Endothelial junctional membrane protrusions serve as hotspots for neutrophil transmigration
Figures
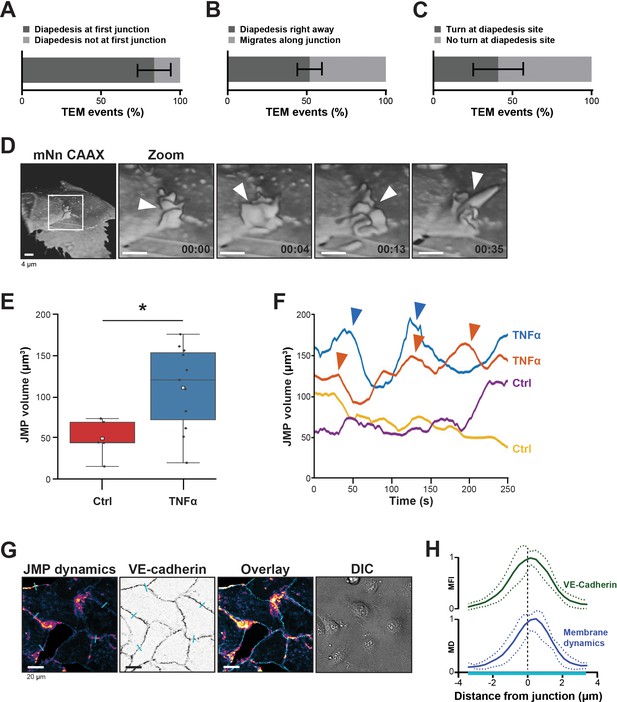
Presence of endothelial junctional membrane protrusions.
(A) Quantification in percentage of diapedesis events at first junction neutrophils encounter (dark bar) versus neutrophils that cross more than one junction before transmigrating (light bar). N=3, on average 13 TEM events per experiment. Mean ± standard deviation (SD). (B) Quantification in percentage of diapedesis events directly upon encountering a junction region (dark bar) versus neutrophils that migrate along such junction region prior to diapedesis (light bar). N=3, on average 13 TEM events per experiment. Mean ± SD. (C) Quantification in percentage of diapedesis events at junction regions for neutrophils that cross straight through (dark bar) versus neutrophils that make a turn at junction regions prior to diapedesis (light bar). N=3, on average 13 TEM events per experiment. Mean ± SD. (D) 3D-view stills from mNeonGreen-CAAX transfected HUVECs showing presence of junctional membrane protrusions, indicated by white arrowheads. Time indicated in seconds in lower right corner. Bar, 4 μm. (E) Boxplots of the volume average of 3D junctional membrane protrusions in time upon TNF-α treatment (blue) versus control (red). White square represents mean. Mann-Whitney U-test: *p=0.0275. (F) Quantification of the dynamics of JMP volume of control (yellow and purple lines) versus 20 hr TNF-α-treated (blue and orange lines) endothelial cells as indicated. two representative lines per condition are shown. JMPs from TNF-treated ECs show more fluctuations in volume than JMPs from control cells as indicated by arrowheads. (G) Junctional membrane dynamics map in pseudo-colors, warm colors indicate high membrane dynamics, cold colors indicate low membrane dynamics (i), VE-cadherin staining (ii), overlay (iii), and DIC (iiii). Turquoise lines indicate sites of line scans for quantification. Bar, 20 μm. (H) Quantification of normalized fluorescence intensity (MFI) of blue lines as indicated in E by line scan analysis of VE-cadherin (green) and normalized value from membrane dynamics map (MD) (dark blue). Green and dark blue lines represent mean of five independent junctions, dotted lines show SD.
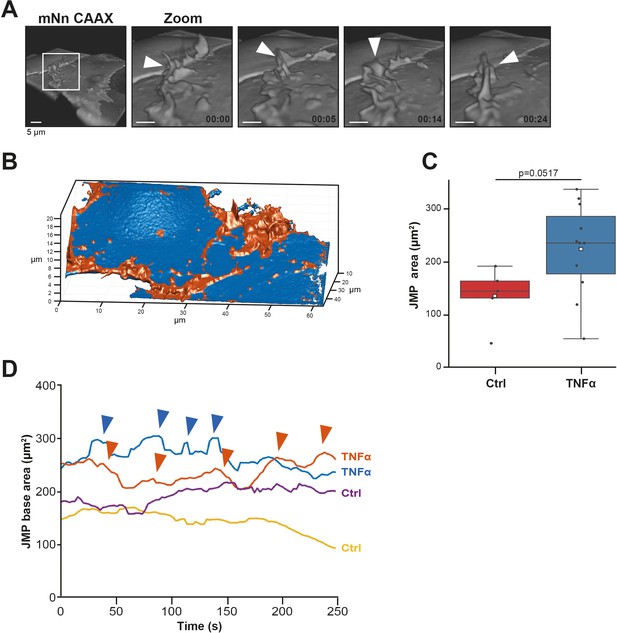
JMP dynamics.
(A) 3D view stills from mNeonGreen-CAAX-transfected HUVECs showing presence of junctional membrane protrusions, indicated by white arrowheads. Time indicated in seconds in lower right corner. Bar, 4 μm. (B) Segmentation of JMPs (orange) identified from ECs (blue) used for the quantification of the LLSM images. Numbers in μm. (C) Boxplots of the JMP area average of 3D junctional membrane protrusions in time upon TNF-α treatment (blue) versus control (red). White square represents mean. Mann-Whitney U-test: p=0.0517. (H) Quantification of the dynamics of JMP volume of control (yellow and purple lines) versus 20 hr TNF-α-treated (blue and orange lines) endothelial cells as indicated. two representative lines per condition are shown. JMPs from TNFα-treated ECs show more fluctuations in volume than JMPs from control cells as indicated by arrowheads.
Transmigration of neutrophils across TNFα-treated endothelial monolayers.
Neutrophils crawl around searching for a spot to transmigrate. Some neutrophils use same TEM spot. Total recording time is 10 min.
JMP dynamics.
3D-view time-lapse of Lattice Light Sheet Microscopy recording from mNeonGreen-CAAX transfected HUVECs showing junctional membrane protrusion activity. 3D reconstruction using Imaris blend function. Total recording time is 5 min. Bar, 5 μm.
JMP volume and area quantification of 20 hr TNFα-treated endothelial cells.
Segmentation of JMPs (orange) identified from ECs (blue) used for the quantification of the LLSM images as described in Materials and methods section.
JMP volume and area quantification of non-treated endothelial cells.
Segmentation of JMPs (orange) identified from ECs (blue) used for the quantification of the LLSM images as described in Materials and methods section.
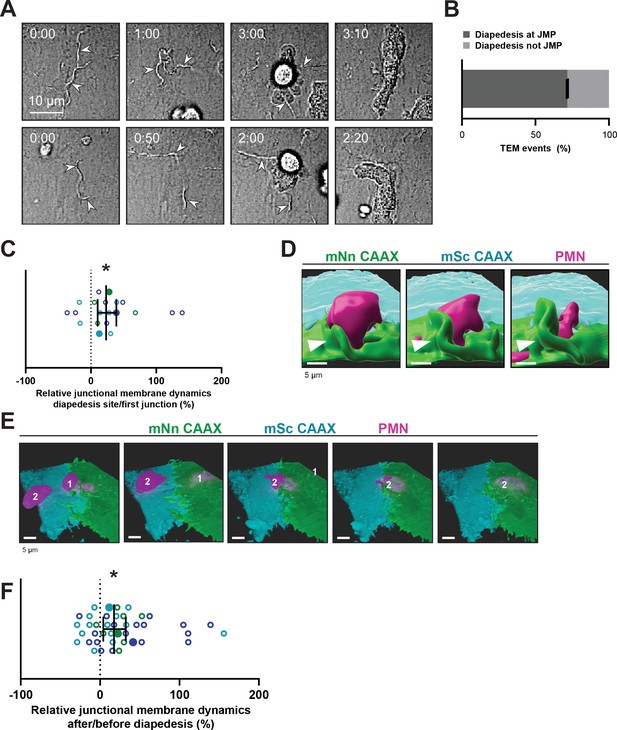
Neutrophils use junctional membrane protrusions as diapedesis site.
(A) Stills from two time-lapse movies showing PMN transendothelial migration (TEM) under flow showing presence of endothelial membrane ruffles, indicated by white arrowheads. The membrane ruffles are present at the site of diapedesis already before PMN adhesion and indicate the spot where the PMN will breach the EC layer. Bar, 10 μm. (B) Quantification of the number of TEM events that show elevated junctional membrane dynamics prior to PMN TEM at site of diapedesis. N=3, on average >10 TEM events per experiment. (C) Ratio membrane dynamics at diapedesis site and membrane dynamics at site where a neutrophil first encounters. Open dots are individual data points from three independent experiments (three colors). Filled dots are means from three experiments. Median with 95% confidence interval (CI) are shown. One-sample Wilcoxon test: *p=0.0305. (D) 3D image stills using Imaris rendering software from two ECs (green/turquoise) and PMN (magenta) showing junctional membrane protrusion at the diapedesis site. Bar, 5 μm. (E) 3D view image from two endothelial cells (green/turquoise) and PMN (magenta, labeled #1) showing a second PMN (labeled #2) transmigrating at the same diapedesis site as the first neutrophil (#1). Bar, 5 μm. (F) Ratio JMPs at the diapedesis after and before TEM showing an increase in endothelial membrane dynamics after diapedesis. Open dots are individual data points from three independent experiments (three colors). Filled dots are means from three experiments. Median with 95% CI is shown. One-sample Wilcoxon test: *p=0.0006.
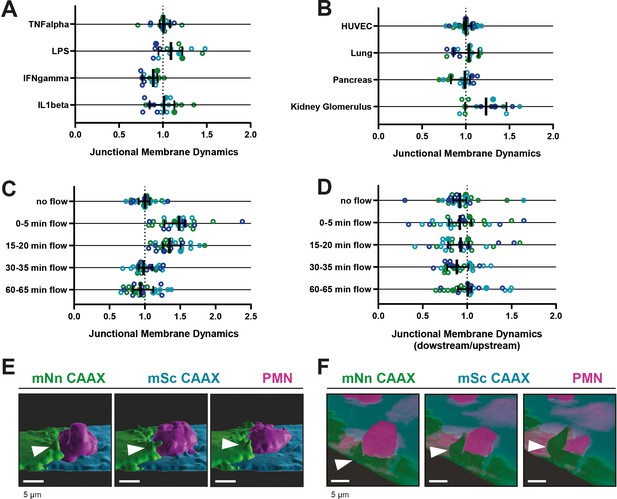
Different stimuli promote JMP dynamics.
(A–D) Junctional membrane dynamics normalized to 20 hr TNFα−stimulated HUVECs cultured on glass without shear flow. Open dots are individual data points from three independent experiments, represented by three different colors. Filled dots are means from three experiments. Median with 95% CI is shown. Dotted line represents a ratio of 1, meaning no difference. (A) JMP dynamics are determined in human umbilical cord-derived, lung-derived, pancreas-derived and kidney glomerulus-derived endothelial cells. (B) JMP dynamics on HUVECs that were treated for 20 hr with inflammatory stimuli as indicated. (C) JMP dynamics on HUVECs that were exposed to flow for different periods of time as indicated. (D) Location of JMP dynamics on HUVECs on upstream or downstream side of flow direction for different periods of time as indicated. (E) 3D-image stills from mNeonGreen-CAAX- and mScarlet-I-CAAX-transfected HUVECs showing transmigrating neutrophil (magenta). 3D reconstruction using Imaris surface rendering function. Bar, 5 μm. (F) 3D-image stills from mNeonGreen-CAAX- and mScarlet-I-CAAX-transfected HUVECs showing transmigrating neutrophil (magenta). 3D reconstruction using Imaris blend function. Bar, 5 μm.
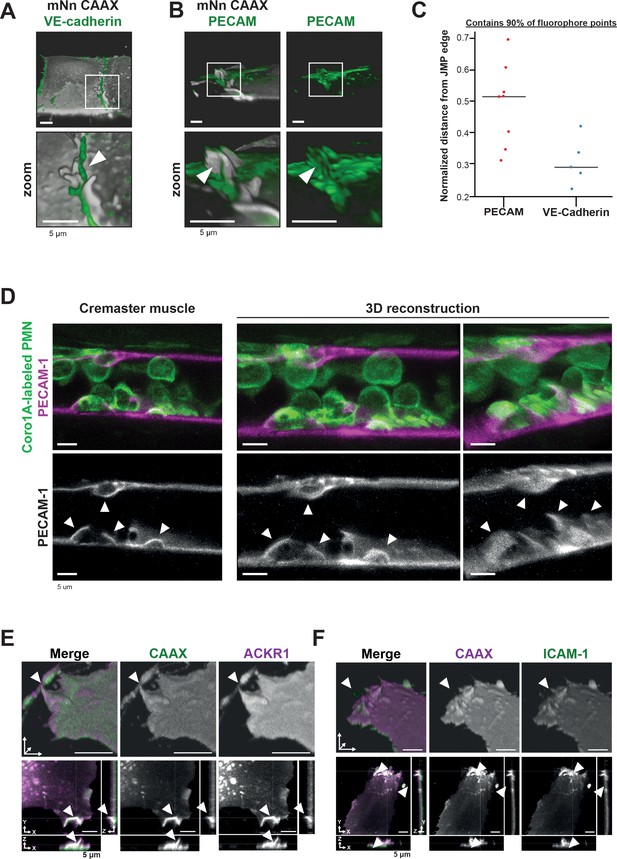
PECAM-1-positive junctional membrane protrusions express ICAM-1, ACKR1 and found in vivo.
(A) LLSM stills from HUVECs expressing membrane-bound CAAX (gray) stained with a directly conjugated VE-cadherin antibody (green). White box indicates zoom region, displayed below. Arrowhead points at JMP that does not overlap with the VE-cadherin staining. Bar, 5 μm. (B) LLSM stills from HUVECs expressing the CAAX membrane label (grays) stained with a directly conjugated PECAM-1antibody (green). White boxes indicate zoom regions. Arrowhead points at JMP that show overlap with the PECAM-1 staining. Bar, 5 μm. (C) Quantification of LLSM images of fluorescent coverage of PECAM-1 and VE-cadherin on CAAX-positive JMPs, based on 90% of all available fluorophore points on the volume measurements of the JMPs. Dots indicate one experiment in which one individual JMP is measured. (D) Confocal intravital microscopy of 20–80 μm diameter cremasteric venules in mice with Coro1A stained neutrophils in green immunostained in vivo for EC junctions by intrascrotal injections of fluorescent-labeled PECAM-1 (red) and stimulated for four hours with IL-1β and TNF-α. Fixed images in top row show presence of PECAM-1-positive membrane protrusions that surround adherent neutrophils in green. Two images on the right show reconstruction. Lower row shows PECAM-1 staining in white only. Arrows show presence of PECAM-1-positive membrane protrusions. Scale bar, 5 µm. (E) Stills from two endothelial cells expressing mScarlet-I-CAAX (green) and mNeonGreen ACKR1 (magenta) showing ACKR1-containing JMPs as indicated by white arrowheads. Of note, the endothelium is grown to a full monolayer but untransfected cells are not detected, as they do not express any FP. Bar, 5 μm. (F) Stills from two endothelial cells expressing mNeonGreen-CAAX (green) and ICAM-1 mScarlet-I (magenta) showing ICAM-1-containing JMPs as indicated by white arrowheads. Bar, 5 µm.
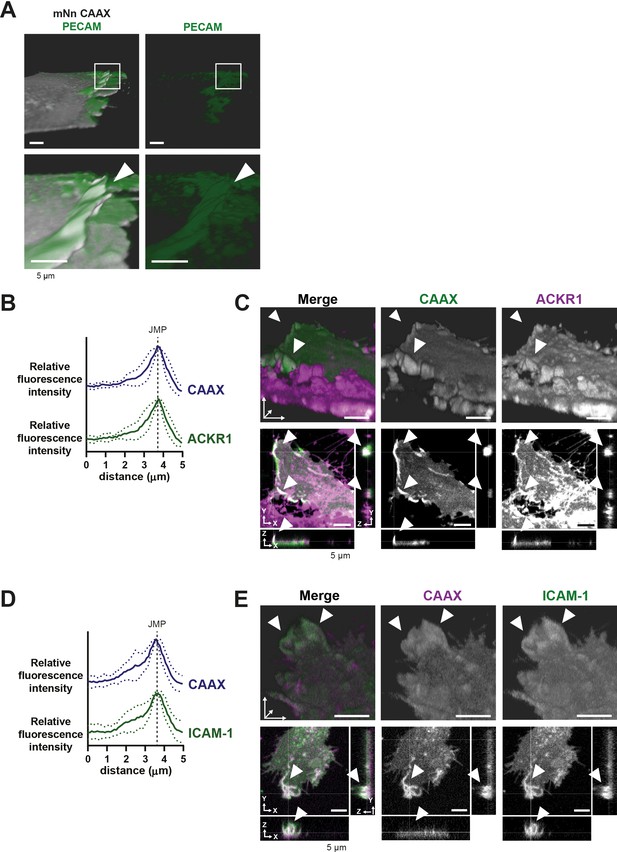
JMPs in vitro and in vivo.
(A) Stills from HUVECs expressing mNeon Green-CAAX membrane label stained with a directly conjugated PECAM-1 antibody (green). 3D reconstruction LLSM using Imaris surface rendering function. White box indicates zoom region. Arrow shows PECAM-1 coverage on JMP. Bar, 5 μm. (B) Quantification of fluorescence intensity by line scan analysis of ACKR1 (green) together with membrane marker CAAX (dark blue). (C) Stills from two endothelial cells expressing mScarlet-I-CAAX (green) and mNeonGreen-ACKR1 (magenta) showing ACKR1-containing JMPs as indicated by white arrowheads. Bar, 5 μm. (D) Quantification of fluorescence intensity by line scan analysis of ICAM-1 (green) together with membrane marker CAAX (dark blue). (E) Stills from two endothelial cells expressing mScarlet-I-CAAX (green) and ICAM-1-GFP (magenta) showing ICAM-1-containing JMPs as indicated by white arrowheads. Bar, 5 μm.

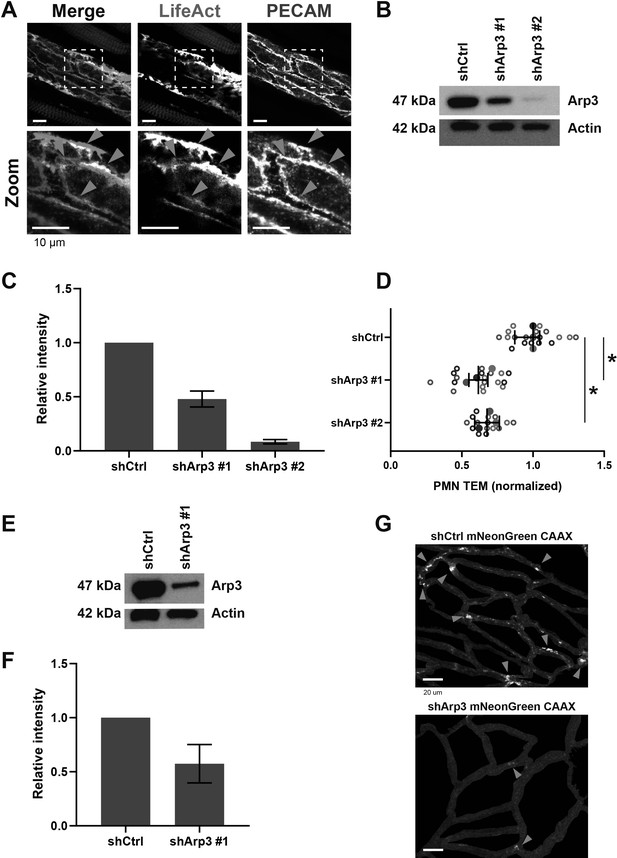
JMP regulation by the actin cytoskeleton.
(A) Stills from two ECs expressing LifeAct-mTurquoise2 showing F-actin-containing JMPs as indicated by white arrowheads. Bar, 10 μm. Images at the right show orthogonal view (XZ direction). (B) Confocal intravital microscopy of cremasteric venules in Lifeact-EGFP mice showing the vasculature after four hours with IL-1β and TNFα stimulation. Leukocyte is indicated by asterisk. Arrowheads show existence of JMPs in vivo. Lower panels show region of interest (ROI) zoom. Bar, 5 µm. (C) JMP dynamics are measured on HUVECs stimulated with TNFα that are treated with the Arp2/3 inhibitor CK-666. Ratio of JMP dynamics after/before CK-666 treatment was calculated. Data points from three independent experiments are shown in three different colors. Median with 95% CI. (D) Example images of membrane dynamics maps upon CK-666 treatment. Control image is from same cells before CK-666 treatment. Blue arrow heads indicate JMPs. Bar, 20 μm. (E) Silencing endothelial Arp3 with two independent shRNAs reduces number of TEM events. Open dots are individual data points from three independent experiments, represented by three different colors. Filled dots are means from three experiments. Median with 95% CI is shown. T-test: Ctrl/sh#1 p=0.0288, Ctrl/sh#2 p=0.0079. (F) Silencing endothelial Arp3 reduces CAAX-positive JMP dynamics. Median with 95% CI is shown. Mann-Whitney U-test: *p=0.0002.
-
Figure 4—source data 1
Arp3 knockdown.
- https://cdn.elifesciences.org/articles/66074/elife-66074-fig4-data1-v2.pdf
Arp3-mediated JMPs.
(A) Confocal intravital microscopy of cremasteric venules in Lifeact-EGFP mice fixed and stained for PECAM-1 (magenta) showing co-localization (blue arrowheads). Lower panels show region of interest (ROI) zoom. Bar, 10 µm. (B) Western blot analysis shows knockdown of Arp3 in EC with two independent shRNA constructs. Actin is shown as loading control. (C) Quantification of western blot in A. T-test: Ctrl/sh#1 p=0.0393, Ctrl/sh#2 p=0.0330. (D) Arp3-knockdown (shArp3 #1 and #2) in ECs reduces neutrophil TEM. Silencing endothelial Arp3 reduced number of TEM events. Data is normalized to control, dashed line represents ratio of 1, meaning no difference. Open dots are individual data points from three independent experiments, represented by three different colors. Filled dots are means from three experiments. Median with 95% CI is shown. T-test: Ctrl/sh#1 p=0.0183, Ctrl/sh#2 p=0.0121. (E) Western blot analysis shows successful knockdown of Arp3 in ECs expressing CAAX. Actin is shown as loading control. (F) Quantification of Western blot in A. (G) JMP dynamics map of ECs that are treated as control or with shRNA Arp3 / mNeonGreen-CAAX. Warm colors indicate increased membrane dynamics, cold colors indicate low membrane dynamics. Blue arrowheads show sites of increased membrane dynamics. Bar, 20 μm.
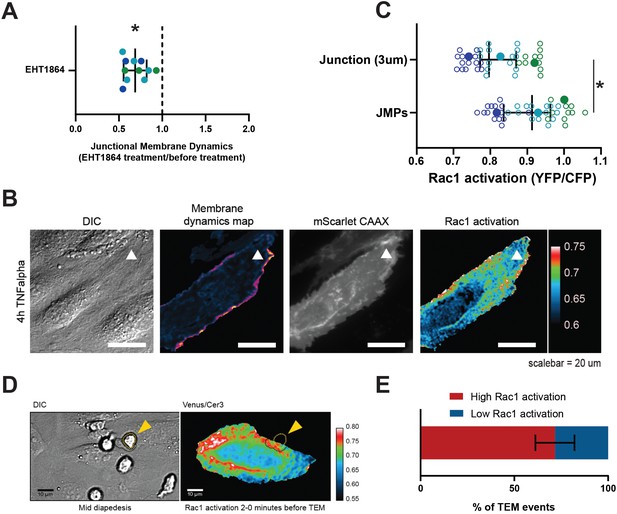
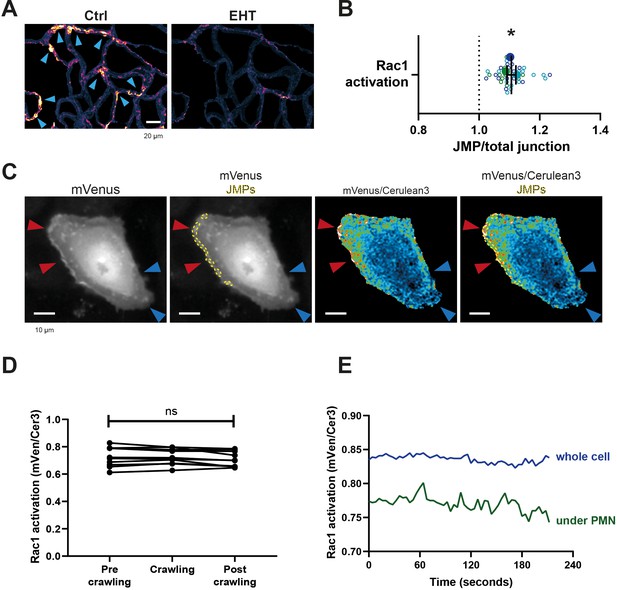
JMPs show high Rac1 activity and function as diapedesis hotspot for neutrophils.
(A) JMP dynamics are measured on HUVECs stimulated with TNFα that are treated with the Rac1 EHT1864 inhibitor. Ratio of JMP dynamics after/before EHT-1864 treatment was calculated. Data points from three independent experiments, represented with three different colors, are shown. Median with 95% CI. One-sample Wilcoxon test: *p<0.0001. (B) DIC ECs (i), membrane dynamics map in pseudo colors (ii), mScarlet-I-CAAX membrane label (iii), FRET-based Rac1 biosensor Venus/Cer3 pseudo color ratio-image (iiii) show that JMPs are correlated with high Rac1 activity (white arrowhead). Scale bar, 20 µm. Calibration bar on the right shows high FRET values in warm colors (red) and low FRET values in cold colors (blue) (C) Quantification of FRET-based Rac1 biosensor activation at JMPs, selected using mScarlet-I-CAAX, compared to the full junction region of 3 µm wide. Open dots are individual data points from three independent experiments, represented with three different colors. Filled dots are means from three experiments. Median with 95% confidence interval (CI) is shown. T-test: *p=0.0087. (D) Still from time-lapse showing PMN TEM under flow. Left image shows DIC of PMN at mid-diapedesis, indicated with yellow arrowhead. Right image shows FRET-based Rac1 biosensor pseudo-color ratio-image. Yellow dotted line indicates part of PMN at the luminal side. Dark dotted line indicates part of PMN at basolateral side. Scale bar, 10 µm. (E) Graph shows quantification of PMNs that transmigrate at high Rac1 regions (red bar) versus low Rac1 regions (blue bar).
Endogenous Rac1 activation promotes asymmetric JMPs.
(A) Example images of membrane dynamics maps upon Rac1 inhibition EHT-1864 treatment. Warm colors indicate increased membrane dynamics, cold colors indicate low membrane dynamics. Blue arrowheads show sites of increased membrane dynamics. Bar, 20 μm. (B) Quantification of FRET-based DORA Rac1 biosensor activation at JMPs versus 3 μm wide junction regions as explained under C. Dotted line indicates ratio of 1, meaning no change. Open dots are individual data points from three independent experiments, represented by three different colors. Filled dots are means from three experiments. Median with 95% CI are shown. One-sample T-test: *p=0.101. (C) Stills from Rac1 biosensor time lapse movie. mVenus (i), mVenus, JMPs indicated by yellow dotted lines (ii), Rac1 biosensor ratio image, warm colors indicate high Rac1 activation (iii), Rac1 biosensor ratio image, JMPs indicated by yellow dotted lines (iiii) Red arrowheads indicate regions of high Rac1 activation and JMPs, blue arrowheads indicate regions of low Rac1 activation and now JMPs. Bar, 10 μm. (D) Quantification of FRET-based Rac1 biosensor activation under the area of crawling PMNs, and the region before and after the PMN has passed. Mann-Whitney U-test. (E) Graph showing example of EC measuring Rac1 activation of whole EC (blue line) and area of EC underneath a crawling neutrophil (green line). Diapedesis event occurs at the end of the lines at 210 s.
Neutrophils prefer spots of high Rac1 activity to transmigrate.
FRET-based DORA Rac1 biosensor shows Rac1 activity in transfected endothelial cells with warm colors (and white color as warmest color) as high FRET efficiency. Neutrophil migration is shown in DIC image on the left and arrow indicate preferred diapedesis site. Note that transfected EC is part of EC monolayer and surrounded by non-transfected ECs.
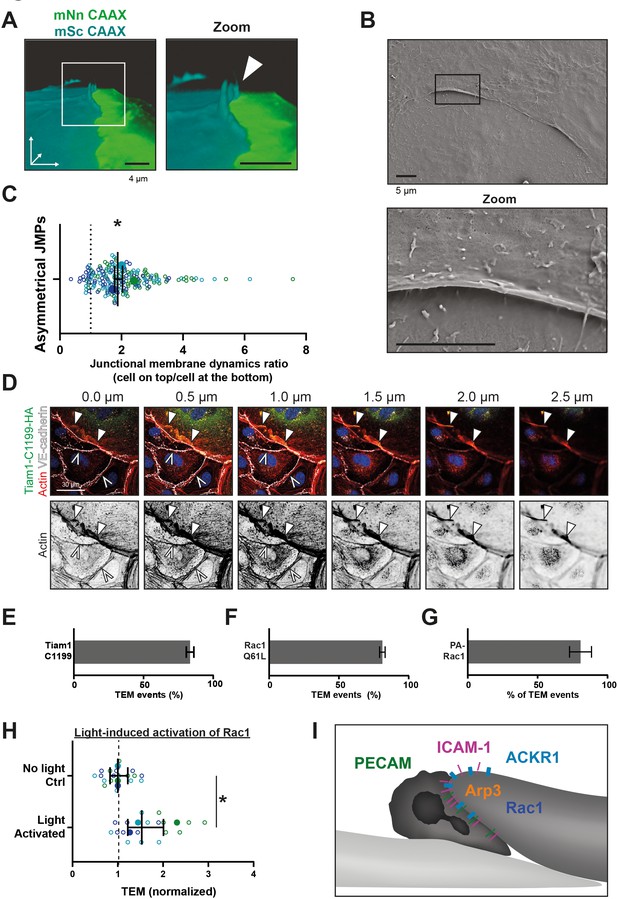
Asymmetric JMPs induced by Rac1 and serve as diapedesis sites.
(A) 3D view still from two endothelial cells showing a turquoise expressing EC and a NeonGreen expressing EC. White box indicates zoom on the right. Arrowhead indicates shows that the turquoise EC displays a membrane protrusion at the junction region, hence, an asymmetric JMP. Bar, 4 μm. (B) Scanning electron microscopy image of asymmetric JMPs. Black box indicates zoom region, displayed on the right. Bar, 5 μm. (C) Ratio of membrane dynamics in EC with JMP and the other EC at a junction region. Open dots are individual data points from three independent experiments, represented by three different colors. Filled dots are means from three experiments. Median with 95% confidence interval (CI) is shown. One-sample T-test: *p=0.344. (D) Immunofluorescent staining for HA (green), F-actin (red), VE-cadherin (white) and DNA (blue) on HUVECs transfected with Tiam1-C1199-HA after overnight TNFα stimulation. Filled white arrowheads indicate the F-actin present at the cell-cell junction between a Tiam1-C1199-HA expressing cell and a control cell, where the open white arrowheads indicate the F-actin present at the junction between two control cells. Panel shows Z-stack from basal (0.0 µm) towards apical (2.5 µm) from left to right, respectively. Arrowheads indicate presence of F-actin rich membrane ruffles in the different focal planes. Scale bar, 30 µm. (E–G) Quantification of diapedesis direction of PMN upon TEM under flow. Majority of the PMNs cross form a wt EC underneath a (E) TIAM1-C1199, (F) Rac1-Q61L, (G) Photoactivatable (PA)-Rac1 EC (dark bar) (H) Quantification of PMN TEM under flow in cells expressing PA-Rac1 either not illuminated (Ctrl) or blue-light illuminated (activated) HUVEC. Data is normalized to control conditions. Upon Rac1 activation, increased number of neutrophils crossed the EC monolayer. Open dots are individual data points from three independent experiments, represented by three different colors. Filled dots are means from three experiments. Median with 95% CI is shown. T-test: p=0.0483. (I) Schematic overview of neutrophil transmigrating from ‘inactive’ EC (bottom left, light) underneath an ‘active’ EC (upper right, dark) showing presence of a JMP. JMP displays Rac1 activity, Arp3, and expression of PECAM-1, ICAM-1, and ACKR1.
-
Figure 6—source data 1
Tiam-mediated Rac activation.
- https://cdn.elifesciences.org/articles/66074/elife-66074-fig6-data1-v2.pdf
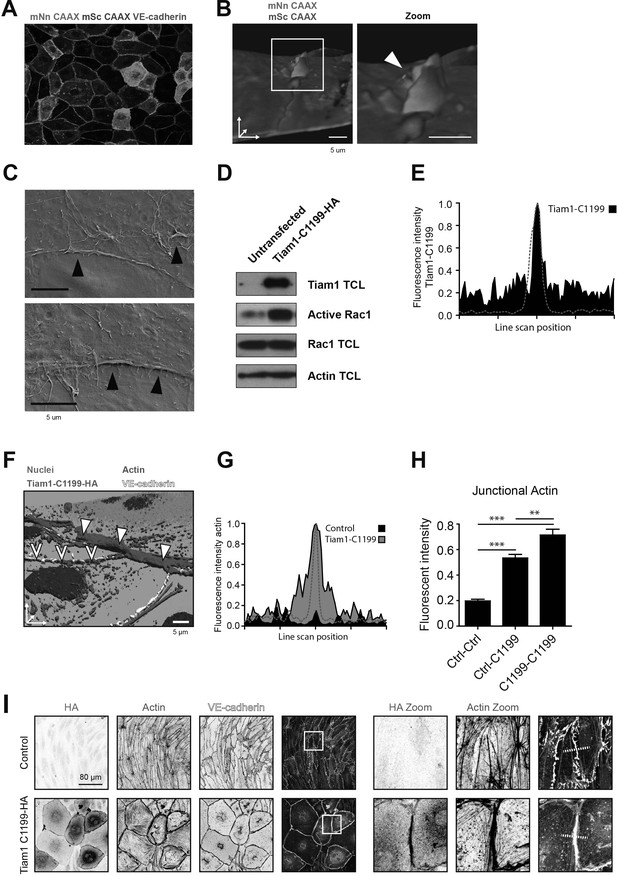
JMPs and exogenous Rac1 activation.
(A) Mixed monolayer of ECs either expressing mNeonGreen-CAAX or mScarlet-I-CAAX (B) 3D image stills from two ECs showing the turquoise cell (right) being on top of the green cell (left). White box indicates zoom region. Arrowhead indicates asymmetric JMP. CAAX- and mScarlet-I-CAAX-transfected HUVECs showing transmigrating neutrophil (magenta). 3D reconstruction using Imaris surface rendering function Bar, 5 μm. (C) Scanning electron microscopy image of asymmetric JMPs in two ECs. Black arrowheads show presence of membrane protrusions. Bar, 5 μm. (D) Rac1 activity upon Tiam1-C1199-HA expression in ECs determined by biochemical Rac1 pulldown assay with biotin-tagged CRIB as bait. Western blot shows Tiam1 expression level, active and total Rac1 as indicated. Actin is shown as loading control. (E) Quantification of Tiam1-C1199-HA staining intensity in which the red dashed line indicates VE-cadherin staining that peaks at the cell-cell junction. Fluorescence intensity was quantified within 7.2 μm from the VE-cadherin positive cell-cell junctions. (F) 3D projection of nuclei (blue), Tiam1-C1199-HA (green), F-actin (red) and VE-cadherin (white) on the junction of a control and Tiam1-C1199-HA expressing EC showing the enrichment and protrusion towards the apical site of F-actin. (G) Quantification of junctional F-actin staining intensity in control cells (black) and Tiam1-C1199-HA cells (gray) in which the red dashed line indicates VE-cadherin staining and that peaks at the cell-cell junction. Fluorescence intensity was quantified within 7.2 μm from the VE-cadherin-positive cell-cell junctions. (H) Quantification of junctional F-actin at a control-control junction (Ctrl-Ctrl), an asymmetric junction of one control cell and one Tiam1-C1199 cell (Ctrl-C1199) and two Tiam1-C1199 cells (C1199-C1199). T-test: p<0.05 (I) Immunofluorescent staining for HA (green), F-actin (red), and VE-cadherin (white) on control ECs and Tiam1-C1199-transfected ECs. White box indicates area of zoom and images are a maximum intensity Z-projection. Scale bar, 80 µm. White dashed line indicates site of line scan for measuring fluorescence intensity indicated in (H).
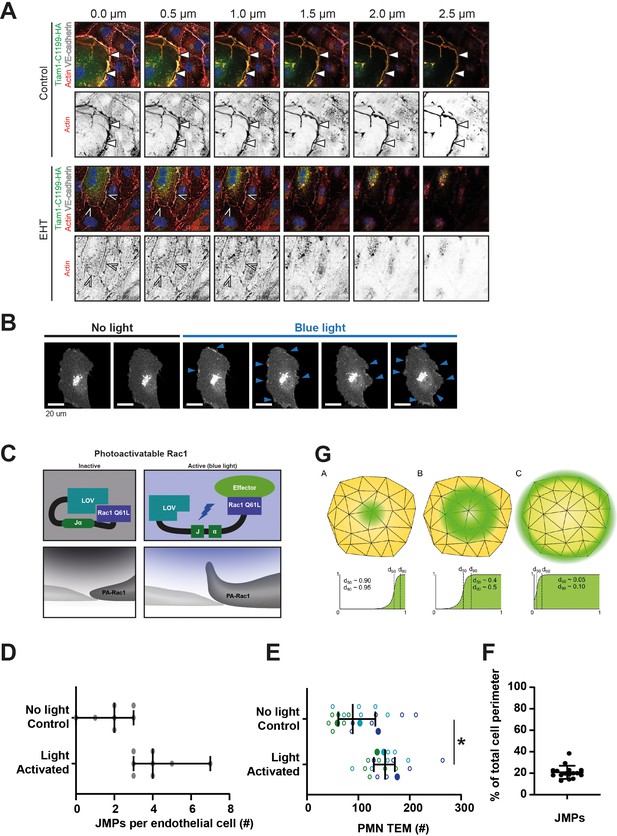
JMPs and exogenous Rac1 activation.
(A) Immunofluorescent staining for Tiam1-C1199-HA (green), F-actin (red) and VE-cadherin (white) on HUVECs after overnight TNFα stimulation. Panel shows Z-stack from basal (0 µm) toward apical (2.5 µm) from left to right, respectively. Arrowheads indicate presence of F-actin-rich membrane ruffles in the different focal planes. Lower panel show cells treated with EHT-1864 (B) Stills from time lapse images of cells expressing PA-Rac1 illuminated with blue light (frame 3–6). (C) Schematic overview of the photo-reactive LOV2 (light-oxygen-voltage) domain sterically blocking Rac1 Q61L interactions until irradiation unwinds a helix (Jα), linking LOV2 to Rac 1 Q61L. This probe can be reversibly and repeatedly activated using 458–473 nm light to generate cell membrane protrusions and ruffling. (D) Quantification of the number of JMPs per endothelial cell expressing PA-Rac1, either illuminated with blue light to activate PA-Rac1, or not illuminated as a control. T-test: *p=0.0035 (E) Quantification of PMN TEM under flow in cells expressing PA-Rac1 either not illuminated (Ctrl) or blue-light illuminated (activated) HUVEC. Open dots are individual data points from three independent experiments, represented by three different colors. Filled dots are means from three experiments. Median with 95% CI is shown. T-test: *p=0.0483. (F) Percentage of the total junction that show JMP activity. Data are from at least 20 different junctions from four different experiments. (G) Stain (VE-Cadherin/PECAM) Coverage Analysis. Three illustrative examples of stain distribution along a JMP region and the resulting coverage analysis. The analysis consists of computing an empirical cumulative distribution (CD) of stained vertices binned by distance from the edge of the JMP region, the distances are normalized by dividing by the maximum distance from the edge. (A) Centrally distributed staining generates a normalized CD with most of the mass near 1, resulting in percentiles that are also close to 1. (B) Broadly distributed staining that is weaker towards the center and edges produces a normalized CD with mass increase around 0.5, percentiles are around 0.5 but spread more broadly. (C) Staining mostly near the edge results in a normalized CD with mass increase near 0, and associated percentiles near 0. Green regions indicate the staining distribution across the JMP mesh. For each panel, the normalized 50th and 90th percentile distances from the edge are shown as vertical lines on the normalized CD and approximate values for each are also given. The normalized percentile values are averaged across time to produce dataset measures of average stain coverage.
Neutrophils prefer spots of Tiam1-induced JMPs to transmigrate.
Tiam1-C1199 is transfected in ECs, and can be identified by enlarged phenotype, in middle of movie. This EC is surrounded by non-transfected ECs. Neutrophils prefer to cross junctions of CTLR-Tiam1 expressing cells. Total recording time is15 minutes.
Neutrophils prefer to migrate across junctions that asymmetrically display JMP.
GFP-Rac1-Q61L (green) and mScarlet-Lifeact are transfected in ECs, separately from each other and show preference for neutrophil TEM. Neutrophils continue to migrate underneath the Rac1 expressing cell. Total recording time is10 min.
Neutrophils prefer to migrate across junctions that asymmetrically display JMP.
Photo-activatable Rac1 (green) is transfected in ECs and photoactivated 5 min prior to the start of the movie. Movie shows the preference for neutrophil TEM. Neutrophils continue to migrate underneath the Rac1-activated cell. Total recording time is 15 min.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Primary cells (Homo-sapiens) | Human Umbilical Vein Endothelial cells (HUVEC) | Lonza | C2519A:0000633426 | passage 2-5 |
| Primary cells (Homo-sapiens) | Human Lung Microvascular Endothelial cells (HMVEC) | Pelo Biotech | PB-CH-150-4011 | passage 4-8 |
| Primary cells (Homo-sapiens) | Human Pancreatic Microvascular Endothelial cells (HMVEC) | Pelo Biotech | PB-CH-147-4011 | passage 4-8 |
| Primary cells (Homo-sapiens) | Human Kidney Glomerulus Microvascular Endothelial cells (HMVEC) | Pelo Biotech | PB-CH-152-5211 | passage 4-8 |
| Primary cells (Homo-sapiens) | Human Embryonic Kidney cells 293T (HEK293T) | ATCC | CRL-3216 | passage 10-30 |
| Transfected construct | CAAX mNeonGreen | van Buul lab | ||
| Transfected construct | CAAX mScarlet-I | van Buul lab | ||
| Transfected construct (Homo-sapiens) | ICAM-1 mScarlet-I | van Buul lab | ||
| Transfected construct (Homo-sapiens) | ACKR1 mNeonGreen | van Buul lab | ||
| Transfected construct | LifeAct mTurquoise2 | van Buul lab | ||
| Transfected construct (Homo-sapiens) | Tiam1-C1199 | Klems et al., 2020 | ||
| Transfected construct | Rac1 DORA biosensor | Timmerman et al., 2015 | ||
| Transfected construct | Photoactivatable Rac1 | Wu et al., 2009 | ||
| Recombinant DNA reagent | pLKO sh Ctrl | Merck | MFCD07785395 | |
| Recombinant DNA reagent | pLKO sh Arp3 | Merck | TRCN0000029381 | |
| Recombinant DNA reagent | pLKO sh Arp3 | Merck | TRCN0000029382 | |
| Antibody | anti human CD144 VE-cadherin AF647 (mouse monoclonal) | BD | Cat #561567 | IF (1:100) |
| Antibody | anti human CD31 PECAM AF647 (mouse monoclonal) | BD | Cat #561654 | IF (1:100) |
| Antibody | anti human actin (mouse monoclonal) | Sigma | Cat #A3853 | WB (1:1000) |
| Antibody | anti human Arp3 (mouse monoclonal) | Merck | cat #A5979 | WB (1:1000) |
| Peptide, recombinant protein | Recombinant Human TNF-alpha | Peprotech | Cat #300-01A | 10 ng/ml |
| Peptide, recombinant protein | Recombinant Human IFN-gamma | R&D | Cat #285-IF | 500 ng/ml |
| Peptide, recombinant protein | IL-1beta | Peprotech | Cat #200-01B | 10 ng/ml |
| Biological compound | LPS | Sigma | Cat #L2880 | 500 ng/ml |
| Chemical compound | Lenti-X concentrator | TaKaRa | Cat #631232 | |
| Chemical compound | CK-666 (Arp2/3 inhibitor) | Sigma | Cat #SML0006 | 100 uM |
| Chemical compound | EHT 1864 (Rac inhibitor) | Tocris | Cat #3872 | 50 uM |





