Post-transcriptional repression of circadian component CLOCK regulates cancer-stemness in murine breast cancer cells
Figures
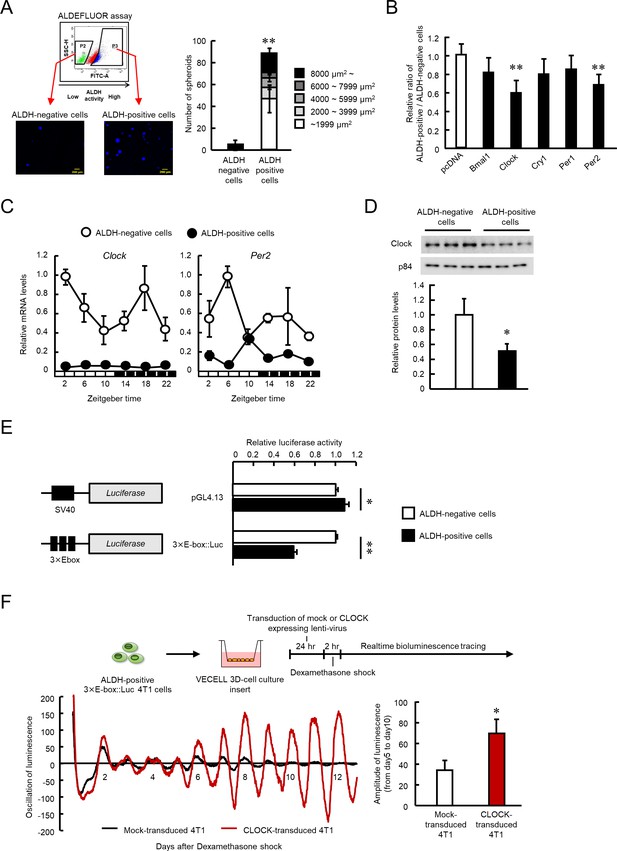
The role of CLOCK in the regulation of ALDH activity in 4T1 mouse breast cancer cells.
(A) Representative photograph of Hoechst-stained tumor spheroids of ALDH-negative or -positive 4T1 cells in agarose. Values are the mean with SD (n = 3). **p<0.01; significant difference from ALDH-negative cells (t6 = 12.598, Student's t-test). (B) Influence of each clock gene expression vector on the ratio of ALDH positive- to negative-cell populations. ALDH-positive 4T1 cells were cultured on 3D scaffold chambers after being transiently transfected with each expression vector using electroporation. ALDH activity was evaluated 5 days after transfection. Values are the mean with SD (n = 6). The mean value of the pcDNA group is set at 1.0. **p<0.01; significant difference from the pcDNA group (F5,30 = 6.807, p<0.001, ANOVA, Dunnett’s post hoc test). (C) The temporal mRNA expression profiles of Clock and Per2 in ALDH-positive and ALDH-negative cells isolated from 4T1 tumor-bearing mice kept under the light/dark cycle (zeightgber time 0. lights on ; zeitgeber time 12. lights off). Values are the mean with SD (n = 3). Data were normalized by β-Actin mRNA levels. (D) Difference in the expression levels of CLOCK protein between ALDH-negative and -positive 4T1 cells. The value of ALDH-negative cells is set at 1.0. Values are the mean with SD (n = 3). *p<0.05; significant difference between two groups (t4 = 3.915, Student’s t-test). (E) Difference in the promoter activities of E-box-driven luciferase reporter in ALDH-negative and -positive 4T1 cells. pGL4.13 or 3 × E-box::Luc reporter vectors were transfected into 4T1 cells, and luciferase assay was performed after cell sorting. Values are the mean with SD (n = 3). The value of ALDH-negative cells is set at 1.0. **p<0.01, *p<0.05; significant difference between two groups (t4 = 2.941 for pGL4.13, t4 = 43.287 for 3 × E-box::Luc, Student’s t-test). (F) The influence of lenti-viral CLOCK transduction on the circadian oscillation of E-box-driven luciferase bioluminescence. Top panel shows the scheme of experimental procedure. Bottom panels show real-time bioluminescence tracing of luciferase activity after dexamethasone synchronization (left) and mean of amplitude of bioluminescence oscillation from day 5 to day 10 after synchronization (n = 3) (right). *p<0.05; significant difference between two groups (t4 = 3.691, Student’s t-test).
-
Figure 1—source data 1
This spreadsheet contains the source for Figure 1.
- https://cdn.elifesciences.org/articles/66155/elife-66155-fig1-data1-v2.xlsx
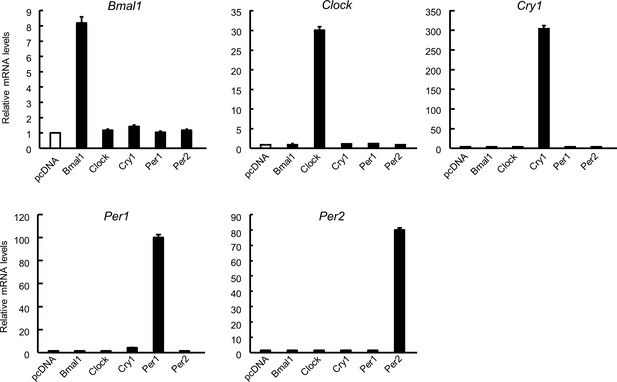
The mRNA expression of clock genes in 4T1 cells transfected with vectors expressing Bmal1, Clock, Cry1, Per1, or Per2.
Data were normalized by 18 s rRNA levels. Values show the mean with SD (n = 3). The value of pcDNA-transfected cells is set at 1.0.
-
Figure 1—figure supplement 1—source data 1
This spreadsheet contains the source for Figure 1—figure supplement 1.
- https://cdn.elifesciences.org/articles/66155/elife-66155-fig1-figsupp1-data1-v2.xlsx
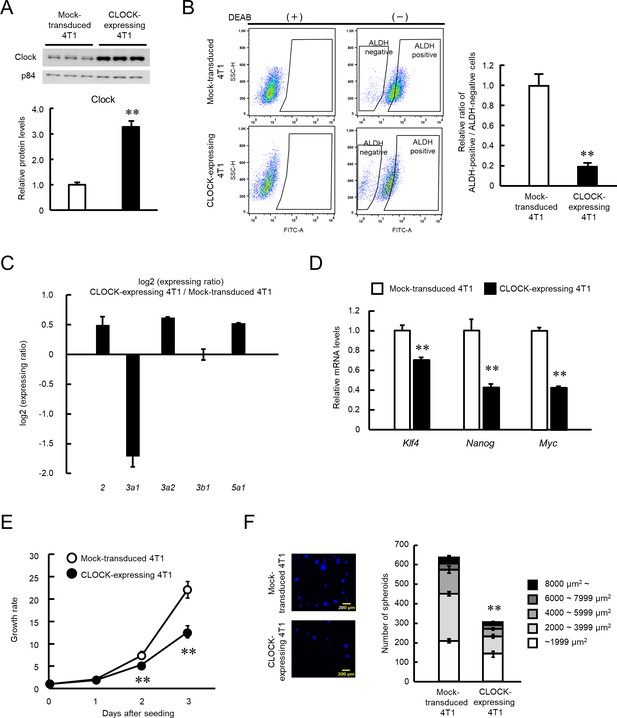
Suppression of stemness properties of 4T1 cells by CLOCK.
(A) CLOCK protein levels in mock-transduced and Clock-expressing lentivirus-infected 4T1 cells. Values show mean with SD (n = 3). **p<0.01; significant difference from mock-transduced 4T1 cells (t4 = 15.736, Student’s t-test). (B) The flow cytometric analysis of ALDEFLUOR assay of CLOCK-expressing 4T1 cells. The population of ALDH-positive cells was defined based on each DEAB group, whose means of fluorescence intensity were set almost the same. The right panel shows the difference in the ratio of ALDH positive- to negative-cell populations between mock-transduced and CLOCK-expressing 4T1 cells. Values show the mean with SD (n = 6). **p<0.01; significant difference from mock-transduced 4T1 cells (t10 = 22.455, Student’s t-test). (C) Difference in the mRNA levels of ALDH isoforms between mock-transduced and CLOCK-expressing 4T1 cells. Values show the mean with SD (n = 3). (D) The mRNA levels of stemness-related genes in ALDH-positive mock-transduced and CLOCK-expressing 4T1 cells. Data were normalized by the18s rRNA levels. Values show the mean with SD (n = 3). The values of the mock-transduced group are set at 1.0. **p<0.01: significant difference from mock-transduced 4T1 cells (t4 = 9.261 for Klf4; t4 = 8.001 for Nanog; t4 = 32.576 for Myc; Student’s t-test). (E) Difference in growth ability between mock-transduced and CLOCK-expressing 4T1 cells. Values show the mean with SD (n = 5–6). Cell viability of seeding day (day 0) are set at 1.0. **p<0.01; significant difference between from mock-transfected 4T1 cells at corresponding time points. (F7, 38 = 425.953, two-way ANOVA with the Tukey–Kramer test). (F) Difference in the spheroid formation ability between mock-transduced and CLOCK-expressing 4T1 cells. The left panel shows a representative photograph of the Hoechst-stained spheroids formed by mock-transduced or CLOCK-expressing 4T1 cells. The right panel shows the number of spheroid and the parcellation of the diameter. Values show the mean with SD (n = 3). **p<0.01; significant difference from mock-transduced 4T1 cells (t4 = 27.067, Student’s t-test, for number of spheroids).
-
Figure 2—source data 1
This spreadsheet contains the source for Figure 2.
- https://cdn.elifesciences.org/articles/66155/elife-66155-fig2-data1-v2.xlsx
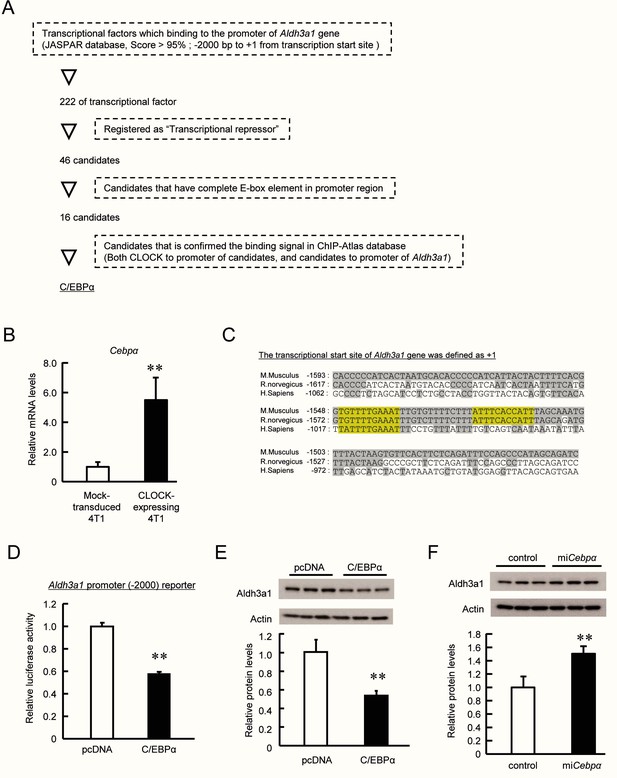
CLOCK regulates the expression of Aldh3a1 through mediation of C/EBPα.
(A) Procedure of selecting transcriptional factors that regulate Aldh3a1 expression in 4T1 cells. Genomic DNA binding prediction database (JASPAR) and ChIP Atlas database were applied to identify factor that mediates CLOCK-regulated Aldh3a1 expression. (B) The expression levels of C/EBPα mRNA in enhanced CLOCK-expressing 4T1 cells. Values are the mean with SD (n = 3). **p<0.01; significant difference between two groups (t4 = 5.703, Student’s t-test). (C) The alignment of promoter sequences of Aldh3a1 genes in mouse (Mus musculus), rat (Ratta norvegicus), and human (Homo sapiens). (D) Repression of Aldh3a1 transcriptional activity by C/EBPα. Luciferase reporter vectors containing −2000 bp upstream region of the mouse Aldh3a1 gene (Aldh3a1 [−2000]::Luc) were constructed. 4T1 cells were transfected with Aldh3a1 [−2000]::Luc and C/EBPα-expressing vectors. Values are the mean with SD (n = 3). The values of pcDNA group are set at 1.0. **p<0.01; significant difference between two groups (t4 = 19.351, Student’s t-test). (E) Repression of Aldh3a1 expression by C/EBPα. 4T1 cells were transfected with C/EBPα-expressing vectors. Values are the mean with SD (n = 3). The values of pcDNA group are set at 1.0. **p<0.01; significant difference between two groups (t4 = 5.730, Student’s t-test). (F) Enhanced expression of Aldh3a1 in C/EBPα-downregulated 4T1 cells. C/EBPα was downregulated by miRNA. Values are the mean with SD (n = 3). The values of control group are set at 1.0. **p<0.01; significant difference between two groups (t4 = 4.892, Student’s t-test).
-
Figure 2—figure supplement 1—source data 1
This spreadsheet contains the source for Figure 2—figure supplement 1.
- https://cdn.elifesciences.org/articles/66155/elife-66155-fig2-figsupp1-data1-v2.xlsx
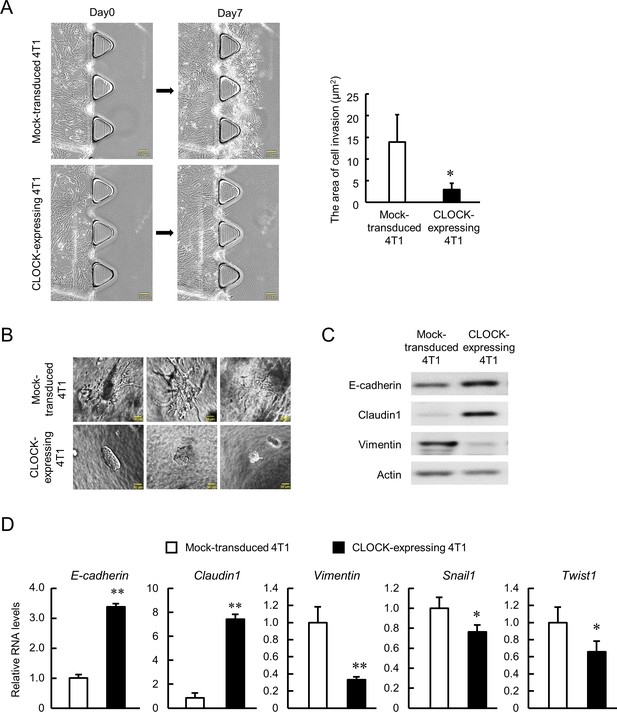
Suppression of invasive potential of 4T1 cells by CLOCK.
(A) Decrease in the invasion ability of CLOCK-expressing 4T1 cells. Microphotographs show invasion of cells into 3D collagen gel. The right panel shows difference in the invasion area between mock-transduced and CLOCK-expressing 4T1 cells. Values show mean with SD (n = 3). *p<0.05; significant difference from mock-transduced 4T1 cells (t4 = 2.968, Student’s t-test). (B) Representative microphotographs of spheroid invasion by mock-transduced and CLOCK-expressing 4T1 cells. Invasive morphology was detected by mock-transduced 4T1 cells. (C) Difference in the expression of adhesion molecules between mock-transduced and CLOCK-expressing 4T1 cells. (D) Differential expression of EMT-related molecules between mock-transduced and CLOCK-expressing 4T1 cells. E-cadherin and Claudin1 indicate the ‘epithelial’ state, and Vimentin, Snail1, and Twist1 indicate the ‘mesenchymal’ state. Data were normalized by the 18s rRNA levels. Values show the mean with SD (n = 3). The values of the mock-transduced group are set at 1.0. **p<0.01, *p<0.05; significant difference compared with mock-transduced 4T1 cells (t4 = 28.550 for E-cadherin; t4 = 18.159 for Claudin1; t4 = 6.233 for Vimentin; t4 = 2.873 for Snail1; t4 = 2.881 for Twist1; Student’s t-test).
-
Figure 3—source data 1
This spreadsheet contains the source for Figure 3.
- https://cdn.elifesciences.org/articles/66155/elife-66155-fig3-data1-v2.xlsx
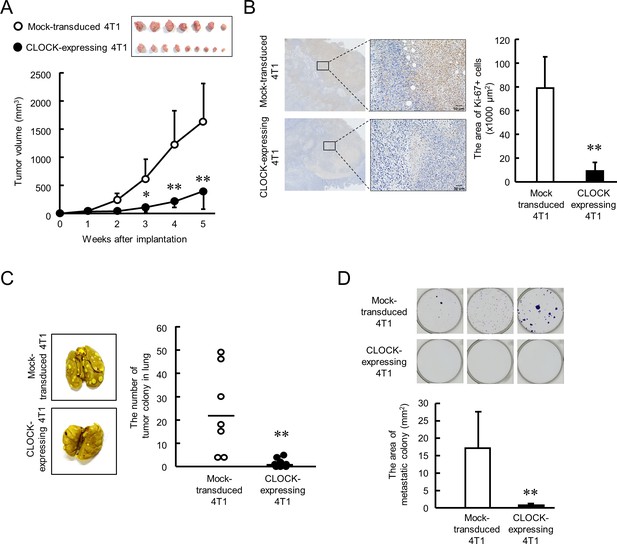
CLOCK-induced suppression of malignancy of 4T1 tumors in mice.
(A) The ability of tumor growth in mice implanted with mock-transduced or CLOCK-expressing 4T1 cells. Top panel shows the photograph of tumor burden isolated from each 4T1 cell implanted mice 6 weeks after implantation. Values are the mean with SD (n = 8–9 animals). **p<0.01; *p<0.05, significant difference compared with mock-transduced 4T1 cell-implanted mice at corresponding time points (F9, 67 = 19.956, p<0.001; two-way ANOVA with the Tukey–Kramer test). (B) Immunohistochemical staining of Ki-67 in mock-transduced or CLOCK-expressing 4T1 tumors. Complexes with Ki-67 and antibodies were visualized by 3, 3’-diaminobenzidine (brown), and nuclei were stained with hematoxylin (blue). Scale bars indicate 50 µm. Values show mean with SD (n = 6 animals). **p<0.01; significant difference from mock-transduced 4T1 cells (t10 = 6.213, Student’s t-test). (C) The number of pulmonary tumor colonies in mice implanted with mock-transduced or CLOCK-expressing 4T1 cells. Pulmonary colonies were assessed 6 weeks after implantation. The left panels show representative photographs of pulmonary tumor colonies of mock-transduced or CLOCK-expressing 4T1 cells implanted in mice. Right panel shows the quantification of the number of tumor colonies in lungs (n = 7–9 animals). **p<0.01; significant difference from mock-transdaced 4T1 (t14 = 3.601; Student’s t-test). (D) The number of metastatic colonies isolated from tumor-bearing mice femora bone marrow. The top panels show the representative photograph of tumor colonies stained by crystalbiolet. The bottom panel shows the quantification of the area of tumor colonies. Values show mean with SD (n = 8 animals). **p<0.01; significant difference from mock-transduced 4T1 cells (t14 = 3.871, Student’s t-test).
-
Figure 4—source data 1
This spreadsheet contains the source for Figure 4.
- https://cdn.elifesciences.org/articles/66155/elife-66155-fig4-data1-v2.xlsx
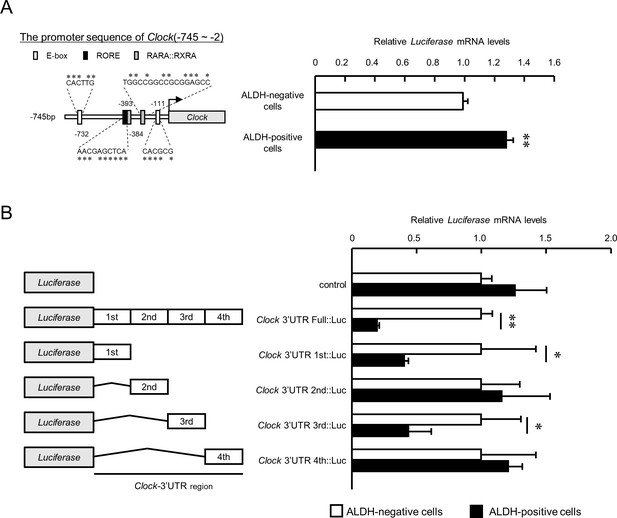
The post-transcriptional regulation of Clock gene expression in ALDH-positive 4T1 cells.
(A) Comparison of the promoter activity of the mouse Clock gene between ALDH-negative and -positive 4T1 cells. Left panel shows the schematic representation of the mouse Clock gene promoter. Numbers near the boxes are nucleotide residues in which E-box, RORE, and RAR/RXR response elements are positioned relative to the transcription start site (+1). Right panel shows the mRNA expression of the Luciferase gene Clock::Luc in ALDH-negative and -positive 4T1 cells. Data were normalized by Neomycin phosphotransferase (Neor) as an internal control. Values are the mean with SD (n = 3).**p<0.01;significant difference between from ALDH-negative cells. The value of ALDH-negative cells is set at 1.0. (B) The mRNA expression of the Luciferase gene in ALDH-negative and -positive 4T1 cells transfected with luciferase reporter constructs containing varying lengths of the 3’UTR of the Clock gene. Data were normalized by the expression levels of hRLuc mRNA. Values are the mean with SD (n = 3). The values of each ALDH-negative cell are set at 1.0. *p<0.05, **p<0.01; significant difference between the two groups (t4 = 7.665 for Clock-3’UTR Full::Luc; t4 = 4.375 for Clock-3’UTR 1 st::Luc; t4 = 2.751 for Clock-3’UTR 3rd::Luc; Student’s t-test).
-
Figure 5—source data 1
This spreadsheet contains the source for Figure 5.
- https://cdn.elifesciences.org/articles/66155/elife-66155-fig5-data1-v2.xlsx
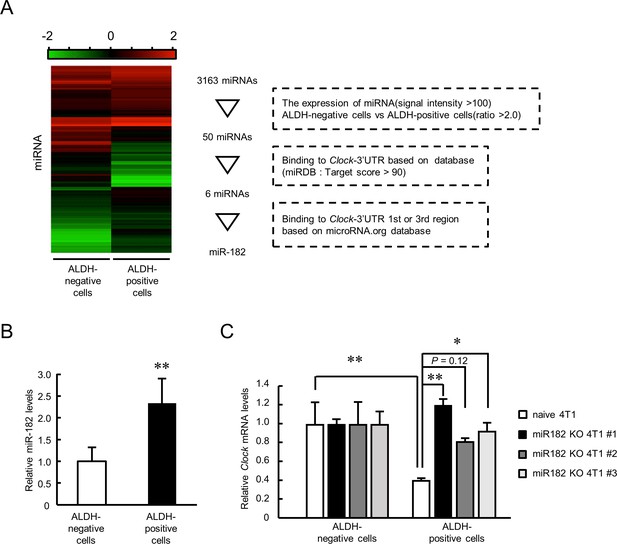
Role of miR-182 in post-transcriptional regulation of Clock gene expression in ALDH-positive 4T1 cells.
(A) Procedure of selecting miRNA that regulates Clock expression in ALDH-positive 4T1 cells. Heatmap shows the differential expression of miRNA between ALDH-negative and -positive 4T1 cells. microRNA target prediction databases (miRDB and microRNA.org) were applied to this selection. (B) The expression levels of mmu-miR-182 in ALDH-negative and -positive cells. Data were normalized by the β-Actin mRNA levels. Values are the mean with SD (n = 5). The value of ALDH-negative cells is set at 1.0. **p<0.01; significant difference from ALDH-negative cells (t8 = 3.828, Student’s t-test). (C) miR-182 negatively regulates the expression of Clock mRNA in ALDH-positive 4T1 cells. Three miR-182 KO clones were selected for this experiment. Data were normalized by the 18 s rRNA levels. Values are the mean with SD (n = 3). The values of ALDH-negative cells are set at 1.0. *p<0.05, **p<0.01; significant difference between two groups (F7,16 = 7.681, p<0.01, ANOVA with the Tukey–Kramer post hoc test).
-
Figure 6—source data 1
This spreadsheet contains the source for Figure 6.
- https://cdn.elifesciences.org/articles/66155/elife-66155-fig6-data1-v2.xlsx
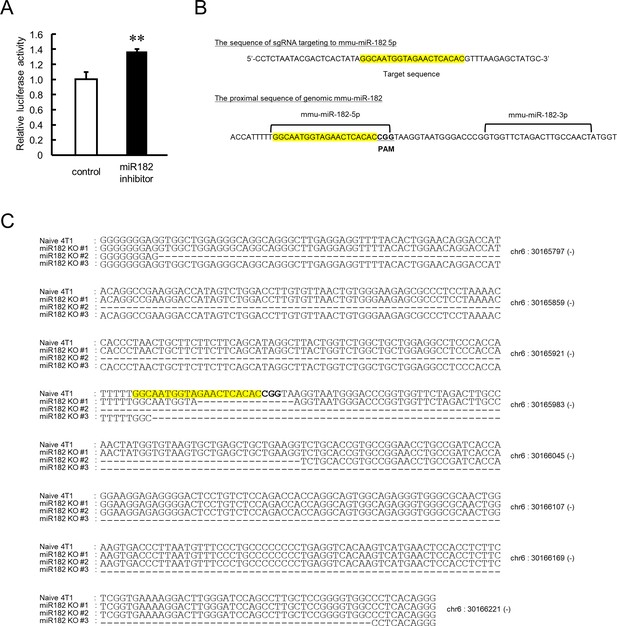
Influence of functional depletion of miR-182 on the regulation of Clock expression.
(A) Influence of the miR-182::hairpin inhibitor on luciferase activity of Clock 3’UTR 1 st::Luc. Data were normalized by the renilla luciferase activity as an internal control. Values are the mean with SD (n = 3). The value of control group is set at 1.0. **p<0.01; significant difference between two groups (t4 = 7.569, p<0.01; Student’s t-test). (B) Scheme of the mmu-miR-182 target region. Yellow shaded areas indicate the sgRNA-targeted sequence. (C) Proximal genomic region around mmu-miR-182 in naive and miR-182 knockout (KO) 4T1 cells clone no. #1 to #3.
-
Figure 6—figure supplement 1—source data 1
This spreadsheet contains the source for Figure 6—figure supplement 1.
- https://cdn.elifesciences.org/articles/66155/elife-66155-fig6-figsupp1-data1-v2.xlsx
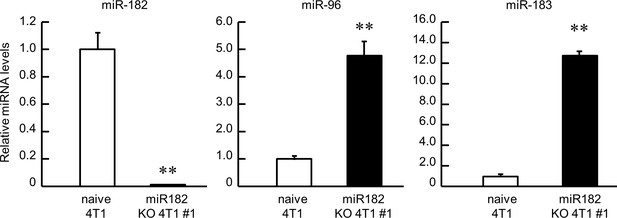
The influence of genomic depletion of miR-182 on the other clustered miRNAs.
Data were normalized by 18 s rRNA levels. Values are the mean with SD (n = 3). The value of naive 4T1 cells is set at 1.0. **p<0.01; significant difference between two groups (t4 = 12.819 for miR182, t4 = 11.439 for miR96, t4 = 55.277 for miR183, Student’s t-test).
-
Figure 6—figure supplement 2—source data 1
This spreadsheet contains the source for Figure 6—figure supplement 2.
- https://cdn.elifesciences.org/articles/66155/elife-66155-fig6-figsupp2-data1-v2.xlsx
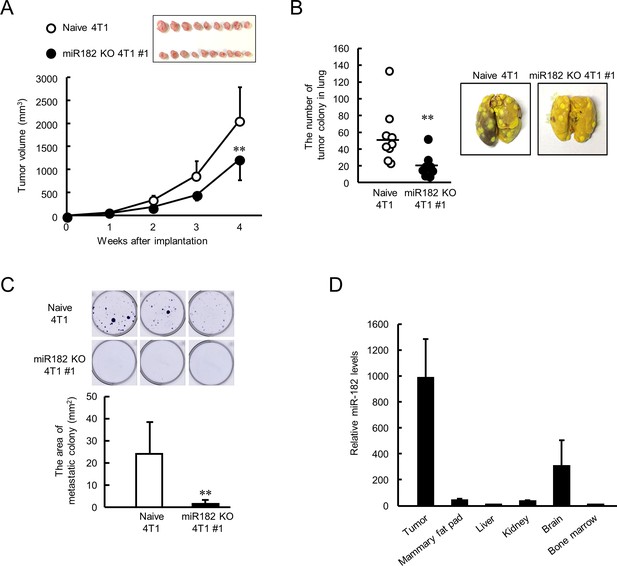
Suppression of malignant properties of 4T1 tumor by knockout of miR-182.
(A) The ability of tumor growth in mice implanted with naive or miR-182 KO 4T1 cells. Values are the mean with SD (n = 9–10 animals). **p<0.01; significant difference compared with naive 4T1 cell-implanted mice at corresponding time points (F7, 63 = 39.494, p<0.001; two-way ANOVA with the Tukey–Kramer test). Top panel shows the photograph of tumors isolated from each 4T1 cell implanted mice 5 weeks after implantation. (B) The number of pulmonary tumor colonies in mice implanted with naive or miR-182 KO 4T1 cells. Left panel shows the quantification of the number of tumor colonies in lungs (n = 9 animals). **p<0.01; significant difference from naive 4T1 group(t16 = 2.993; Student’s t-test). The right panels show representative photographs of pulmonary tumor colonies of naive or miR-182 KO 4T1 cells implanted in mice. Pulmonary tumor colonies were assessed 5 weeks after implantation. (C) The number of metastatic colonies isolated from tumor-bearing mice femora bone marrow. The top panels show the representative photograph of tumor colonies stained by crystalbiolet. The bottom panel shows the quantification of the area of tumor colonies. Values show mean with SD (n = 6 animals). **p<0.01; significant difference from naive 4T1 cells (t10 = 3.354, Student’s t-test). (D) The organ distribution of miR-182 in naive 4T1-bearing mice. Data were normalized by the 18 s rRNA levels. Values are the mean with SD (n = 3 animals). The value of bone marrow is set at 1.0.
-
Figure 7—source data 1
This spreadsheet contains the source for Figure 7.
- https://cdn.elifesciences.org/articles/66155/elife-66155-fig7-data1-v2.xlsx
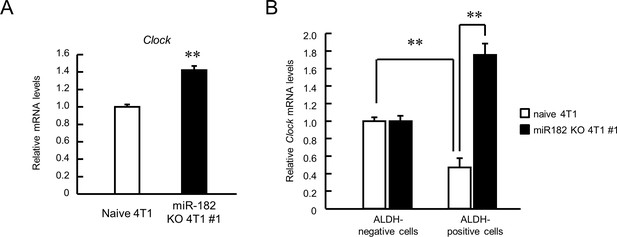
The expression of CLOCK in miR-182 KO 4T1 cell #1-bearing mice tumor.
(A) The mRNA expression levels of Clock derived from whole tumor. Values are the mean with SD (n = 3 animals). The value of naive 4T1 group is set at 1.0. **p<0.01; significant difference between two groups (t4 = 14.173, Student’s t-test). (B) The mRNA expression levels of Clock derived from each sorted ALDH-negative cells or ALDH-positive cells. Values are the mean with SD (n = 3 animals). The value of each ALDH-negative cells group is set at 1.0. **p<0.01; significant difference between two groups (F3,11 = 98.647, p<0.01, ANOVA with the Tukey–Kramer post hoc test).
-
Figure 7—figure supplement 1—source data 1
This spreadsheet contains the source for Figure 7—figure supplement 1.
- https://cdn.elifesciences.org/articles/66155/elife-66155-fig7-figsupp1-data1-v2.xlsx
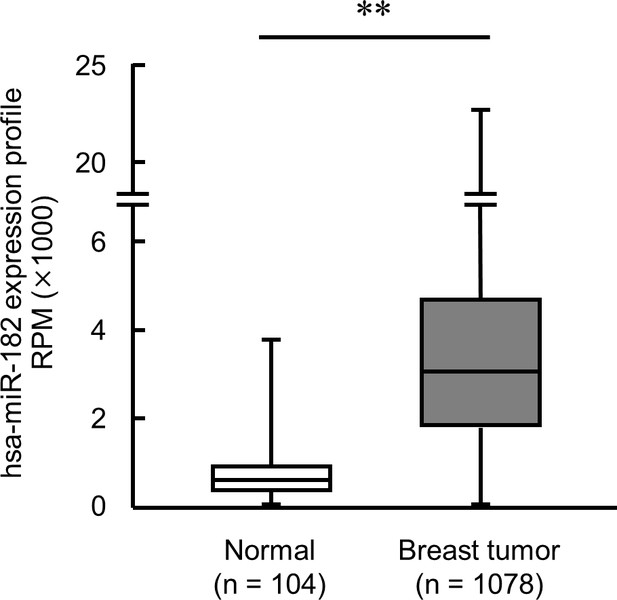
The expression levels of hsa-miR-182 in breast tumor and normal mammary gland derived from patients.
Data were obtained from miR-TV database. **p<0.01; significant difference between two groups (Student’s t-test).
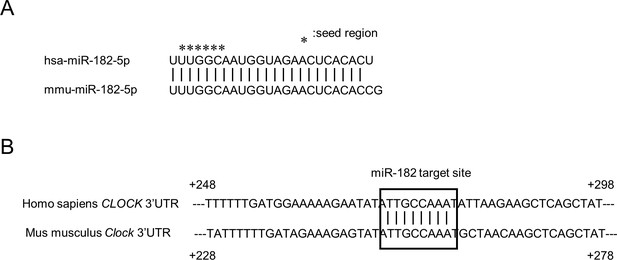
Sequence conservation of miR-182.
(A) The alignment of hsa-miR-182–5 p and mmu-miR-182–5 p. * shows seed region of each miRNA. (B) The alignment of human CLOCK 3’UTR and mouse Clock 3’UTR. The nucleotide immediately after the stop codon in exon 23 of each CLOCK gene was defined as +1.
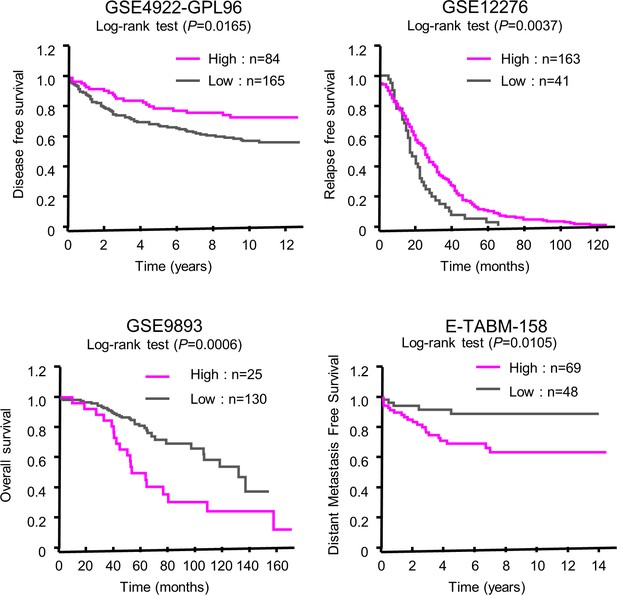
Database analysis of clinical relevance of CLOCK expression to patients' outcome.
The data were obtained from the PrognoScan database (http://www.prognoscan.org/). Association of gene expression with patient survival was evaluated by minimum p-value approach. Patients were divided into high- or low-CLOCK expression groups at all possible cutoff points. The differences of any two groups were estimated by the log-rank test.
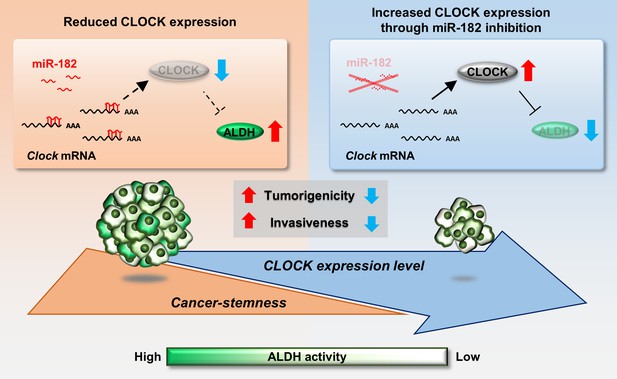
Schematic diagrams indicating the role of CLOCK in the regulation of malignancy of breast cancer stem-like cells.
mmu-miR-182 post-transcriptionally suppresses the expression of Clock in ALDH-positive 4T1 cells, leading to maintenance of their malignancy. Increased CLOCK levels attenuate the stemness properties of ALDH-positive cells.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Mus musculus) | Clock | NCBI Gene Database | NCBI Gene: 12753 | |
| Cell line (Mus musculus) | 4T1 | ATCC | Cat #: CRL-2539; RRID:CVCL_0125 | |
| Cell line (Homo sapiens) | Lenti-X 293T | Clonetech | Cat #: 632180 | |
| Transfected construct (Mus musculus) | miRDIAN Hairpin Inhibitor | Horizon Discovery | Cat #: IH-310436–08 | |
| Antibody | Rabbit polyclonal anti-CLOCK | Abcam | Cat #: ab3517 RRID:AB_303866 | WB (1:4000) |
| Antibody | Rabbit polyclonal anti-p84 (THOC1) | Proteintech | Cat #: 10920–1-AP RRID:AB_2202239 | WB (1:1500) |
| Antibody | Goat polyclonal anti-E-Cadherin | R and D systems | Cat #: AF748 RRID:AB_355568 | WB (1:1000) |
| Antibody | Rabbit polyclonal anti-Claudin 1 | Proteintech | Cat #: 13050–1-AP RRID:AB_2079881 | WB (1:2000) |
| Antibody | Mouse monoclonal anti-Vimentin | R and D systems | Cat #: MAB21052 RRID:AB_2832972 | WB (1:1000) |
| Antibody | Goat polyclonal anti-Actin-HRP | Santa Cruz Biotechnology | Cat #: sc-1616 | WB (1:5000) |
| Antibody | Mouse monoclonal anti-Ki-67 | Agilent Technology | Cat #: M7240 RRID:AB_2142367 | IHC (1:200) |
| Recombinant DNA reagent | pcDNA3.1(+) (plasmid) | Invitrogen | Cat #: V79020 | |
| Recombinant DNA reagent | pGL4.18 (plasmid) | Promega | Cat #: E6731 | |
| Recombinant DNA reagent | pGL4.13 (plasmid) | Promega | Cat #: E6681 | |
| Recombinant DNA reagent | pRL-SV40 (plasmid) | Promega | Cat #: E2231 | |
| Sequence-based reagent | Primer for sgRNA synthesis | This paper | CCTCTAATA CGACTCACT ATAGGCAAT GGTAGAACT CACACGTTT AAGAGCTAT GC | |
| Sequence-based reagent | Mouse Clock promoter F | This paper | PCR primers for construction of reporter vector | ATACTCGAGA GGTCACTTG GGTCGT |
| Sequence-based reagent | Mouse Clock promoter R | This paper | PCR primers for construction of reporter vector | ATAAGATCT CCTTCCCCT CCTCCACG |
| Peptide, recombinant protein | Recombinant Cas9 | Clontech | Cat #: Z2641N | |
| Peptide, recombinant protein | Recombinant Mouse TGF-β1 | R and D systems | Cat #: 7666 MB | |
| Commercial assay or kit | ALDEFLUOR Kit | StemCell Technologies | Cat #: ST-01700 | |
| Commercial assay or kit | Lipofectamine LTX and Plus Regent | ThermoFisher SCIENTIFIC | Cat #: 15338100 | |
| Commercial assay or kit | Dual-Luciferase reporter assay system | Promega | Cat #: E1910 | |
| Commercial assay or kit | ReverTra Ace qPCR RT Kit | Toyobo | Cat #: FSQ-201 | |
| Commercial assay or kit | THUNDERBIRD SYBR qPCR Mix | Toyobo | Cat #: QPS-201 | |
| Commercial assay or kit | Taqman MicroRNA Reverse Transcription Kit | Applied Biosystems | Cat #: 4366596 | |
| Commercial assay or kit | Taqman MicroRNA Assay | Applied Biosystems | Cat #: 4427975 | |
| Commercial assay or kit | Guide-it sgRNA in vitro transcription kit | Clontech | Cat #: Z2635N | |
| Commercial assay or kit | Lentiviral High Titer Packaging Mix with pLVSIN series | Takara bio | Cat #: 6952 | |
| Commercial assay or kit | CellTiter-Glo Luminescent cell viability assay | Promega | Cat #: 7572 | |
| Chemical compound, drug | 6-Thioguanine | Fujifilm | Cat #: 203–03771 | |
| Software, algorithm | BD FACS Diva | BD Biosciences | RRID:SCR_001456 | |
| Software, algorithm | FlowJo | BD Biosciences | RRID:SCR_008520 | |
| Software, algorithm | Lumicycle analysis software | Actimetrics | ||
| Software, algorithm | ImageQuant LAS 3000 | GE Healthcare | RRID:SCR_014246 | |
| Software, algorithm | BZ analyzer software | KEYENCE | RRID:SCR_017205 | |
| Software, algorithm | Off-Spotter | Thomas Jefferson University | RRID:SCR_015739 | |
| Software, algorithm | JMP | Statistical Discovery | RRID:SCR_014242 | |
| Other | Hoechst33342 | Dojindo laboratories | Cat #: 346–07951 |
Additional files
-
Supplementary file 1
Primer sequences for quantitative PCR analysis.
- https://cdn.elifesciences.org/articles/66155/elife-66155-supp1-v2.xlsx
-
Supplementary file 2
The mRNA levels of transcription factors estimated to bind to genomic.
Clock promoter regions in microarray analysis of ALDH-positive and -negative 4T1 cells.
- https://cdn.elifesciences.org/articles/66155/elife-66155-supp2-v2.xlsx
-
Supplementary file 3
The expression of miRNAs reported as regulating.
Clock expression in ALDH-positive and -negative 4T1 cells in microarray analysis.
- https://cdn.elifesciences.org/articles/66155/elife-66155-supp3-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/66155/elife-66155-transrepform-v2.docx





