PomX, a ParA/MinD ATPase activating protein, is a triple regulator of cell division in Myxococcus xanthus
Figures
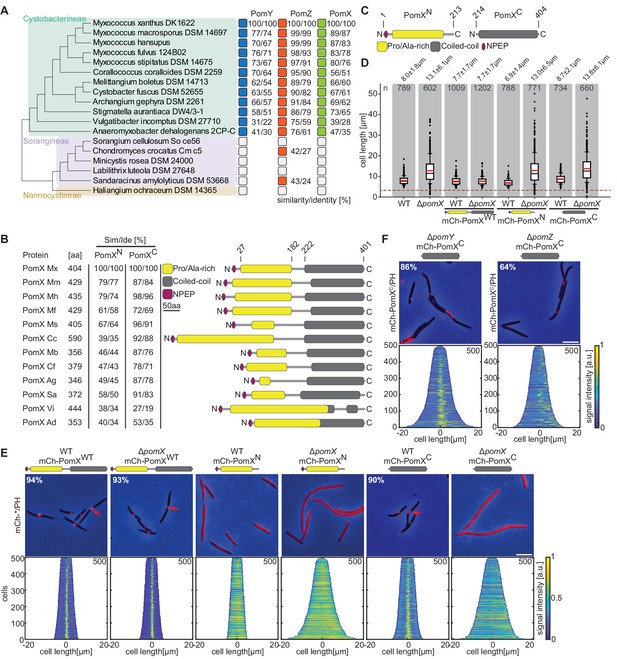
PomX consists of two domains that are both required for function.
(A) Similarity and identity analysis of PomX, PomY, and PomZ homologs. The three Myxococcales suborders are indicated. An open box indicates that a homolog is not present. (B) Similarity and identity of PomX domains in different PomX homologs. Similarity and identity were calculated based on the domains of M. xanthus PomX shown in C. (C) PomX truncations used in this study. Numbers on top indicate the start and stop positions of the truncations relative to full-length PomXWT. (D) Cell length distribution of cells of indicated genotypes. Cells below stippled line are minicells. Numbers indicate mean cell length±STDEV. In the boxplots, boxes include the 25th and the 75th percentile, whiskers data points between the 10% and 90% percentile, outliers are shown as black dots. Black and red lines indicate the median and mean, respectively. Number of analyzed cells is indicated. In the complementation strains, pomX alleles were expressed from plasmids integrated in a single copy at the attB site. (E) Fluorescence microscopy of cells of indicated genotypes. Phase-contrast and fluorescence images of representative cells were overlayed. Numbers indicate fraction of cells with fluorescent clusters. Demographs show fluorescence signals of analyzed cells sorted according to length and with off-center signals to the right. Numbers in upper right indicate number of cells used to create demographs. Scale bar, 5 µm. (F) Fluorescence microscopy of cells of indicated genotypes. Images of representative cells and demographs were created as in (E). Scale bar, 5 µm. For experiments in D, E and F similar results were obtained in two independent experiments.
-
Figure 1—source data 1
Source Data for Figure 1D.
- https://cdn.elifesciences.org/articles/66160/elife-66160-fig1-data1-v2.xlsx
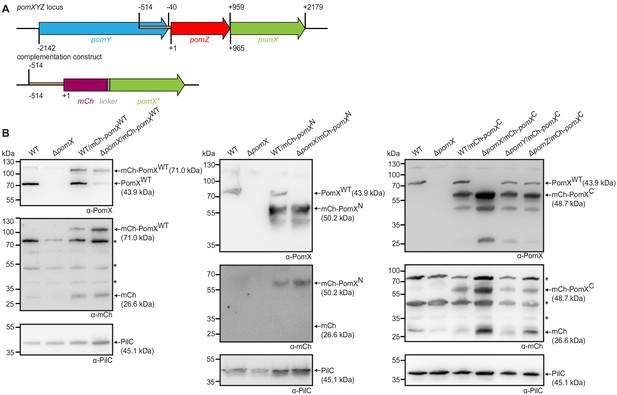
PomX variants accumulate in M.xanthus.
(A) Schematic of pomXYZ locus (upper panel) and the construct used for ectopic expression of mCh-pomX and its variants from the attB site (lower panel). The brown region upstream of pomZ was used as a promoter for the expression of mCh-pomX variants. All coordinates are relative to the first nucleotide in pomZ start codon (+1). (B) Western blot analysis of mCh-PomX (71.0 kDa), mCh-PomXN (50.2 kDa), and mCh-PomXC (48.7 kDa) accumulation in indicated strains. Protein from the same number of cells was loaded per lane. Molecular mass markers are indicated on the left and analyzed proteins on the right including calculated MW. The same blots were sequentially analyzed with α-PomX (top panel), α-mCh (middle panel), and α-PilC (lower panel). PilC was used as a loading control. Note PomXWT (43.9 kDa) does not migrate at the expected size in SDS-PAGE but as a protein of a molecular weight of ~72 kDa. Similarly, mCh-PomXWT, mCh-PomXN, and mCh-PomXC migrate at ~110 kDa, ~60 kDa, and ~62 kDa, respectively. Note that the three bands labeled * in the right and left α-mCh western blot of (B) are unspecific bands that sometimes appear in the western blots with α-mCh antibodies.
-
Figure 1—figure supplement 1—source data 1
Source data for Figure 1—figure supplement 1B.
- https://cdn.elifesciences.org/articles/66160/elife-66160-fig1-figsupp1-data1-v2.xlsx
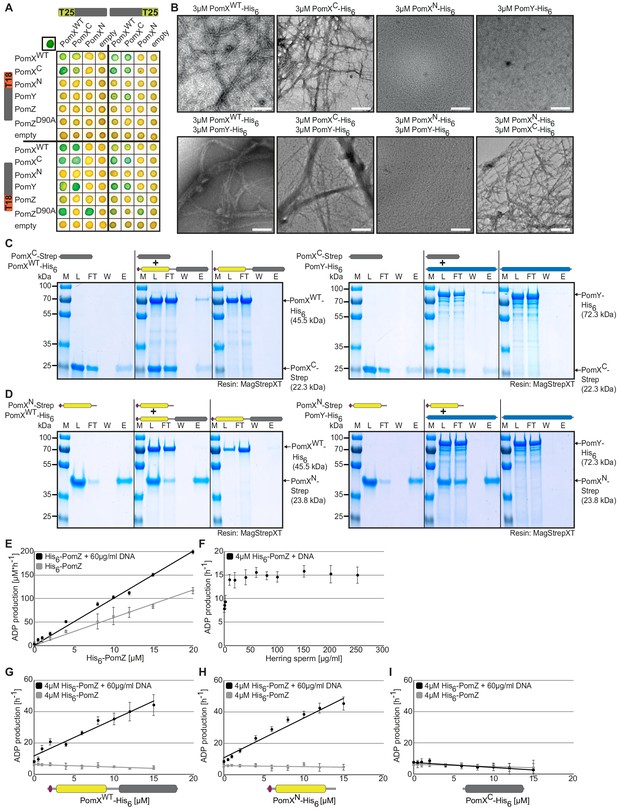
PomXC interacts with PomX and PomY while PomXN stimulates PomZ ATPase activity.
(A) BACTH analysis of interactions between Pom proteins. The indicated protein fragments were fused to T18 and T25 as indicated. Blue colony indicates an interaction, white no interaction. Positive control in upper left corner, leucine zipper of GCN4 fused to T25 and T18. For negative controls, co-transformations with empty plasmids were performed. Images show representative results and were performed in three independent experiments. (B) TEM images of negatively stained purified proteins. Proteins were applied to the EM grids alone or after mixing in a 1:1 molar ratio as indicated before staining. Scale bar, 200 nm. Images show representative results of several independent experiments. (C, D) In vitro pull-down experiments with purified PomXC-Strep, PomXN-Strep, PomXWT-His6, and PomY-His6. Instant Blue-stained SDS-PAGE shows load (L), flow-through (FL), wash (W), and elution (E) fractions using MagStrep XT beads in pull-down experiments with 10 µM of indicated proteins alone or pre-mixed as indicated on top. Molecular size markers are shown on the left and proteins analyzed on the right together with their calculated MW. Note that PomXWT-His6 (Schumacher et al., 2017) and PomXN-Strep migrate aberrantly and according to a higher MW. All samples in a panel were analyzed on the same gel and black lines are included for clarity. Experiments were repeated in two independent experiments with similar results. (E–I) His6-PomZ ATPase activity. ADP production rate was determined in an NADH-coupled photometric microplate assay in the presence of 1 mM ATP at 32°C. DNA and PomX variants were added as indicated. Spontaneous ATP hydrolysis and NADH consumption was accounted for by subtracting the measurements in the absence of His6-PomZ. Data points show the mean±STDEV calculated from six independent measurements.
-
Figure 2—source data 1
Source data for Figure 2A.
- https://cdn.elifesciences.org/articles/66160/elife-66160-fig2-data1-v2.xlsx
-
Figure 2—source data 2
Source data for Figure 2B.
- https://cdn.elifesciences.org/articles/66160/elife-66160-fig2-data2-v2.xlsx
-
Figure 2—source data 3
Source data for Figure 2C.
- https://cdn.elifesciences.org/articles/66160/elife-66160-fig2-data3-v2.xlsx
-
Figure 2—source data 4
Source data for Figure 2D.
- https://cdn.elifesciences.org/articles/66160/elife-66160-fig2-data4-v2.xlsx
-
Figure 2—source data 5
Source data for Figure 2E.
- https://cdn.elifesciences.org/articles/66160/elife-66160-fig2-data5-v2.xlsx
-
Figure 2—source data 6
Source data for Figure 2F.
- https://cdn.elifesciences.org/articles/66160/elife-66160-fig2-data6-v2.xlsx
-
Figure 2—source data 7
Source data for Figure 2G.
- https://cdn.elifesciences.org/articles/66160/elife-66160-fig2-data7-v2.xlsx
-
Figure 2—source data 8
Source data for Figure 2H.
- https://cdn.elifesciences.org/articles/66160/elife-66160-fig2-data8-v2.xlsx
-
Figure 2—source data 9
Source data for Figure 2I.
- https://cdn.elifesciences.org/articles/66160/elife-66160-fig2-data9-v2.xlsx
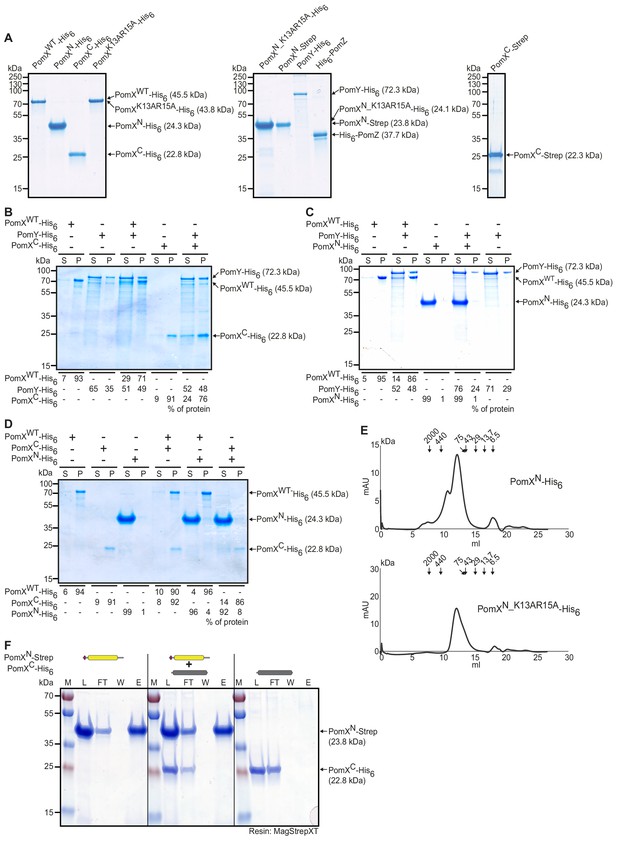
Purification and analysis of Pom proteins.
(A) SDS-PAGE analysis of purified proteins used in this study. Molecular size markers are shown on the left and the purified proteins including calculated MW on the right. Two µg per protein was loaded. Note that PomXWT-His6, PomXN-His6, PomXK12AR15A-His6, PomXN_K13AR15A-His6, and PomXN-Strep do not separate according to their calculated MW. (B–D) Sedimentation assays with indicated proteins. The indicated proteins were mixed at final concentrations of 3 µM as indicated. Following high-speed ultracentrifugation, the supernatant (S) and pellet (P) fractions were separated by SDS-PAGE. Molecular size markers are shown on the left and analyzed proteins on the right. Numbers below show the quantification of indicated protein in the different fractions in %. Similar results were observed in two independent experiments. (E) Size exclusion chromatography elution profile of PomXN-His6 and PomXN_K13AR15A-His6. The elution pattern of PomXN-His6 and PomXN_K13AR15A-His6 from a Superdex 200 10/300 GL gel filtration column was measured at 280 nm. Arrows indicate elution maxima of protein standards of the indicated size in kDa. The same results were observed in two independent experiments. (F) In vitro pull-down experiments with purified PomXN-Strep and PomXC-His6. Instant Blue-stained SDS-PAGE shows load (L), flow-through (FL), wash (W), and elution (E) fractions using MagStrep XT beads in pull-down experiments with 10 µM of indicated proteins alone or pre-mixed as indicated on top. Molecular size markers are shown on the left and proteins analyzed on the right together with their calculated MW. All samples in a panel were analyzed on the same gel and black lines are included for clarity. Experiments were repeated in two independent experiments with similar results.
-
Figure 2—figure supplement 1—source data 1
Source data for Figure 2—figure supplement 1A.
- https://cdn.elifesciences.org/articles/66160/elife-66160-fig2-figsupp1-data1-v2.xlsx
-
Figure 2—figure supplement 1—source data 2
Source data for Figure 2—figure supplement 1B.
- https://cdn.elifesciences.org/articles/66160/elife-66160-fig2-figsupp1-data2-v2.xlsx
-
Figure 2—figure supplement 1—source data 3
Source data for Figure 2—figure supplement 1C.
- https://cdn.elifesciences.org/articles/66160/elife-66160-fig2-figsupp1-data3-v2.xlsx
-
Figure 2—figure supplement 1—source data 4
Source data for Figure 2—figure supplement 1D.
- https://cdn.elifesciences.org/articles/66160/elife-66160-fig2-figsupp1-data4-v2.xlsx
-
Figure 2—figure supplement 1—source data 5
Source data for Figure 2—figure supplement 1E.
- https://cdn.elifesciences.org/articles/66160/elife-66160-fig2-figsupp1-data5-v2.xlsx
-
Figure 2—figure supplement 1—source data 6
Source data for Figure 2—figure supplement 1F.
- https://cdn.elifesciences.org/articles/66160/elife-66160-fig2-figsupp1-data6-v2.xlsx
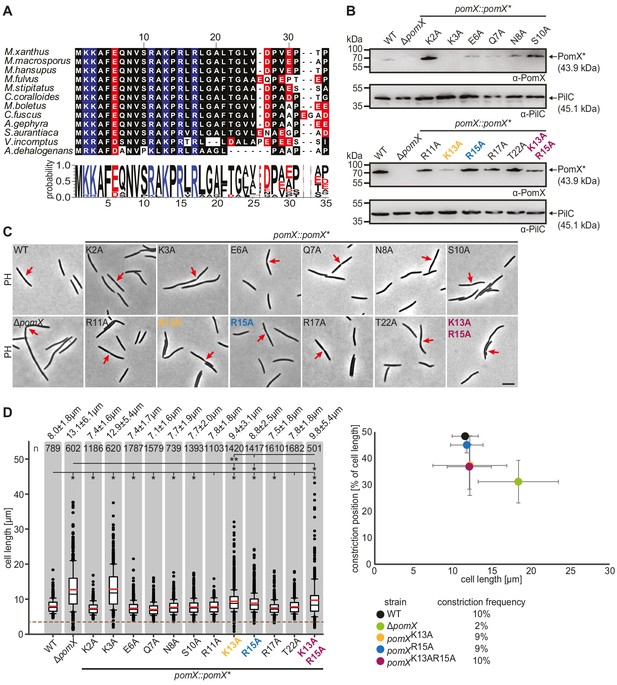
PomXN harbors a conserved N-terminal peptide crucial for cell division site positioning at midcell.
(A) Multiple sequence alignment of the conserved PomX N-terminus. Black background indicates similar amino acids. Positively and negatively charged residues are indicated in blue and red, respectively. Weblogo consensus sequence is shown below. (B) Western blot analysis of accumulation of PomX variants. Protein from the same number of cells was loaded per lane. Molecular mass markers are indicated on the left. PilC was used as a loading control. (C) Phase-contrast microscopy of strains of indicated genotypes. Representative cells are shown. Red arrows indicate cell division constrictions. Scale bar, 5 µm. (D) Analysis of cell length distribution and cell division constrictions of cells of indicated genotypes. Left panel, boxplot is as in Figure 1D. Number of cells analyzed is indicated at the top. *p<0.001; **p<0.05 in Mann-Whitney test. Right panel, cell division position in % of cell length is plotted as a function of cell length. Dots represent mean ± STDEV. Numbers below indicate cell division constriction frequency. In B, C, and D, similar results were obtained in two independent experiments.
-
Figure 3—source data 1
Source data for Figure 3B.
- https://cdn.elifesciences.org/articles/66160/elife-66160-fig3-data1-v2.xlsx
-
Figure 3—source data 2
Source data for Figure 3D.
- https://cdn.elifesciences.org/articles/66160/elife-66160-fig3-data2-v2.xlsx
PomX homologs are highly conserved.
Alignment of PomX homologs from other fully sequenced genomes of Myxobacteria. Sequences were aligned with MUSCLE and color-coded by homology using Bioedit. Black and white backgrounds indicate similar/homologous amino acids and no conservation, respectively.
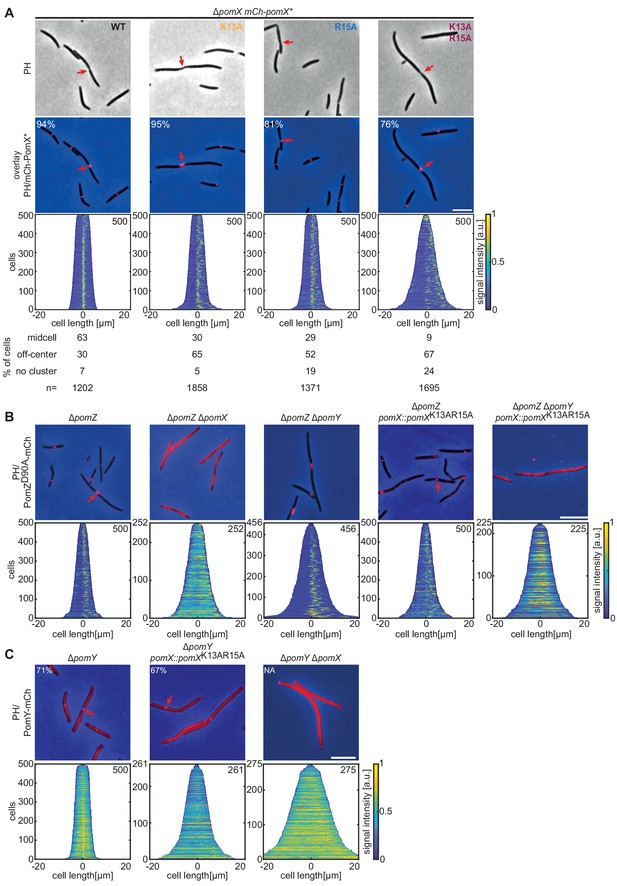
PomXK13AR15A forms clusters and interacts with PomY but not with PomZ in vivo.
(A-C) Fluorescence microscopy of cells of indicated genotypes. Phase-contrast (PH) images and/or overlays of fluorescence images and PH of representative cells. Red arrows indicate division constrictions. Scale bar, 5 µm. In A, numbers in overlays indicate fraction of cells with a cluster and numbers below indicate localization patterns in % and number of cells analyzed. Demographs are as in Figure 1E. Similar results were observed in two independent experiments.
-
Figure 4—source data 1
Source data for Figure 4A.
- https://cdn.elifesciences.org/articles/66160/elife-66160-fig4-data1-v2.xlsx
-
Figure 4—source data 2
Source data for Figure 4B.
- https://cdn.elifesciences.org/articles/66160/elife-66160-fig4-data2-v2.xlsx
-
Figure 4—source data 3
Source data for Figure 4C.
- https://cdn.elifesciences.org/articles/66160/elife-66160-fig4-data3-v2.xlsx
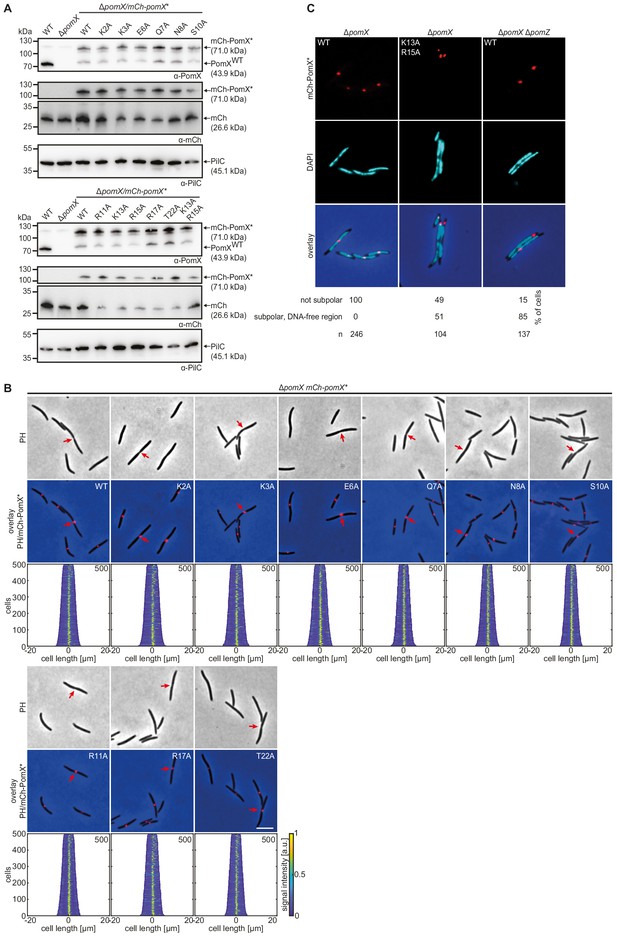
The PomXK13AR15A variant is impaired in function.
(A) Western blot analysis of the accumulation of mCh-PomX variants in indicated strains. Protein from the same number of cells was loaded per lane. Molecular mass marker is shown on the left and analyzed proteins on the right. The same blots were sequentially analyzed with α-PomX (top panel), α-mCh (middle panel), and α-PilC (lower panel) antibodies. PilC was used as a loading control. Note PomX (43.9 kDa) does not migrate at the expected size in SDS-PAGE but instead as a protein of a molecular weight of 72 kDa. Similarly, mCh-PomX migrates at ~110 kDa. Similar results were obtained in two independent experiments. (B) Fluorescence microscopy of indicated mCh-PomX variants. Phase-contrast and fluorescence images of representative cells and the overlay are shown. Red arrows indicate cell division constrictions. Scale bar, 5 µm. Demographs were created as in Figure 1E. Experiments were repeated in two independent experiments with similar results. (C) Fluorescence microscopy of mCh-PomX variants in DAPI-stained cells of the indicated genotype. The mCh signal (first panel), DAPI signal (second panel), and the overlay (third panel) show representative cells. Amino acid substitutions are indicated in white in the mCh images. Scale bar, 5 µm. Quantification of mCh-PomX* localization patterns in % and the number of analyzed cells is shown below the images. Images show representative cells. Similar results were obtained in two independent experiments.
-
Figure 4—figure supplement 1—source data 1
Source data for Figure 4—figure supplement 1A.
- https://cdn.elifesciences.org/articles/66160/elife-66160-fig4-figsupp1-data1-v2.xlsx
-
Figure 4—figure supplement 1—source data 2
Source data for Figure 4—figure supplement 1C.
- https://cdn.elifesciences.org/articles/66160/elife-66160-fig4-figsupp1-data2-v2.xlsx
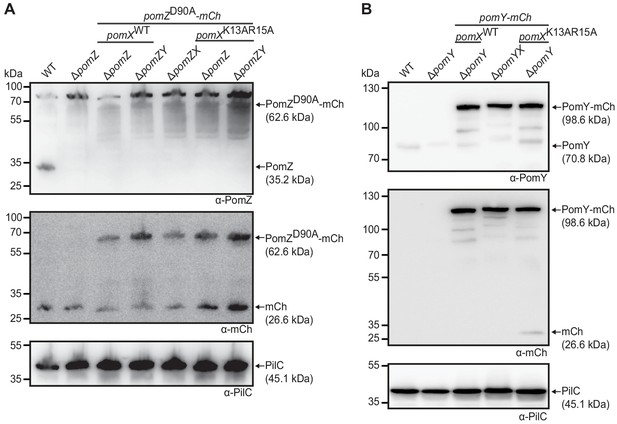
Western blot analysis of PomY-mCh and PomZD90A-mCh accumulation.
(A) Western blot analysis of PomZD90A-mCh accumulation in indicated strains. Protein from the same number of cells was loaded per lane. Molecular mass markers are indicated on the left and analyzed proteins including MW on the right. The same blots were sequentially analyzed with α-PomZ (top panel), α-mCh (middle panel), and α-PilC (lower panel). PilC was used as a loading control. The same results were observed in two independent experiments. (B) Western blot analysis of PomY-mCh accumulation in indicated strains. Blots were done as in (A), but α-PomY antibodies were used instead of α-PomZ antibodies. The same results were observed in two independent experiments.
-
Figure 4—figure supplement 2—source data 1
Source data for Figure 4—figure supplement 2A.
- https://cdn.elifesciences.org/articles/66160/elife-66160-fig4-figsupp2-data1-v2.xlsx
-
Figure 4—figure supplement 2—source data 2
Source data for Figure 4—figure supplement 2B.
- https://cdn.elifesciences.org/articles/66160/elife-66160-fig4-figsupp2-data2-v2.xlsx
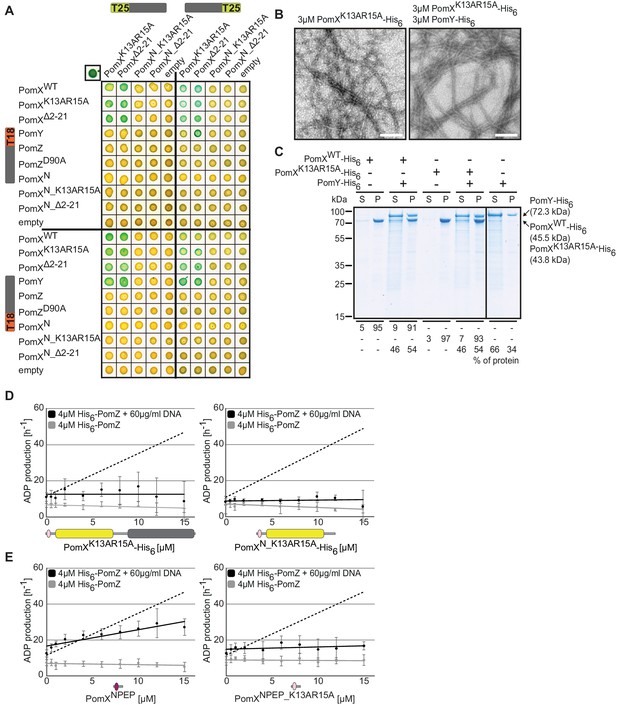
PomX AAP activity resides in PomXNPEP.
(A) BACTH analysis of interactions between Pom proteins and PomX variants. Experiments were performed in parallel with those in Figure 2A. For presentation purposes, the results for PomXWT and PomXN T25 fusion proteins and their interaction with PomZ and PomZD90A T18 fusion proteins were not included but are included in Figure 2A. Images show representative results and similar results were obtained in three independent experiments. (B) TEM images of negatively stained purified proteins. Experiments were done as in Figure 2B. Scale bar, 200 nm. Images show representative results of several independent experiments. (C) Sedimentation assays with indicated purified proteins. Proteins were analyzed at a concentration of 3 µM alone or in combination. After high-speed centrifugation, proteins in the supernatant (S) and pellet (P) fractions were separated by SDS-PAGE and stained with Instant Blue. Molecular size markers are shown on the left and analyzed proteins on the right including their calculated MW. Numbers below indicate % of proteins in different fractions. Similar results were obtained in two independent experiments. All samples were analyzed on the same gel; the black line indicates that lanes were removed for presentation purposes. (D, E) His6-PomZ ATPase activity. Experiments were done and analyzed as in Figure 2E–I in the presence or absence of DNA and the indicated proteins and peptides. Data points show the mean±STDEV calculated from six independent measurements. In (D), stippled lines indicate the regression of the ADP production rate in the presence of PomXWT-His6 (left, Figure 2G) and PomXN-His6 (right, Figure 2H). In E, the stippled line indicates the regression of the ADP production rate in the presence of PomXWT-His6.
-
Figure 5—source data 1
Source data for Figure 5A.
- https://cdn.elifesciences.org/articles/66160/elife-66160-fig5-data1-v2.xlsx
-
Figure 5—source data 2
Source data for Figure 5B.
- https://cdn.elifesciences.org/articles/66160/elife-66160-fig5-data2-v2.xlsx
-
Figure 5—source data 3
Source data for Figure 5C.
- https://cdn.elifesciences.org/articles/66160/elife-66160-fig5-data3-v2.xlsx
-
Figure 5—source data 4
Source data for Figure 5D.
- https://cdn.elifesciences.org/articles/66160/elife-66160-fig5-data4-v2.xlsx
-
Figure 5—source data 5
Source data for Figure 5E.
- https://cdn.elifesciences.org/articles/66160/elife-66160-fig5-data5-v2.xlsx
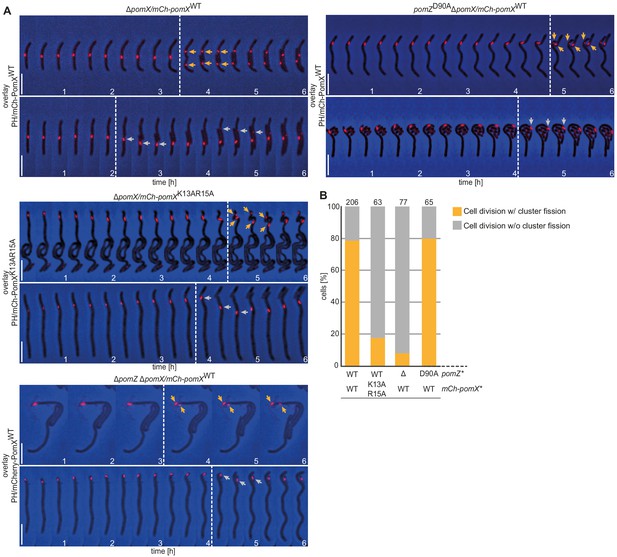
The PomX/PomZ interaction is important for cluster fission during division.
(A) Fluorescence time-lapse microscopy of mCh-PomX variants in cells of indicated genotypes. Overlays of representative mCh images and PH are shown in 20 min intervals. Stippled lines indicate cell division events. Orange and gray arrows mark mCh-PomX clusters in daughter cells after cell division with cluster fission and without cluster fission, respectively. Scale bar, 5 µm. (B) Quantification of cluster fission during cell division in cells of indicated genotypes. Cell division events were divided into those with (orange) and without (gray) cluster fission. Number of analyzed cell divisions is shown on top. The same results were obtained in two independent experiments.
-
Figure 6—source data 1
Source data for Figure 6B.
- https://cdn.elifesciences.org/articles/66160/elife-66160-fig6-data1-v2.xlsx
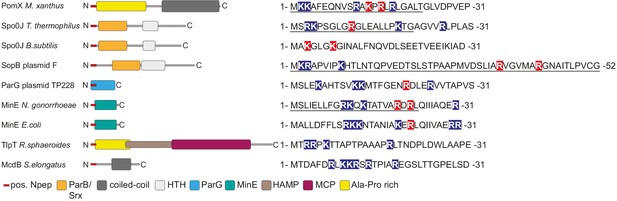
AAPs of MinD/ParA ATPases are diverse but share common features.
Left, domain analysis of known and predicted AAPs of ParA/MinD ATPases with key below. Sequences on the right, N-terminus of indicated proteins. Positively charged amino acids are indicated on blue, and positively charged residues experimentally demonstrated to be important for AAP activity on red. Underlined sequences indicate peptides experimentally demonstrated to have AAP activity. Spo0J of T. thermophilus and B. subtilis, and SopB of plasmid F are ParB homologs.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (M. xanthus) | pomX | NCBI | mxan_0636 new locus tag MXAN_RS03090 | |
| Gene (M. xanthus) | pomY | NCBI | mxan_0634 new locus tag MXAN_RS03080 | |
| Gene (M. xanthus) | pomZ | NCBI | mxan_0635 new locus tag MXAN_RS03085 | |
| Strain, strain background (E. coli) | Arctic Express DE3 RP | Agilent Technologies | E. coli B F– ompT hsdS(rB– mB–) dcm+TetR gal λ(DE3) endA Hte [cpn10 cpn60 GentR] | Used for protein expression |
| Strain, strain background (E. coli) | Rosetta 2 DE3 | Novagen | F- ompT hsdSB(rB- mB-) gal dcm (DE3) pRARE2 (CamR) | Used for protein expression |
| Strain, strain background (E. coli) | NiCo21(DE3) | New England Biolabs | can::CBD fhuA2 [lon] ompT gal (λ DE3) [dcm] arnA::CBD slyD::CBD glmS6Ala ∆hsdS λ DE3 = λ sBamHIo ∆EcoRI-B int::(lacI::PlacUV5::T7 gene1) i21 ∆nin5 | Used for protein expression |
| Strain, strain background (E. coli) | NEB Turbo | New England Biolabs | F' proA+B+ lacIq(∆lacZM15/fhuA2 ∆(lac-proAB) glnV galK16 galE15 R(zgb-210::Tn10) TetS endA1 thi-1 ∆(hsdS-mcrB)5) | Used for cloning |
| Strain, strain background (M. xanthus) | DK1622 | DOI: 10.1073/pnas.76.11.5952 | Wildtype | |
| Strain, strain background (M. xanthus) | SA3108 | DOI: 10.1111/mmi.12094 | ΔpomZ | Strain with an in-frame deletion in pomZ |
| Strain, strain background (M. xanthus) | SA3146 | DOI: 10.1111/mmi.12094 | ΔpomZ; attB::Pmxan0635 pomZD90A-mCh (pKA43) | Strain expressing PomZD90A-mCh in a ΔpomZ background |
| Strain, strain background (M. xanthus) | SA4223 | https://doi.org/10.1016/j.devcel.2017.04.011 | ΔpomX | Strain with an in-frame deletion in pomX |
| Strain, strain background (M. xanthus) | SA4252 | https://doi.org/10.1016/j.devcel.2017.04.011 | ΔpomX; attB::Pmxan0635 mCh-pomX (pAH53) | Strain expressing mCh-PomX in a ΔpomX background |
| Strain, strain background (M. xanthus) | SA4703 | https://doi.org/10.1016/j.devcel.2017.04.011 | ΔpomY | Strain with an in-frame deletion in pomY |
| Strain, strain background (M. xanthus) | SA4712 | https://doi.org/10.1016/j.devcel.2017.04.011 | ΔpomY; attB::PpilA pomY-mCh (pDS7) | Strain expressing PomY-mCh in a ΔpomY background |
| Strain, strain background (M. xanthus) | SA4797 | https://doi.org/10.1016/j.devcel.2017.04.011 | ΔmglA; ΔpomX; attB::Pmxan0635 mCh-pomX (pAH53) | Strain expressing mCh-PomX in a non-motile ΔpomX background |
| Strain, strain background (M. xanthus) | SA4297 | this study | Wild-type; attB::Pmxan0635 mCh-pomX (pAH53) | Strain expressing mCh-PomX in WT background |
| Strain, strain background (M. xanthus) | SA6100 | this study | pomX::pomXK13AR15A | Strain with a replacement of pomX with the pomXK13AR15A allele |
| Strain, strain background (M. xanthus) | SA7014 | this study | ΔpomX; ΔpomZ; attB::Pmxan0635 pomZD90A-mCh (pKA43) | Strain expressing PomZD90A-mCh in a pomX and pomZ deletion background |
| Strain, strain background (M. xanthus) | SA7061 | this study | ΔmglA; ΔpomZ; ΔpomX; attB::Pmxan0635 mCh-pomX (pAH53) | Strain expressing mCh-PomX in a non-motile pomX and pomZ deletion background |
| Strain, strain background (M. xanthus) | SA7063 | this study | ΔpomZ; ΔpomX; attB::Pmxan0635 mCh-pomX (pAH53) | Strain expressing mCh-PomX in a pomX and pomZ deletion background |
| Strain, strain background (M. xanthus) | SA8240 | this study | pomX::pomXK13AR15A; ΔpomZ; attB::Pmxan0635 pomZD90A-mCh (pKA43) | Strain expressing PomZD90A-mCh in a pomZ deletion background with pomXK13AR15A mutation. |
| Strain, strain background (M. xanthus) | SA8250 | this study | pomX::pomXK13AR15A; ΔpomY; attB::PpilA pomY-mCh (pDS7) | Strain expressing PomZD90A-mCh in a pomXK13AR15A background |
| Strain, strain background (M. xanthus) | SA8268 | this study | pomX::pomXK13AR15A; ΔpomY; ΔpomZ; attB::Pmxan0635 pomZD90A-mCh (pKA43) | Strain expressing PomZD90A-mCh in a pomZ and pomY deletion background with pomXK13AR15A mutation |
| Strain, strain background (M. xanthus) | SA9700 | this study | pomX::pomXE6A | Strain with a replacement of pomX with the pomXE6A allele |
| Strain, strain background (M. xanthus) | SA9701 | this study | pomX::pomXQ7A | Strain with a replacement of pomX with the pomXQ7A allele |
| Strain, strain background (M. xanthus) | SA9702 | this study | pomX::pomXN8A | Strain with a replacement of pomX with the pomXN8A allele |
| Strain, strain background (M. xanthus) | SA9714 | this study | pomX::pomXR11A | Strain with a replacement of pomX with the pomXR11A allele |
| Strain, strain background (M. xanthus) | SA9715 | this study | pomX::pomXK3A | Strain with a replacement of pomX with the pomXK3A allele |
| Strain, strain background (M. xanthus) | SA9716 | this study | pomX::pomXR17A | Strain with a replacement of pomX with the pomXR17A allele |
| Strain, strain background (M. xanthus) | SA9717 | this study | pomX::pomXT22A | Strain with a replacement of pomX with the pomXT22A allele |
| Strain, strain background (M. xanthus) | SA9718 | this study | pomX::pomXK2A | Strain with a replacement of pomX with the pomXK2A allele |
| Strain, strain background (M. xanthus) | SA9719 | this study | pomX::pomXR15A | Strain with a replacement of pomX with the pomXR15A allele |
| Strain, strain background (M. xanthus) | SA9720 | this study | ΔpomY; ΔpomZ; attB::Pmxan0635 pomZD90A-mCh (pKA43) | Strain expressing PomZD90A-mCh in a pomZ and pomY deletion background |
| Strain, strain background (M. xanthus) | SA9721 | this study | ΔpomX; ΔpomY; attB::PpilA pomY-mCh (pDS7) | Strain expressing PomY-mCh in a pomY and pomX deletion background |
| Strain, strain background (M. xanthus) | SA9726 | this study | ΔpomX; attB::Pmxan0635 mCh-pomXN (pDS252) | Strain expressing mCh-PomXN in a pomX deletion background |
| Strain, strain background (M. xanthus) | SA9727 | this study | Wild-type; attB::Pmxan0635 mCh-pomXN (pDS252) | Strain expressing mCh-PomXN in a WT background |
| Strain, strain background (M. xanthus) | SA9731 | this study | pomX::pomXK13A | Strain with a replacement of pomX with the pomXK13A allele |
| Strain, strain background (M. xanthus) | SA9732 | this study | pomX::pomXS10A | Strain with a replacement of pomX with the pomXS10A allele |
| Strain, strain background (M. xanthus) | SA9739 | this study | ΔpomX; attB::Pmxan0635 mCh-pomXQ7A (pDS317) | Strain expressing mCh-PomXQ7A in a pomX deletion background |
| Strain, strain background (M. xanthus) | SA9740 | this study | ΔpomX; attB::Pmxan0635 mCh-pomXN8A (pDS318) | Strain expressing mCh-PomXN8A in a pomX deletion background |
| Strain, strain background (M. xanthus) | SA9741 | this study | ΔpomX; attB::Pmxan0635 mCh-pomXR17A (pDS323) | Strain expressing mCh-PomXR17A in a pomX deletion background |
| Strain, strain background (M. xanthus) | SA9742 | this study | ΔpomX; attB::Pmxan0635 mCh-pomXT22A (pDS324) | Strain expressing mCh-PomXT22A in a pomX deletion background |
| Strain, strain background (M. xanthus) | SA9743 | this study | ΔpomX; attB::Pmxan0635 mCh-pomXS10A (pDS319) | Strain expressing mCh-PomXS10A in a pomX deletion background |
| Strain, strain background (M. xanthus) | SA9744 | this study | ΔpomX; attB::Pmxan0635 mCh-pomXR11A (pDS320) | Strain expressing mCh-PomXR11A in a pomX deletion background |
| Strain, strain background (M. xanthus) | SA9747 | this study | ΔpomX; attB::Pmxan0635 mCh-pomXK13A (pDS321) | Strain expressing mCh-PomXK13A in a pomX deletion background |
| Strain, strain background (M. xanthus) | SA9748 | this study | ΔpomX; attB::Pmxan0635 mCh-pomXR15A (pDS322) | Strain expressing mCh-PomXR15A in a pomX deletion background |
| Strain, strain background (M. xanthus) | SA9749 | this study | ΔpomX; attB::Pmxan0635 mCh-pomXK2A (pDS314) | Strain expressing mCh-PomXK2A in a pomX deletion background |
| Strain, strain background (M. xanthus) | SA9750 | this study | ΔpomX; attB::Pmxan0635 mCh-pomXK3A (pDS315) | Strain expressing mCh-PomXK3A in a pomX deletion background |
| Strain, strain background (M. xanthus) | SA9751 | this study | ΔpomX; attB::Pmxan0635 mCh-pomXE6A (pDS316) | Strain expressing mCh-PomXE6A in a pomX deletion background |
| Strain, strain background (M. xanthus) | SA9752 | this study | ΔpomX; attB::Pmxan0635 mCh-pomXK13AR15A (pDS325) | Strain expressing mCh-PomXK13AR15A in a pomX deletion background |
| Strain, strain background (M. xanthus) | SA9753 | this study | ΔmglA; ΔpomX; attB::Pmxan0635 mCh-pomXK13AR15A (pDS325) | Strain expressing mCh-PomXK13AR15A in a non-motile pomX deletion background |
| Strain, strain background (M. xanthus) | SA9754 | this study | ΔmglA; ΔpomZ; ΔpomX; Pmxan0635 mCh-pomX (pAH53); mxan18-19::Pmxan0635 pomZD90A (pDS80) | Strain expressing mCh-PomX in a non-motile pomX and pomZ deletion background that expresses PomZD90A. |
| Strain, strain background (M. xanthus) | SA9755 | this study | Wild-type; attB::Pmxan0635 mCh-pomXC (pDS329) | Strain expressing mCh-PomXC in a WT background |
| Strain, strain background (M. xanthus) | SA9756 | this study | ΔpomY; attB::Pmxan0635 mCh-pomXC (pDS329) | Strain expressing mCh-PomXC in a pomY deletion background |
| Strain, strain background (M. xanthus) | SA9757 | this study | ΔpomZ; attB::Pmxan0635 mCh-pomXC (pDS329) | Strain expressing mCh-PomXC in a pomZ deletion background |
| Strain, strain background (M. xanthus) | SA9762 | this study | ΔpomX; attB::Pmxan0635 mCh-pomXC (pDS329) | Strain expressing mCh-PomXC in a pomX deletion background |
| Recombinant DNA reagent | pAH27 (plasmid) | https://doi.org/10.1016/j.devcel.2017.04.011 | Construct for in-frame deletion of pomX, KmR | |
| Recombinant DNA reagent | pAH53 (plasmid) | https://doi.org/10.1016/j.devcel.2017.04.011 | Pmxan0635 mCh-pomX, Mx8 attB, KmR | |
| Recombinant DNA reagent | pDS1 (plasmid) | https://doi.org/10.1016/j.devcel.2017.04.011 | Construct for in-frame deletion of pomY, KmR | |
| Recombinant DNA reagent | pDS7 (plasmid) | https://doi.org/10.1016/j.devcel.2017.04.011 | PpilA pomY-mCh, Mx8 attB, KmR | |
| Recombinant DNA reagent | pDS16 (plasmid) | https://doi.org/10.1016/j.devcel.2017.04.011 | Construct for in-frame deletion of pomY and pomZ, KmR | |
| Recombinant DNA reagent | pDS80 (plasmid) | https://doi.org/10.1016/j.devcel.2017.04.011 | Pmxan0635 pomZD90A, mxan_18–19 intergenic region, TcR | |
| Recombinant DNA reagent | pEMR3 (plasmid) | https://doi.org/10.1016/j.devcel.2017.04.011 | Overexpression of PomX-His6, KmR | |
| Recombinant DNA reagent | pKA1 (plasmid) | DOI: 10.1111/mmi.12094 | Construct for in-frame deletion of pomZ, KmR | |
| Recombinant DNA reagent | pKA3 (plasmid) | DOI: 10.1111/mmi.12094 | Overexpression of His6-PomZ, KmR | |
| Recombinant DNA reagent | pKA43 (plasmid) | DOI: 10.1111/mmi.12094 | Pmxan0635 pomZD90A-mCh, Mx8 attB, TcR | |
| Recombinant DNA reagent | pMAT12 (plasmid) | https://doi.org/10.1016/j.devcel.2017.04.011 | Construct for in-frame deletion of pomZ and pomX, KmR | |
| Recombinant DNA reagent | pAH152 (plasmid) | this study | Overexpression of PomXC-His6, KmR | |
| Recombinant DNA reagent | pSL16 (plasmid) | DOI: 10.1038/emboj.2011.291 | Construct for in-frame deletion of mglA, KmR | |
| Recombinant DNA reagent | pUT18 (plasmid) | https://doi.org/10.1073/pnas.95.10.5752 | BACTH plasmid | |
| Recombinant DNA reagent | pUT18C (plasmid) | https://doi.org/10.1073/pnas.95.10.5752 | BACTH plasmid | |
| Recombinant DNA reagent | pKT25 (plasmid) | https://doi.org/10.1073/pnas.95.10.5752 | BACTH plasmid | |
| Recombinant DNA reagent | pKNT25 (plasmid) | https://doi.org/10.1073/pnas.95.10.5752 | BACTH plasmid | |
| Recombinant DNA reagent | pAH154 (plasmid) | this study | Pmxan0635 mCh-pomXN, Mx8 attB, KmR | |
| Recombinant DNA reagent | pAH157 (plasmid) | this study | Overexpression of PomXN-His6, KmR | |
| Recombinant DNA reagent | pAH165 (plasmid) | this study | Overexpression of PomXN_K13AR15A-His6, KmR | |
| Recombinant DNA reagent | pDS100 (plasmid) | this study | BACTH plasmid for pomZ (pUT18C), AmpR | |
| Recombinant DNA reagent | pDS103 (plasmid) | this study | BACTH plasmid for pomX (pUT18C), AmpR | |
| Recombinant DNA reagent | pDS106 (plasmid) | this study | BACTH plasmid for pomX (pKT25), KmR | |
| Recombinant DNA reagent | pDS109 (plasmid) | this study | BACTH plasmid for pomZ (pUT18), AmpR | |
| Recombinant DNA reagent | pDS110 (plasmid) | this study | BACTH plasmid for pomX (pUT18), AmpR | |
| Recombinant DNA reagent | pDS114 (plasmid) | this study | BACTH plasmid for pomX (pKNT25) KmR | |
| Recombinant DNA reagent | pDS115 (plasmid) | this study | BACTH plasmid for pomZD90A (pUT18C), AmpR | |
| Recombinant DNA reagent | pDS117 (plasmid) | this study | BACTH plasmid for pomZD90A (pUT18), AmpR | |
| Recombinant DNA reagent | pDS120 (plasmid) | this study | BACTH plasmid for pomY (pUT18C), AmpR | |
| Recombinant DNA reagent | pDS122 (plasmid) | this study | BACTH plasmid for pomY (pUT18), AmpR | |
| Recombinant DNA reagent | pDS184 (plasmid) | this study | BACTH plasmid for pomXΔ2-21 (pUT18), AmpR | |
| Recombinant DNA reagent | pDS185 (plasmid) | this study | BACTH plasmid for pomXΔ2-21 (pUT18C), AmpR | |
| Recombinant DNA reagent | pDS186 (plasmid) | this study | BACTH plasmid for pomXΔ2-21 (pKT25), KmR | |
| Recombinant DNA reagent | pDS187 (plasmid) | this study | BACTH plasmid for pomXΔ2-21 (pKNT25) KmR | |
| Recombinant DNA reagent | pDS188 (plasmid) | this study | BACTH plasmid for pomXC (pUT18), AmpR | |
| Recombinant DNA reagent | pDS189 (plasmid) | this study | BACTH plasmid for pomXC (pUT18C), AmpR | |
| Recombinant DNA reagent | pDS190 (plasmid) | this study | BACTH plasmid for pomXC (pKT25), KmR | |
| Recombinant DNA reagent | pDS191 (plasmid) | this study | BACTH plasmid for pomXC (pKNT25) KmR | |
| Recombinant DNA reagent | pDS192 (plasmid) | this study | BACTH plasmid for pomXN (pUT18), AmpR | |
| Recombinant DNA reagent | pDS193 (plasmid) | this study | BACTH plasmid for pomXN (pUT18C), AmpR | |
| Recombinant DNA reagent | pDS194 (plasmid) | this study | BACTH plasmid for pomXN (pKT25), KmR | |
| Recombinant DNA reagent | pDS195 (plasmid) | this study | BACTH plasmid for pomXN (pKNT25) KmR | |
| Recombinant DNA reagent | pDS232 (plasmid) | this study | Overexpression of PomXN-Strep, KmR | |
| Recombinant DNA reagent | pDS252 (plasmid) | this study | Pmxan0635 mCh-pomXN, Mx8 attB, KmR | |
| Recombinant DNA reagent | pDS253 (plasmid) | this study | BACTH plasmid for pomXN_K13AR15A (pUT18), AmpR | |
| Recombinant DNA reagent | pDS254 (plasmid) | this study | BACTH plasmid for pomXN_K13AR15A (pUT18C), AmpR | |
| Recombinant DNA reagent | pDS255 (plasmid) | this study | BACTH plasmid for pomXN_K13AR15A (pKT25), KmR | |
| Recombinant DNA reagent | pDS256 (plasmid) | this study | BACTH plasmid for pomXN_K13AR15A (pKNT25) KmR | |
| Recombinant DNA reagent | pDS257 (plasmid) | this study | BACTH plasmid for pomXN_Δ2-21 (pUT18), AmpR | |
| Recombinant DNA reagent | pDS258 (plasmid) | this study | BACTH plasmid for pomXN_Δ2-21 (pUT18C), AmpR | |
| Recombinant DNA reagent | pDS259 (plasmid) | this study | BACTH plasmid for pomXN_Δ2-21 (pKT25), KmR | |
| Recombinant DNA reagent | pDS260 (plasmid) | this study | BACTH plasmid for pomXN_Δ2-21 (pKNT25) KmR | |
| Recombinant DNA reagent | pDS303 (plasmid) | this study | nat. site codon exchange for pomXK2A, KmR | |
| Recombinant DNA reagent | pDS304 (plasmid) | this study | nat. site codon exchange for pomXK3A, KmR | |
| Recombinant DNA reagent | pDS305 (plasmid) | this study | nat. site codon exchange for pomXE6A, KmR | |
| Recombinant DNA reagent | pDS306 (plasmid) | this study | nat. site codon exchange for pomXQ7A, KmR | |
| Recombinant DNA reagent | pDS307 (plasmid) | this study | nat. site codon exchange for pomXN8A, KmR | |
| Recombinant DNA reagent | pDS308 (plasmid) | this study | nat. site codon exchange for pomXS10A, KmR | |
| Recombinant DNA reagent | pDS309 (plasmid) | this study | nat. site codon exchange for pomXR11A, KmR | |
| Recombinant DNA reagent | pDS310 (plasmid) | this study | nat. site codon exchange for pomXK13A, KmR | |
| Recombinant DNA reagent | pDS311 (plasmid) | this study | nat. site codon exchange for pomXR15A, KmR | |
| Recombinant DNA reagent | pDS312 (plasmid) | this study | nat. site codon exchange for pomXR17A, KmR | |
| Recombinant DNA reagent | pDS313 (plasmid) | this study | nat. site codon exchange for pomXT22A, KmR | |
| Recombinant DNA reagent | pDS314 (plasmid) | this study | Pmxan0635 mCh-pomXK2A, Mx8 attB, KmR | |
| Recombinant DNA reagent | pDS315 (plasmid) | this study | Pmxan0635 mCh-pomXK3A, Mx8 attB, KmR | |
| Recombinant DNA reagent | pDS316 (plasmid) | this study | Pmxan0635 mCh-pomXE6A, Mx8 attB, KmR | |
| Recombinant DNA reagent | pDS317 (plasmid) | this study | Pmxan0635 mCh-pomXQ7A, Mx8 attB, KmR | |
| Recombinant DNA reagent | pDS318 (plasmid) | this study | Pmxan0635 mCh-pomXN8A, Mx8 attB, KmR | |
| Recombinant DNA reagent | pDS319 (plasmid) | this study | Pmxan0635 mCh-pomXS10A, Mx8 attB, KmR | |
| Recombinant DNA reagent | pDS320 (plasmid) | this study | Pmxan0635 mCh-pomXR11A, Mx8 attB, KmR | |
| Recombinant DNA reagent | pDS321 (plasmid) | this study | Pmxan0635 mCh-pomXK13A, Mx8 attB, KmR | |
| Recombinant DNA reagent | pDS322 (plasmid) | this study | Pmxan0635 mCh-pomXR15A, Mx8 attB, KmR | |
| Recombinant DNA reagent | pDS323 (plasmid) | this study | Pmxan0635 mCh-pomXR17A, Mx8 attB, KmR | |
| Recombinant DNA reagent | pDS324 (plasmid) | this study | Pmxan0635 mCh-pomXT22A, Mx8 attB, KmR | |
| Recombinant DNA reagent | pDS325 (plasmid) | this study | Pmxan0635 mCh-pomXK13AR15A, Mx8 attB, KmR | |
| Recombinant DNA reagent | pDS329 (plasmid) | this study | Pmxan0635 mCh-pomXC, Mx8 attB, KmR | |
| Recombinant DNA reagent | pDS333 (plasmid) | this study | Overexpression of PomXC-Strep, KmR | |
| Recombinant DNA reagent | pEMR1 (plasmid) | this study | Overexpression of PomY-His6, KmR | |
| Recombinant DNA reagent | pSH1 (plasmid) | this study | nat. site codon exchange for pomXK13AR15A, KmR | |
| Recombinant DNA reagent | pSH36 (plasmid) | this study | BACTH plasmid for pomXK13AR15A (pKNT25) KmR | |
| Recombinant DNA reagent | pSH37 (plasmid) | this study | BACTH plasmid for pomXK13AR15A (pKT25), KmR | |
| Recombinant DNA reagent | pSH38 (plasmid) | this study | BACTH plasmid for pomXK13AR15A (pUT18), AmpR | |
| Recombinant DNA reagent | pSH39 (plasmid) | this study | BACTH plasmid for pomXK13AR15A (pUT18C), AmpR | |
| Recombinant DNA reagent | pSH58 (plasmid) | this study | Overexpression of PomXK13AR15A-His6, KmR | |
| Sequence-based reagent | pomX BTH fwd XbaI | this study | PCR primer | 5’-GCGTCTAGAGATGAAGAAAGCCTTTGAAC-3’ |
| Sequence-based reagent | pomX BTH rev KpnI | this study | PCR primer | 5’-GCGGGTACCCGGCGCACCGTGGCCTGAC-3’ |
| Sequence-based reagent | pomY BTH fwd XbaI | this study | PCR primer | 5’-GCGTCTAGAGGTGAGCGACGAGCGTCCG-3’ |
| Sequence-based reagent | pomY BTH rev KpnI | this study | PCR primer | 5’-GCGGGTACCCGAGCGGCGAAGTATTTGTG-3’ |
| Sequence-based reagent | pomZ BTH fwd XbaI | this study | PCR primer | 5’-GCGTCTAGAGATGGAAGCGCCGACGTAC-3’ |
| Sequence-based reagent | pomZ BTH rev KpnI | this study | PCR primer | 5’-GCGGGTACCCGGCCGGCCTGCTGGGTGCC-3’ |
| Sequence-based reagent | pomXΔ2–21 BTH fwd XbaI | this study | PCR primer | 5’-GCGTCTAGAGATGACGGGCCTCGTCGACCCC-3’ |
| Sequence-based reagent | pomXC BTH fwd XbaI | this study | PCR primer | 5’-GCGTCTAGAGATGGCCACCGTGGCGGAGGCG-3’ |
| Sequence-based reagent | pomXN BTH rev KpnI | this study | PCR primer | 5’-GCGGGTACCCGGGGCAGCGGCTCCGGGCG-3’ |
| Sequence-based reagent | 0636 up fwd | this study | PCR primer | 5’-GCGGGATCCGTCACCCCAAGCCATTC-3’ |
| Sequence-based reagent | PomX K2A rev native | this study | PCR primer | 5’-CAAAGGCTTTCGCCATGGTTCTCAG-3’ |
| Sequence-based reagent | PomX K2A fwd native | this study | PCR primer | 5’-CTGAGAACCATGGCGAAAGCCTTTG-3’ |
| Sequence-based reagent | 0636 HindIII rev stop | this study | PCR primer | 5’-GCGAAGCTTTCAGCGCACCGTGGCCTGAC-3’ |
| Sequence-based reagent | PomX K3A rev native | this study | PCR primer | 5’-CTGTTCAAAGGCCGCCTTCATGGTTC-3’ |
| Sequence-based reagent | PomX K3A fwd native | this study | PCR primer | 5’-GAACCATGAAGGCGGCCTTTGAACAG-3’ |
| Sequence-based reagent | PomX E6A rev | this study | PCR primer | 5’-GGACACGTTCTGCGCAAAGGCTTTCTT-3’ |
| Sequence-based reagent | PomX E6A fwd | this study | PCR primer | 5’-AAGAAAGCCTTTGCGCAGAACGTGTCC-3’ |
| Sequence-based reagent | PomX Q7A rev | this study | PCR primer | 5’-GCGGGACACGTTCGCTTCAAAGGCTTT-3’ |
| Sequence-based reagent | PomX Q7A fwd | this study | PCR primer | 5’-AAAGCCTTTGAAGCGAACGTGTCCCGC-3’ |
| Sequence-based reagent | PomX N8A rev | this study | PCR primer | 5’-GGCGCGGGACACCGCCTGTTCAAAGGC-3’ |
| Sequence-based reagent | PomX N8A fwd | this study | PCR primer | 5’-GCCTTTGAACAGGCGGTGTCCCGCGCC-3’ |
| Sequence-based reagent | PomX S10A rev | this study | PCR primer | 5’-CGGCTTGGCGCGCGCCACGTTCTGTTC-3’ |
| Sequence-based reagent | PomX S10A fwd | this study | PCR primer | 5’-GAACAGAACGTGGCGCGCGCCAAGCCG-3’ |
| Sequence-based reagent | PomX R11A rev | this study | PCR primer | 5’-GCGCGGCTTGGCCGCGGACACGTTCTG-3’ |
| Sequence-based reagent | PomX R11A fwd | this study | PCR primer | 5’-CAGAACGTGTCCGCGGCCAAGCCGCGC-3’ |
| Sequence-based reagent | PomX K13A rev | this study | PCR primer | 5’-GCGGAGGCGCGGCGCGGCGCGGGACAC-3’ |
| Sequence-based reagent | PomX K13A fwd | this study | PCR primer | 5’-GTGTCCCGCGCCGCGCCGCGCCTCCGC-3’ |
| Sequence-based reagent | PomX R15A rev | this study | PCR primer | 5’-GCCCAGGCGGAGCGCCGGCTTGGCGCG-3’ |
| Sequence-based reagent | PomX R15A fwd | this study | PCR primer | 5’-CGCGCCAAGCCGGCGCTCCGCCTGGGC-3’ |
| Sequence-based reagent | PomX R17A rev | this study | PCR primer | 5’-CAGCGCGCCCAGCGCGAGGCGCGGCTT-3’ |
| Sequence-based reagent | PomX R17A fwd | this study | PCR primer | 5’-AAGCCGCGCCTCGCGCTGGGCGCGCTG-3’ |
| Sequence-based reagent | PomX T22A rev | this study | PCR primer | 5’-GTCGACGAGGCCCGCCAGCGCGCCCAG-3’ |
| Sequence-based reagent | PomX T22A fwd | this study | PCR primer | 5’-CTGGGCGCGCTGGCGGGCCTCGTCGAC-3’ |
| Sequence-based reagent | PomX K13AR15A rev | this study | PCR primer | 5’-CAGAACGTGTCCCGCGCCGCGCCGGCCCTCCGCCTGGGCGCGCTG-3’ |
| Sequence-based reagent | PomX K13AR15A fwd | this study | PCR primer | 5’-CAGCGCGCCCAGGCGGAGGGCCGGCGCGGCGCGGGACACCTTCTG-3’ |
| Sequence-based reagent | mCherry XbaI fwd | this study | PCR primer | 5’-GCGTCTAGAGTGAGCAAGGGCGAGGAG-3’ |
| Sequence-based reagent | PomX K2A rev | this study | PCR primer | 5’-TTCAAAGGCTTTCGCCATGGCTCCGCC-3’ |
| Sequence-based reagent | PomX K2A fwd | this study | PCR primer | 5’-GGCGGAGCCATGGCGAAAGCCTTTGAA-3’ |
| Sequence-based reagent | KA348 | this study | PCR primer | 5’-GCCAAGCTTTCAGCGCACCGTGGCCTG-3’ |
| Sequence-based reagent | PomX K3A fwd | this study | PCR primer | 5’-GGAGCCATGAAGGCGGCCTTTGAACAG-3’ |
| Sequence-based reagent | PomX K3A rev | this study | PCR primer | 5’-CTGTTCAAAGGCCGCCTTCATGGCTCC-3’ |
| Sequence-based reagent | AH142 | this study | PCR primer | 5’-GGAATTCCATATGGCCACCGTGGCGGAGGCG-3’ |
| Sequence-based reagent | KA346 | this study | PCR primer | 5’-GCCAAGCTTGCGCACCGTGGCCTGACTC-3’ |
| Sequence-based reagent | AH143 | this study | PCR primer | 5’-CCCAAGCTTGGGCAGCGGCTCCGGGCG-3’ |
| Sequence-based reagent | NdeI PomX fwd | this study | PCR primer | 5’-GGAATTCCATATGAAGAAAGCCTTTGAACAG-3’ |
| Sequence-based reagent | AH144 | this study | PCR primer | 5’-GCCAAGCTTTCAGGGCAGCGGCTCCGGGCG-3’ |
| Sequence-based reagent | KA384 | this study | PCR primer | 5’-GCGGGATCCGGCGGAGCCATGAAGAAAGCCTTTGAACAG-3’ |
| Sequence-based reagent | DS276 | this study | PCR primer | 5’-GCGAAGCTTACTTCTCGAACTGTGGGTGACTCCAGCGCACCGTGGCCTGAC-3’ |
| Sequence-based reagent | DS277 | this study | PCR primer | 5’-GCGCCATGGCCACCGTGGCGGAGGCG-3’ |
| Sequence-based reagent | PomX BspHI fwd | this study | PCR primer | 5’-GCGTCATGAAGAAAGCCTTTGAACAGAACG-3’ |
| Sequence-based reagent | PomXN rev strep-tag | this study | PCR primer | 5’-GCGAAGCTTACTTCTCGAACTGTGGGTGACTCCAGGGCAGCGGCTCCGGGCG-3’ |
| Sequence-based reagent | NdeI-PomY fwd | this study | PCR primer | 5’-GGAATTCCATATGAGCGACGAGCGTCCGGAC-3’ |
| Sequence-based reagent | PomY C-term his rev | this study | PCR primer | 5’-CGGAAGCTTAGCGGCGAAGTATTTGTGC-3’ |
| Sequence-based reagent | AH141 | this study | PCR primer | 5’-GCGGGATCCGGCGGAGCCGCCACCGTGGCGGAGGCG-3’ |
| Antibody | α-PomX (rabbit, polyclonal) | https://doi.org/10.1016/j.devcel.2017.04.011 | Western Blot (1:15000) | |
| Antibody | α-PomY (rabbit, polyclonal) | https://doi.org/10.1016/j.devcel.2017.04.011 | Western Blot (1:15000) | |
| Antibody | α-PomZ (rabbit, polyclonal) | DOI: 10.1111/mmi.12094 | Western Blot (1:10000) | |
| Antibody | α-PilC (rabbit, polyclonal) | DOI:10.1111/j.1365–2958.2009.06891.x | Western Blot (1:3000) | |
| Antibody | α-mCherry (rabbit, polyclonal) | Biovision | Cat# 5993 | Western Blot (1:10000) |
| Antibody | horseradish-conjugated α-rabbit immunoglobulin G (goat,polyclonal) | Sigma-Aldrich | Cat# A0545-1ML | Western Blot (1:25000) |
| Peptide, recombinant protein | PomXNPEP | Thermo Scientific | MKKAFEQNVSRAKPRLRLGALT | |
| Peptide, recombinant protein | PomXNPEPK13AR15A | Thermo Scientific | MKKAFEQNVSRAAPALRLGALT | |
| Software, algorithm | Metamorph_v 7.5 | Molecular Devices | ||
| Software, algorithm | Oufti | DOI: 10.1111/mmi.13264 | http://www.oufti.org/ | |
| Software, algorithm | Matlab R2018a | MathWorks | ||
| Commercial assay or kit | Luminata Forte | Fisher scientific | Cat# 10394675 |





