Uniparental nuclear inheritance following bisexual mating in fungi
Figures
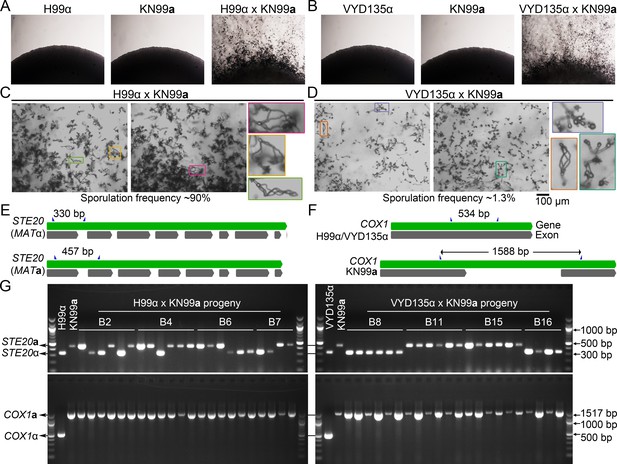
Chromosome shuffled strain exhibits unusual sexual reproduction.
(A, B) Images of cultures for the individual strains H99α, KN99a, and VYD135α, showing no self-filamentation on mating medium. Magnification=10×. (C, D) Light microscopy images showing robust sporulation in the H99α×KN99a cross, whereas the VYD135α×KN99a cross exhibited robust hyphal development but infrequent sporulation events. The inset images in colored boxes show examples of basidia observed in each of the crosses. Scale bar, 100 µm. (E, F) A scheme showing the MATα (H99α and VYD135α) and MATa (KN99a) alleles at the STE20 (E) and COX1 (F) loci. Primers used for PCR analysis are marked by blue triangles. (G) Gel images showing PCR amplification of STE20 and COX1 alleles in the progeny obtained from four different basidia for both H99α×KN99a and VYD135α×KN99a crosses. PCR analysis for the parental strains is also shown, and key bands for DNA marker are labeled. PCR, polymerase chain reaction.
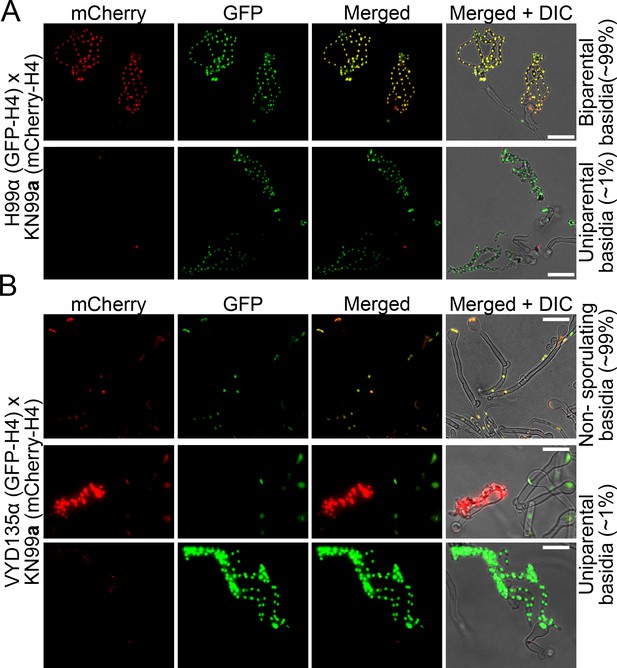
Fluorescence microscopy reveals uniparental nuclear inheritance in the wild-type crosses.
(A) Crosses of GFP-H4 tagged H99α and mCherry-H4 tagged KN99a revealed the presence of both fluorescent markers in most spore chains along with uniparental nuclear inheritance in rare cases (~1%). In these few sporulating basidia, only one of the fluorescent signals was observed in the spore chains, reflecting the presence of only one parental nucleus in these basidia. (B) Crosses involving GFP-H4 tagged VYD135α and mCherry-H4 tagged KN99a revealed the presence of spore chains with only one fluorescent color. In the majority of basidia that have both parental nuclei, marked by both GFP and mCherry signals, spore chains are not produced, consistent with a failure of meiosis in these basidia. Scale bar, 10 µm.
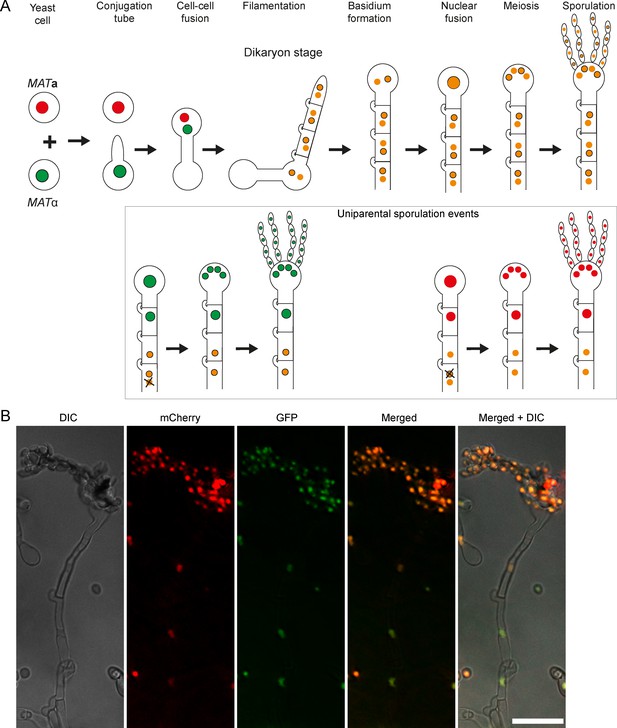
Dynamics of sexual reproduction and sporulation analyzed with C. neoformans strains expressing nuclear-localized fluorescent reporter proteins.
(A) A cartoon depicting various stages of sexual reproduction in C. neoformans, from the formation of conjugation tube to sporulation, and possible dynamics of the nuclei at these different stages. After the cell-cell fusion, tagged proteins assort into both nuclei and yield a yellow/orange fluorescence color as a result of the mixing of the green and red signals. Cartoons in the box show hypothetical scenarios where uniparental nuclear inheritance occurs after the loss of one parental nucleus. (B) Direct fluorescence microscopy images showing the status of GFP-H4 tagged and mCherry-H4 tagged nuclei in post-mating hyphae as well as in spores. Both GFP and mCherry fluorescent colors were observed in hyphae and spores as hypothesized in (A). Scale bar, 10 µm.
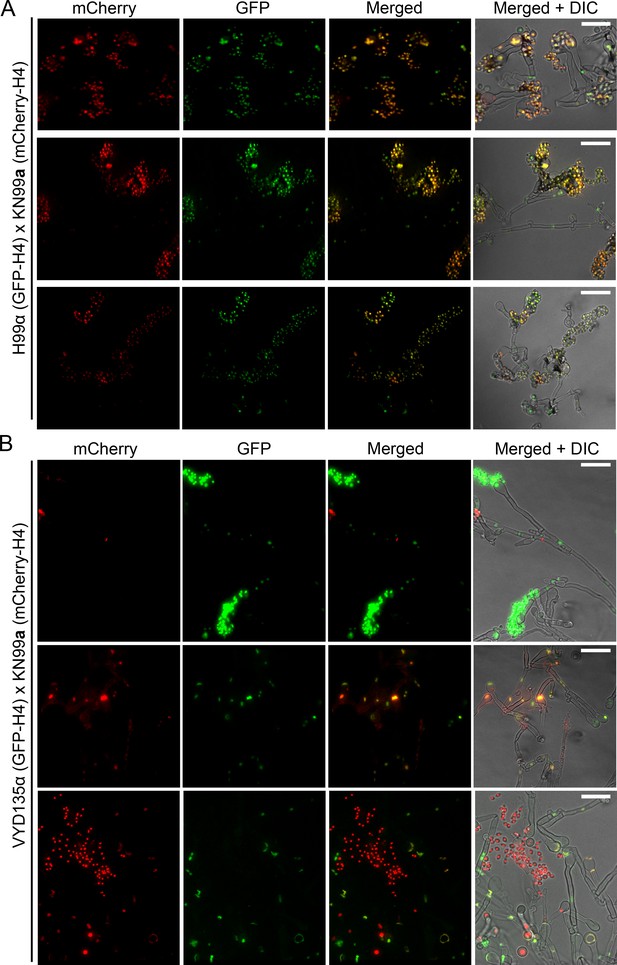
Nuclear dynamics during sporulation in the wild-type and VYD135α crosses.
GFP-H4 and mCherry-H4 tagging revealed different localization patterns in the (A) wild-type H99α×KN99a and (B) VYD135α×KN99a crosses. Wild-type spore chains mostly harbored both the nuclear stains as a result of bisexual meiosis. On the other hand, basidia with only one of the parental nuclei produced spores in VYD135α×KN99a crosses; basidia with both nuclei failed to produce spore chains and, as a result, remained as bald basidia. Scale bar, 10 µm.
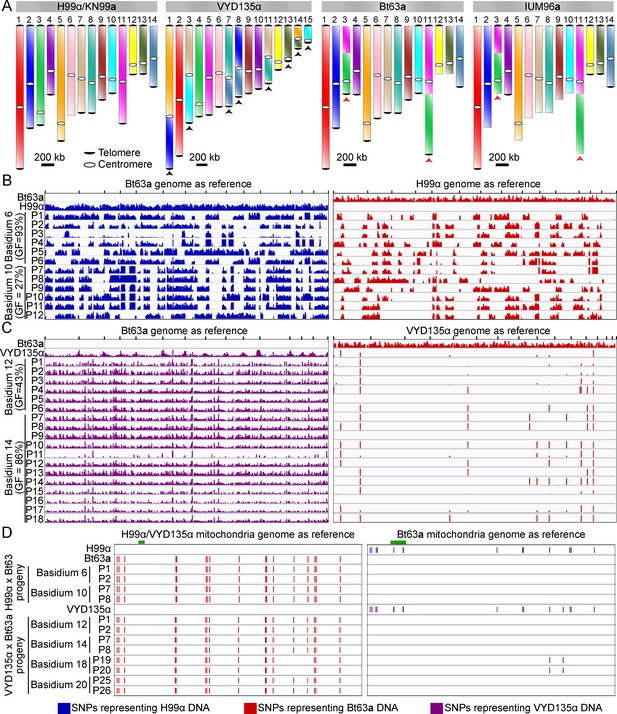
VYD135α progeny exhibit strict uniparental nuclear inheritance and lack the signature of meiotic recombination.
(A) Chromosome maps for H99α/ΚN99a, VYD135α, Bt63a, and IUM96a showing the karyotype variation. The genome of the wild-type strain H99α served as the reference. Black arrowheads represent chromosome translocations between VYD135α and H99α whereas red arrowheads mark chromosomes with a translocation between H99α and Bt63a or IUM96a. (B) Whole-genome sequencing, followed by SNP identification, of H99α×Bt63a progeny revealed evidence of meiotic recombination in all of the progeny. The left panel shows SNPs with respect to the Bt63a genome whereas the right panel depicts SNPs against the H99α genome. H99α and Bt63a Illumina sequencing data served as controls for SNP calling. (C) SNP analysis of VYD135α ×Bt63a progeny revealed no contribution of the Bt63a parental genome in the progeny as evidenced by the presence of SNPs only against Bt63a (left panel) but not against the VYD135α genome (right panel). The presence of a few SNPs observed in VYD135α, as well as all VYD135α×Bt63a progeny, are within nucleotide repeat regions. GF stands for germination frequency and P stands for progeny. (D) SNP analysis of H99α×Bt63a and VYD135α×Bt63a progeny using mitochondrial DNA as the reference revealed that mitochondrial DNA is inherited from Bt63a in all of the progeny. Progeny obtained from VYD135α×Bt63a basidium 18 also revealed recombination between the two parental mitochondrial genomes as marked by the absence or presence of two SNPs when mapped against VYD135α and Bt63a mitochondrial genomes, respectively. The green bar in each panel depicts the locus used for PCR analysis of the mitochondrial genotype in the progeny. PCR, polymerase chain reaction; SNP, single nucleotide polymorphism.
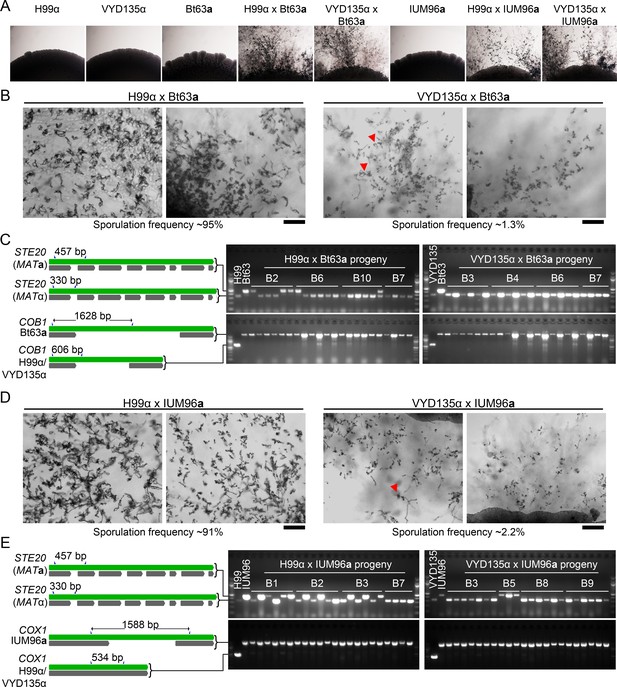
Pseudosexual reproduction occurs in natural isolates, Bt63a and IUM96a.
(A) Images of the mating spots showing filamentation when two strains of opposite mating types are crossed. No filamentation is observed without the presence of a mating partner. (B, D) Light microscopy images showing sporulation frequency in crosses involving Bt63a (B) and IUM96a (D). Scale bar, 100 µm. (C, E) Schemes depicting the STE20 alleles used for MAT locus and COB1 (for Bt63a) and COX1 (for IUM96a) alleles for mitochondrial genotyping, respectively. Gel images show the PCR analysis on progeny from four basidia and the parental strains for all crosses as mentioned. PCR, polymerase chain reaction.
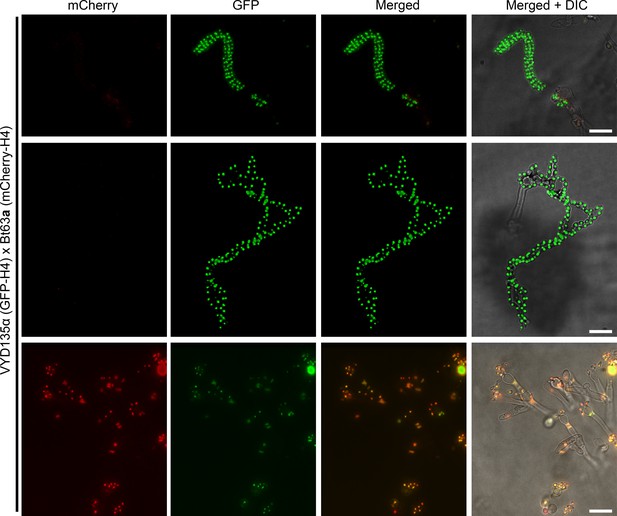
Bt63a fluorescence microscopy revealed pseudosexual reproduction events.
GFP-H4 tagged VYD135α crossed with mCherry-H4 tagged Bt63a showed only VYD135α sporulation events as also observed in spore dissection analysis. Scale bar, 10 µm.

VYD135α×Bt63a progeny lack signatures of meiotic recombination.
SNP analysis on VYD135α×Bt63a progeny revealed no contribution of the Bt63a parental genome in the progeny as evidenced by the presence of SNPs only against Bt63a (left panel) but not against VYD135α genome (right panel). The few SNPs observed in VYD135α as well as all VYD135α×Bt63a progeny lie within nucleotide repeat regions. GF stands for germination frequency and P stands for progeny. SNP, single nucleotide polymorphism.
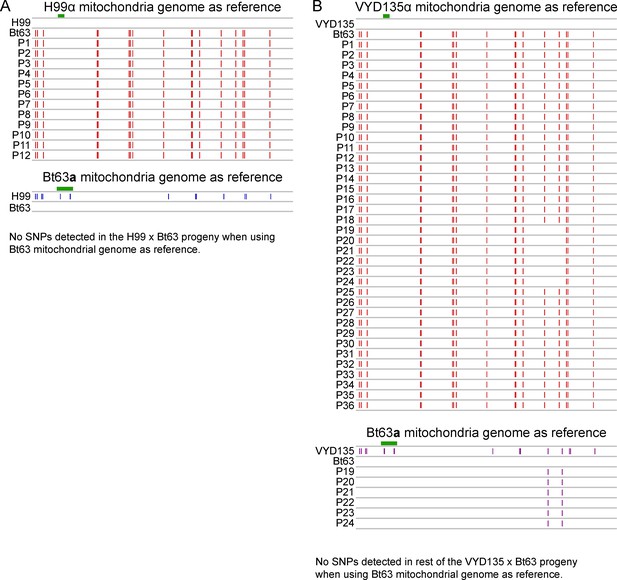
Mitochondria are inherited from MATa parent in all of the progeny.
(A) A map of SNPs detected in H99α×Bt63a progeny when using H99α mitochondrial DNA (upper panel) and Bt63a mitochondrial DNA (lower panel) as the reference. (B) SNP analysis revealed variants in all the progeny when using VYD135α mitochondrial DNA as the reference but not when using Bt63a mitochondrial DNA. The two SNPs detected against Bt63a DNA in progeny P19–24 (basidium 18) suggest recombination of two parental mitochondrial DNA during mating. The green bar in each panel depicts the fragment used for PCR analysis in Figure 3—figure supplement 1. P stands for progeny. PCR, polymerase chain reaction; SNP, single nucleotide polymorphism.
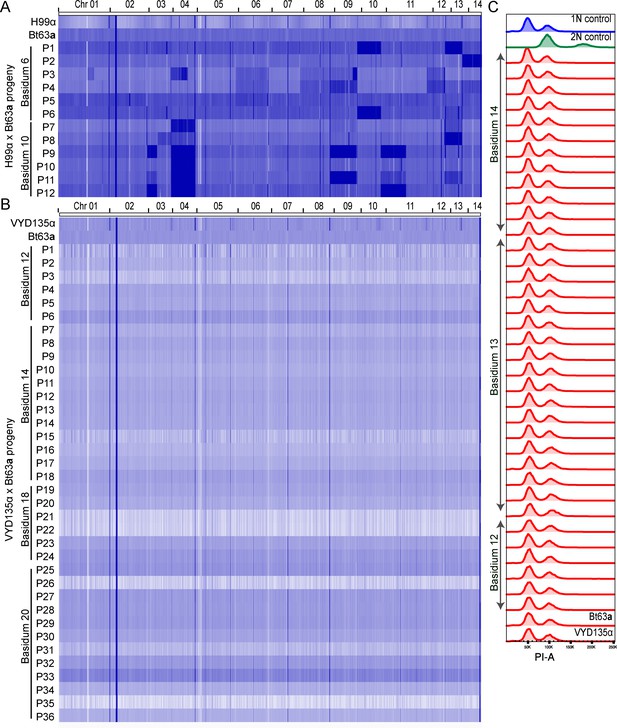
VYD135α×Bt63a progeny are haploid.
(A) Whole-genome sequencing of the H99α×Bt63a progeny revealed extensive aneuploidy in the progeny. Each progeny seemed to exhibit aneuploidy for at least one chromosome. (B) Whole-genome sequencing data revealed that the progeny obtained from VYD135α×Bt63a 5-week-old crosses are euploid in nature as they show a uniform level of genomic content when mapped to the Bt63 genome. VYD135α and Bt63a whole-genome sequencing data were also mapped as controls. Each lane represents one strain, and the difference in intensity correlates with the number of reads obtained per sample. (C) Flow-cytometry analysis on progeny obtained from three basidia confirmed that all the germinating progeny are haploid. While progeny from B12 and B14 are the same as used for the whole-genome sequencing, progeny from B3 were subjected to only flow cytometry analysis. Bt63a and VYD135α were also analyzed as controls for this experiment. P stands for progeny.

IUM96a exhibits meiotic recombination in progeny with H99α but not with the genome shuffle strain VYD135α.
(A) The left panel depicts SNPs with respect to the IUM96a genome whereas the right panel shows SNPs against the H99α genome. Whole-genome sequencing, followed by SNP analysis, for the H99α×IUM96a progeny (basidia 3 and 4) revealed evidence of meiotic recombination in the progeny. Basidium 7 from the H99α×IUM96a cross produced uniparental progeny, which was confirmed by SNP analysis on a subset of these progeny. The progeny exhibited SNPs only against the IUM96a genome but not against the H99α genome. (B) SNP analysis from two different basidia revealed the inheritance of only one set of parental nuclear DNA in the progeny from VYD135α×IUM96a cross. Basidium 3 progeny possessed DNA from only the VYD135α parent, while basidium 5 progeny inherited nuclear DNA from IUM96a alone. The results obtained from this analysis are congruent with mating-type PCR results shown in Supplementary file 1b. GF stands for germination frequency and P stands for progeny. PCR, polymerase chain reaction; SNP, single nucleotide polymorphism.
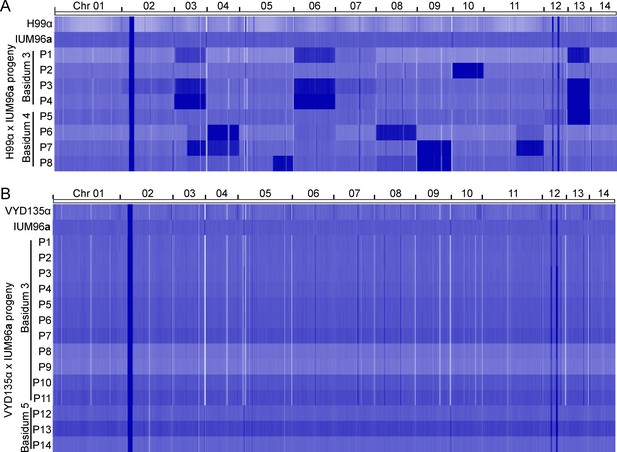
Ploidy analysis of IUM96a progeny reveals haploid uniparental progeny.
Whole-genome sequencing analysis revealed the presence of multiple aneuploidies in the (A) H99α×IUM96a progeny, but a completely euploid genome for the (B) VYD135α ×IUM96a progeny. P stands for progeny.
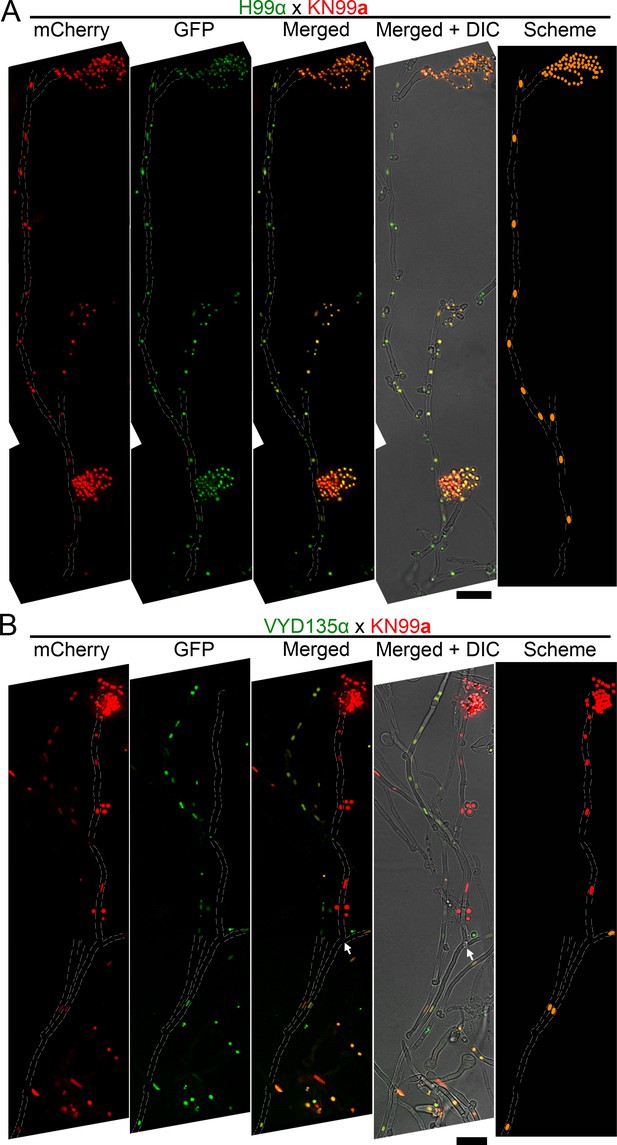
Pan-hyphal microscopy reveals the loss of one parental nucleus during pseudosexual reproduction.
Spore-producing long hyphae were visualized in both (A) wild-type H99α×KN99a and (B) VYD135α×KN99a crosses to study the dynamics of nuclei in hyphae. Both nuclei were present across the hyphal length in the wild-type and resulted in the production of recombinant spores. On the other hand, one of the nuclei was lost during hyphal branching in the VYD135α×KN99a cross and resulted in uniparental nuclear inheritance in the spores that were produced. The arrow in (B) marks the hyphal branching point after which only one of the parental nuclei is present (also see Figure 4—figure supplement 1A). The images were captured as independent sections and assembled to obtain the final presented image. Scale bar, 10 µm.
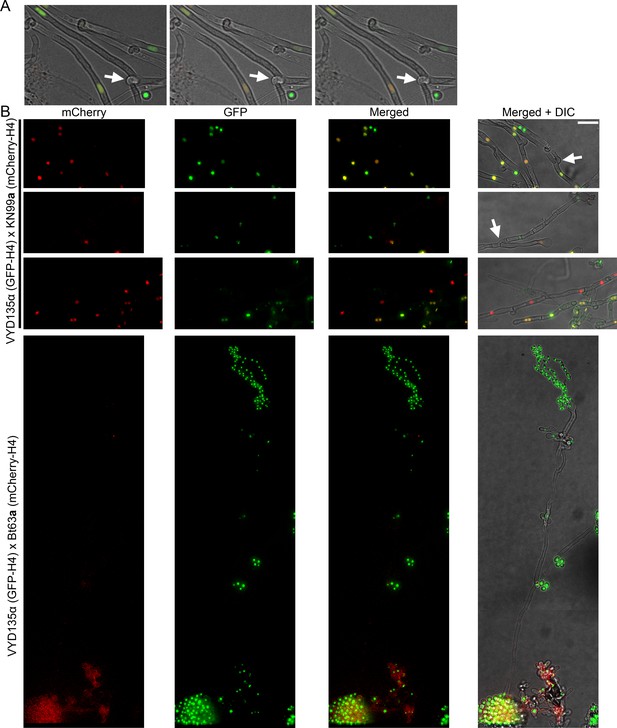
Hyphal branches act as a gateway for nuclear separation during pseudosexual reproduction.
(A) Individual z-sections showing the hyphal branching (marked by arrow) where the two parental nuclei segregate in the Figure 4B. (B) Images showing hyphal branching points where one of the parental nuclei separates from the main hyphae with two parental nuclei (top two panels). The branch point is marked with the arrow. The lower two panels show the long hyphae with only one of the parental nuclei in them. The third panel shows other hyphae with both parental nuclei suggesting that separation occurred at an early stage. The fourth panel exhibits the same between VYD135α×Bt63a but also has a sporulating basidium on it. Scale bar, 10 µm.
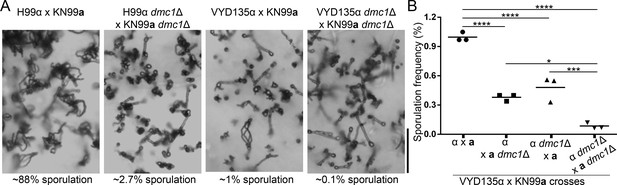
Meiotic recombinase Dmc1 is required for pseudosexual reproduction.
(A) Light microscopy images showing the impact of dmc1 mutation on sexual and pseudosexual reproduction in C. neoformans. Scale bar, 100 µm. (B) A graph showing quantification (n=3) of sporulation events in multiple crosses with dmc1Δ mutants. At least 3000 basidia were counted in each experiment.
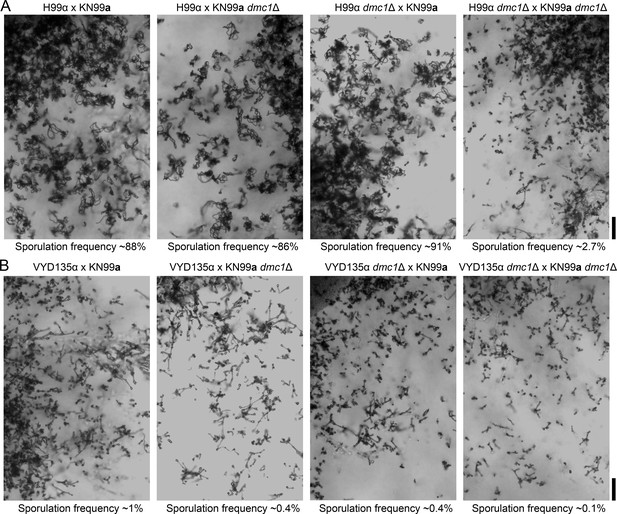
Dmc1 deletion leads to severe sporulation defects in both sexual and pseudosexual reproduction.
Light microscopy images showing the phenotype of DMC1 deletion in (A) H99α×KN99a unilateral crosses as well as bilateral mutant crosses and (B) VYD135α×KN99a dmc1Δ unilateral and bilateral crosses. The deletion of DMC1 led to a reduction in sporulating basidia in bilateral mutant crosses.
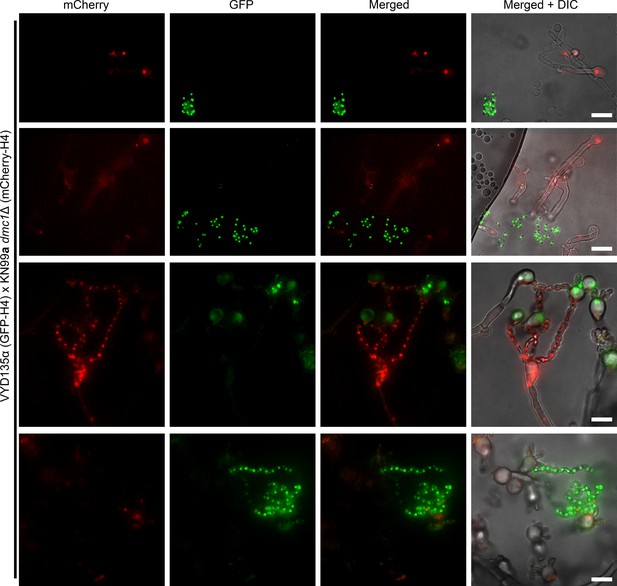
Meiotic regulator Dmc1 is required for pseudosexual reproduction.
A cross between a GFP-H4 tagged VYD135α strain and an mCherry-H4 tagged KN99a dmc1Δ mutant revealed that Dmc1 is required for pseudosexual reproduction events. The majority of the KN99a dmc1Δ nucleus-containing basidia failed to produce spore chains (top two rows and bottom rows). While all 11 observed basidia with VYD135α nuclei produced spores, only 2 out of 19 observed basidia with KN99a dmc1Δ nuclei produced spores. One of these two is represented in the third row. Scale bar, 10 µm.
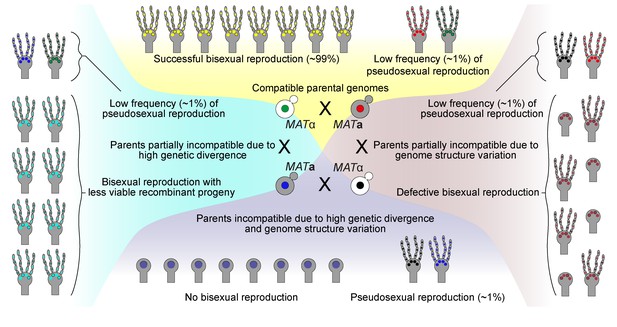
Model for the role of pseudosexual reproduction in C. neoformans ecology.
Scenarios showing possible roles for pseudosexual reproduction under various hypothetical mating conditions. Except for one condition where the two parents are completely compatible with each other, pseudosexual reproduction could play a significant role in survival and dissemination despite its occurrence at a low frequency.
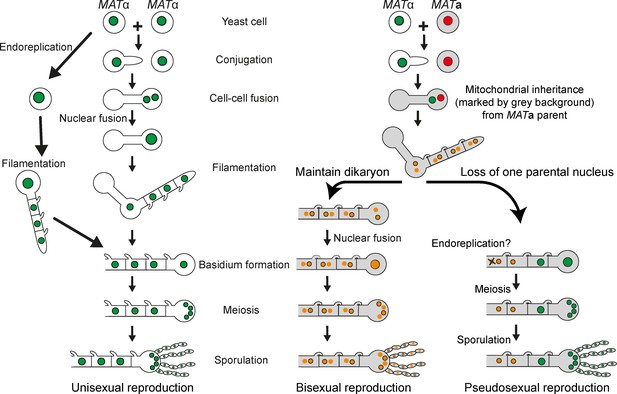
Unisexual, bisexual, and pseudosexual reproduction in C. neoformans.
A diagram depicting various types of sexual reproduction in Cryptococcus species. C. deneoformans exhibits unisexual reproduction in which two cells of the same mating-type fuse or a single cell undergoes endoreplication followed by the production of clonal progeny. Both C. neoformans and C. deneoformans show bisexual reproduction in which two cells of opposite mating types fuse with each other and produce recombinant progeny. Pseudosexual reproduction, as proposed in this study, arises from bisexual mating but generates clonal progeny of one of the parents after the other parental nucleus is lost during development. While both unisexual and pseudosexual reproduction produce clonal progeny, they differ with respect to the inheritance of mitochondrial DNA (marked by gray color cell background in the illustration).
Tables
Genotype analysis of basidia-specific spores germinated from H99α×KN99a and VYD135α×KN99a crosses.
| Basidia # | H99α×KN99a cross | VYD135α×KN99a cross | ||||||
|---|---|---|---|---|---|---|---|---|
| Spores germinated/ dissected | % Germinated | MAT | Mito | Spores germinated/ dissected | % Germinated | MAT | Mito | |
| 1 | 5/14 | 36 | 4α+1a | a | 12/24 | 50 | All α | a |
| 2 | 14/14 | 100 | 7α+7a | a | 6/10 | 60 | All α | a |
| 3 | 12/14 | 86 | 2α+7a+3a/α | a | 15/15 | 100 | All a | a |
| 4 | 10/14 | 71 | 4α+6a | a | 22/27 | 81 | All a | a |
| 5 | 7/13 | 54 | 6a+1a/α | a | 3/12 | 25 | All α | a |
| 6 | 13/14 | 93 | 6α+7a | a | 25/27 | 93 | All α | a |
| 7 | 11/14 | 79 | 6α+5a | a | 4/4 | 100 | All α | a |
| 8 | 14/14 | 100 | 12α+2a | a | 10/13 | 77 | All α | a |
| 9 | 10/14 | 71 | 4α+6a | a | 13/15 | 87 | All α | a |
| 10 | 14/14 | 100 | 7α+7a | a | 31/61 | 51 | All α | a |
| 11 | 14/14 | 100 | 10α+4a | a | 10/10 | 100 | All a | a |
| 12 | 12/14 | 86 | 8α+4a | a | 4/5 | 80 | All a | a |
| 13 | 4/11 | 36 | All a | a | 24/28 | 86 | All a | a |
| 14 | 13/13 | 100 | 8α+5a | a | 16/28 | 57 | All a | a |
| 15 | 14/14 | 100 | 7α+7a | a | 11/11 | 100 | All a | a |
| 16 | 14/14 | 100 | 6α+8a | a | 10/22 | 45 | All α | a |
-
Mito refers to Mitochondria.
Additional files
-
Supplementary file 1
Genotyping of progeny obtained, strains and primers used for this study.
(a). The genotype of basidia-specific spores dissected from H99α×Bt63a and VYD135α×Bt63a crosses. (b). The genotype of basidia-specific spores dissected from H99α×IUM96-2828a and VYD135α×IUM96-2828a crosses. (c). Genotype analysis of basidia-specific progeny from H99α dmc1Δ×KN99a dmc1Δ and VYD135α dmc1Δ×KN99a dmc1Δ crosses. (d). Strains used in this study. (e). Primers used in this study.
- https://cdn.elifesciences.org/articles/66234/elife-66234-supp1-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/66234/elife-66234-transrepform-v2.docx





