calculated from the fit parameters and the fraction Cdc45 labeled.
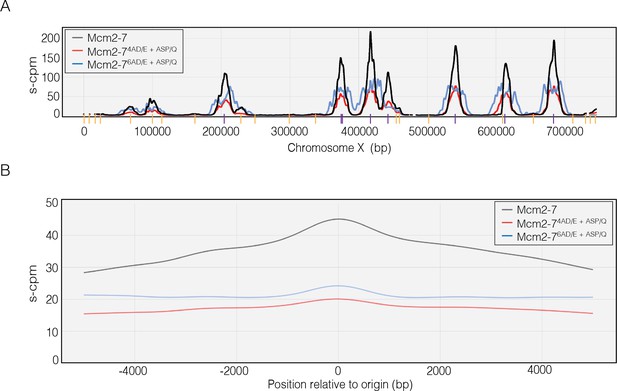
Dbf4-dependent kinase (DDK)-dependent phosphorylation sites on the N-terminal tails of Mcm4 and Mcm6 promote origin initiation.
(A) Impact of Mcm4 and Mcm6 N-terminal DDK phosphosite mutations at specific origins of replication. Normalized sequence read numbers are reported for 5-bromo-2-deoxyuridine (BrdU)-labeled DNAs across the length of Saccharomyces cerevisiae chromosome X for strains expressing wild-type Mcm2–7 (yML514), Mcm2-74AD/E+ASP/Q (yML512), or Mcm2-76AD/E+ASP/Q (yML513). Known origins of replication are represented by vertical lines at the bottom of the plot. Early origins are represented with purple vertical lines (origins with time of replication <20 min; Yabuki et al., 2002; Siow et al., 2012), whereas non-early origins are represented by yellow vertical lines. (B) Impact of Mcm4 and Mcm6 N-terminal DDK phosphorylation site mutations across all origins. The average BrdU sequence read depth (s-cpm) for strains expressing wild-type Mcm2–7, Mcm2-74AD/E+ASP/Q, or Mcm2-76AD/E+ASP/Q is reported for DNA sequences flanking all origins of replication. Origins were aligned with respect to the midpoint of each origin.
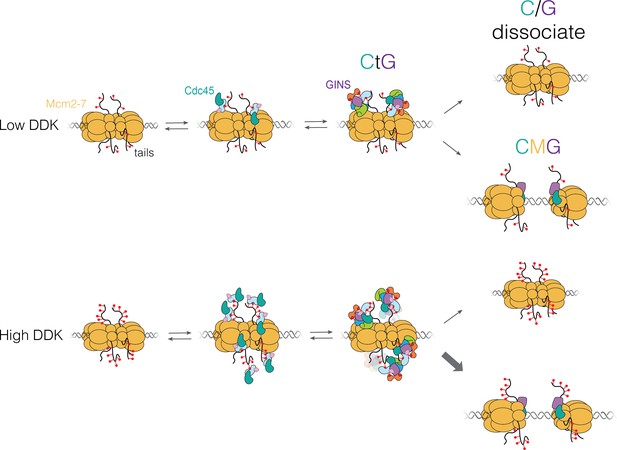
Proposed model for Cdc45-Mcm2-7-GINS (CMG) formation.
Dbf4-dependent kinase (DDK) levels control the amount of Mcm4 and Mcm6 N-terminal tail phosphorylation. These modifications indirectly (via Sld3/7) recruit Cdc45 and GINS, forming the Cdc45-tail-GINS (CtG) intermediate. The CtG intermediate can then follow one of two paths: (i) dissociation or (ii) deposition onto the structured core of the Mcm2-7 helicase to form the CMG. We hypothesize that the rate of this conversion is the same for any CtG, but it is infrequent. Thus, having more CtGs (e.g., at higher DDK levels) increases the probability of CMG formation, which we assume is irreversible. Relevant proteins are labeled in the illustration; red lollipops represent phosphorylations. C/G: Cdc45-GINS.
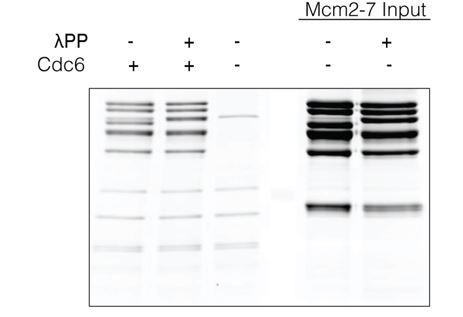
Lambda phosphatase (λPP) does not affect Mcm2-7 loading.
Ensemble Mcm2-7 loading assays were conducted in the presence and absence of λPP. A loading control in the absence of Cdc6 is included as well as inputs for Mcm2-7. Note that the Mcm2 protein migrates more slowly after dephosphorylation.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Saccharomyces cerevisiae) | yCKR006 | This study | MATa ade2-1 can1-100 pep4Δ(unmarked) bar1Δ(unmarked) HIS3::pSKM004 (GAL1,10-MCM2,Flag-MCM3), URA3::pALS1 (GAL1,10-CDT1, GAL4), LYS2::pST022 (GAL1,10-MCM4-SNAP, MCM5), TRP1::pCKR006 (GAL1,10-MCM6-N-termΔ, MCM7) | Mcm2-74SNAP-6ΔN expression strain. Additional information in Materials and methods. Available upon requestfrom Bell Lab. |
| Strain, strain background (Saccharomyces cerevisiae) | yCKR009 | This study | MATa ade2-1 can1-100 pep4Δ(unmarked) bar1Δ(unmarked)HIS3::pSKM004 (GAL1,10-MCM2, Flag-MCM3), URA3::pALS1 (GAL1,10-CDT1, GAL4), LYS2::pST022 (GAL1,10-MCM4-SNAP, MCM5),TRP1::pCKR009 (GAL1,10-MCM6-AD/E+ASP/Q, MCM7) | Mcm2-74SNAP-6AD/E+ASP/Q expression strain. Additional information in Materials and methods. Available upon request from Bell Lab. |
| Strain, strain background (Saccharomyces cerevisiae) | yCKR047 | This study | MATa ade2-1 can1-100 pep4Δ(unmarked) bar1Δ(unmarked) TRP1::pSKM003 (GAL1,10-MCM6,MCM7), URA3::pALS1 (GAL1,10-DT1,GAL4),HIS3::pST048 (GAL1,10-MCM2-LPETGG, Flag-MCM3), LYS2::pCKR022 (GAL1,10-MCM4-AD/E+ASP/Q, MCM5) | Mcm2-72SORT-4AD/E+ASP/Q expression strain. Additional information in Materials and methods. Available upon request from Bell Lab. |
| Strain, strain background (Saccharomyces cerevisiae) | yMM034 | This study | MATa ade2-1, trp1-1, leu2-3,112, his3-11,15 ura3-1, can1-100, bar1Δ (unmarked), LYS2:: HisG, pep4Δ (unmarked), LEU2:: (CDC45-3xFlag-LPETGG) | Cdc45SORT expression strain. Additional information in Materials and methods. Available upon request from Bell Lab. |
| Strain, strain background (Saccharomyces cerevisiae) | yST179 | Ticau et al., 2017 | MATa ade2-1 can1-100 pep4Δ(unmarked) bar1Δ(unmarked)TRP1::pSKM003 (GAL1,10-MCM6,MCM7),URA3::pALS1 (GAL1,10-Cdt1,GAL4), HIS3::pST036 (GAL1,10-SORT-MCM2,Flag-MCM3), LYS2::pSKM002 (GAL1,10-MCM4, MCM5) | Mcm2-72SORT expression strain. |
| Strain, strain background (Saccharomyces cerevisiae) | yST147 | Ticau et al., 2017 | MATa ade2-1 can1-100 pep4Δ(unmarked) bar1Δ(unmarked) TRP1::pSKM003 (GAL1,10-MCM6,MCM7), HIS3::pSKM004 (GAL1,10-MCM2,Flag-MCM3), LYS2::pST022 (GAL1,10-fSNAP-MCM4, MCM5), URA3::pALS1 (GAL1,10-CDT1, GAL4) | Mcm2-74SNAP expression strain. |
| Strain, strain background (Saccharomyces cerevisiae) | yML512 | This study, Randell et al., 2010 | MatA, ade2-1, ura3-11, his3-11,15, can-100, mcm4Δ::hisG TRP1::pJR140 (MCM4-HA/HIS-AD/E ASP/Q) LEU2::p405-BrdU-Inc | Additional information in Materials and methods. Available upon request from Bell Lab. |
| Strain, strain background (Saccharomyces cerevisiae) | yML513 | This study, Randell et al., 2010 | MatA, ade2-1, ura3-11, his3-11,15, can-100, mcm4Δ::hisG TRP1::pJR149 (MCM6-HA/HIS-AD/E ASP/Q) LEU2::p405-BrdU-Inc | Additional information in Materials and methods. Available upon requestfrom Bell Lab. |
| Strain, strain background (Saccharomyces cerevisiae) | yML514 | This study, Randell et al., 2010 | MatA, ade2-1, ura3-11, his3-11,15, can-100, mcm4Δ::hisG TRP1::pJR121 (MCM4-HA/HIS-AD/E ASP/Q)LEU2::p405-BrdU-Inc | Additional information in Materials and methods. Available upon request from Bell Lab. |
| Recombinant DNA reagent | pSNAP-tag (T7)-2 | NEB | N9181S, RRID:Addgene_90314 | Vector that can be used for expression of SNAP-protein alone (20 kDa). |
| Recombinant DNA reagent | pUC19-ARS1 | Heller et al., 2011 | pUC19 (ARS1) | Used in ensemble CMG formation assay and to construct the SM templates. |
| Recombinant DNA reagent | pCKR006 | This study | pSK003 (GAL1,10-MCM6-N-termΔ, MCM7) | Additional information in Materials and methods. Available upon request from Bell Lab. |
| Recombinant DNA reagent | pCKR009 | This study | pSKM003 (GAL1,10-MCM6-AD/E+ASP/Q, MCM7) | Additional information in Materials and methods. Available upon request from Bell Lab. |
| Recombinant DNA reagent | pCKR022 | This study | pSKM002 (GAL1,10-MCM4-AD/E+ASP/Q, MCM5) | Additional information in Materials and methods. Available upon requestfrom Bell Lab. |
| Recombinant DNA reagent | pCKR032 | This study | pMM033 (CDC45-3xFlag-LPETGG) | Additional information in Materials and methods. Available upon request from Bell Lab. |
| Recombinant DNA reagent | pLDK03 | This study | pFJD5 (6XHIS-PreScission Protease site-Psf3-LPETGG) | Used to express GINSSORT649 in bacteria. Additional information in Materials and methods. Available upon requestfrom Bell Lab. |
| Recombinant DNA reagent | pLDK04 | This study | pSNAP-tag (T7)−2 (8xARG-Mcm4 N-terminal tail-SNAP) | Used to express the Mcm4 N-terminal tail fused to the SNAP tag in bacteria. Additional information in Materials and methods. Available upon requestfrom Bell Lab. |
| Recombinant DNA reagent | p405-BrdU-Inc | Viggiani and Aparicio, 2006 | p405-BrdU-Inc (LEU2, BrdU Incorporation), RRID:Addgene_71791 | Plasmid that allows yeast cells to incorporate BrdU. |
| Sequence-based reagent | Oligo for 1.2 kb circular template | IDT | 5′-AATTAGCGGCCGCAAGGC/iBiodT/GATTAAGTT-3′ | NotI site in bold. |
| Sequence-based reagent | Oligo for 1.2 kb circular template | IDT | 5′-ATTAAGCGGCCGCAGCGGA/iAlexa488N/TAACAATTT-3′ | NotI site in bold. |
| Sequence-based reagent | Oligo for 1.1 kb unwinding template | IDT | 5′- /5Cy5/TACGCCAAGCTTGCATGCGGATGTTGC −3′ | Part of NotI sticky end in bold. |
| Sequence-based reagent | Oligo for 1.1 kb unwinding template | IDT | 5′- /5Phos/GGCCGCAACATCCGCATGCAAGCTTGGCGTA/3BHQ2/ −3′ | Part of NotI sticky end in bold. |
| Peptide, recombinant protein | Peptide for coupling to dyes for Sortase labeling | Ticau et al., 2017 | NH2-GGGHHHHHHHHHHC-COOH | |
| Chemical compound, drug | DY549-P1 | Dyomics | Dyomics: 549P1-03 | Maleimide-coupled fluorescent dye |
| Chemical compound, drug | DY649-P1 | Dyomics | Dyomics: 649P1-03 | Maleimide-coupled fluorescent dye |
| Chemical compound, drug | SNAP-Surface 549 | NEB | Fluorescent substrate used to label SNAP-tag fusion proteins (Mcm2-) | |
| Antibody | Anti-Mcm2-7, rabbit polyclonal | Bell Lab | UM174 | (1:10,000) |
| Antibody | Anti-Cdc45, rabbit polyclonal | Lõoke et al., 2017 | HM7135 | (1:2000) |
| Antibody | Anti-GINS, rabbit polyclonal | Lõoke et al., 2017 | HM7128 | (1:2000) |
| Gene (Saccharomyces cerevisiae) | MCM2 | Saccharomyces Genome Database | SGD: S000000119 | |
| Gene (Saccharomyces cerevisiae) | MCM3 | Saccharomyces Genome Database | SGD: S000000758 | |
| Gene (Saccharomyces cerevisiae) | MCM4 | Saccharomyces Genome Database | SGD: S000006223 | |
| Gene (Saccharomyces cerevisiae) | MCM5 | Saccharomyces Genome Database | SGD: S000004264 | |
| Gene (Saccharomyces cerevisiae) | MCM6 | Saccharomyces Genome Database | SGD: S000003169 | |
| Gene (Saccharomyces cerevisiae) | MCM7 | Saccharomyces Genome Database | SGD: S000000406 | |
| Gene (Saccharomyces cerevisiae) | CDC45 | Saccharomyces Genome Database | SGD: S000004093 | |
| Gene (Saccharomyces cerevisiae) | SLD5 | Saccharomyces Genome Database | SGD: S000002897 | |
| Gene (Saccharomyces cerevisiae) | PSF1 | Saccharomyces Genome Database | SGD: S000002420 | |
| Gene (Saccharomyces cerevisiae) | PSF2 | Saccharomyces Genome Database | SGD: S000003608 | |
| Gene (Saccharomyces cerevisiae) | PSF3 | Saccharomyces Genome Database | SGD: S000005506 | |
| Software, algorithm | Matlab | Mathworks | The 'intervals' files are readable by the imscroll program: (https://github.com/gelles-brandeis/CoSMoS_Analysis; Gelles and Friedman, 2021; copy archived at swh:1:rev:3eec2cbfa54018389fc1905b54c4b062723a5a7f) |
Additional files
-
Supplementary file 1
Flourescence-intensity histogram fit parameters.
Supplementary file 1a. Fit parameters for Cdc45 fluorescence-intensity histograms. Supplementary file 1b. Fit parameters for GINS fluorescence-intensity histograms.
- https://cdn.elifesciences.org/articles/65471/elife-65471-supp1-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/65471/elife-65471-transrepform-v2.pdf