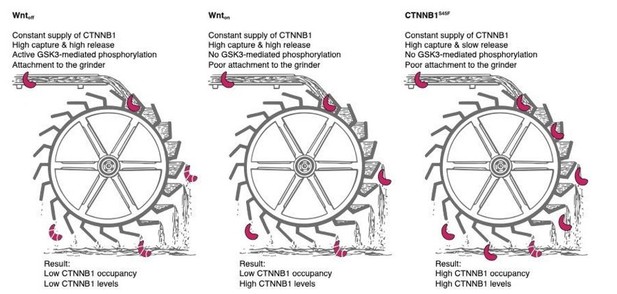
Quantitative live-cell imaging and computational modeling shed new light on endogenous WNT/CTNNB1 signaling dynamics
Figures
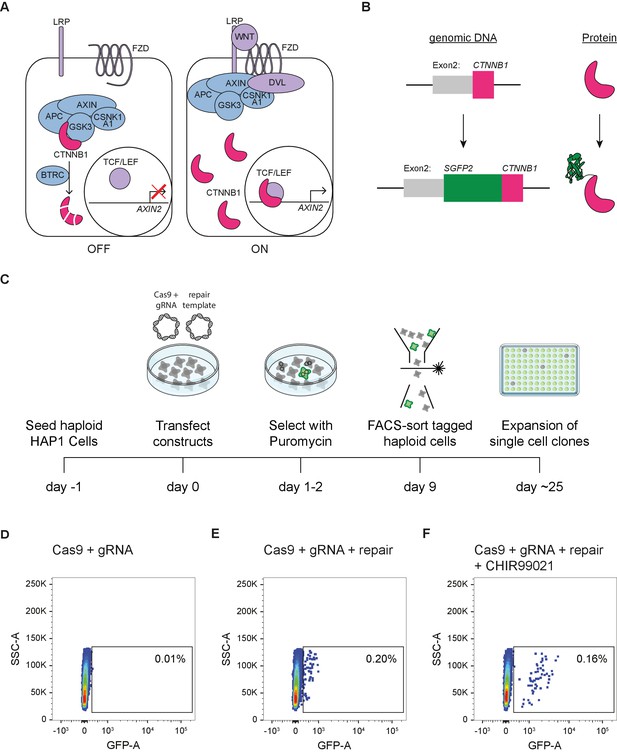
Generation of HAP1SGFP2-CTNNB1 cell lines.
(A) Cartoon depicting the current model of the WNT/CTNNB1 pathway. In the absence of WNT ligands (left, ‘OFF’), free cytoplasmic CTNNB1 is captured by the destruction complex consisting of AXIN, APC, CSNK1A1, and GSK3, which leads to its phosphorylation, BTRC-mediated ubiquitination and subsequent proteasomal degradation, resulting in low levels of CTNNB1 in the cytoplasm and nucleus. Binding of the WNT protein (right, ‘ON’) to the FZD and LRP receptors inhibits the destruction complex through DVL. CTNNB1 accumulates in the cytoplasm and subsequently translocates to the nucleus, where it promotes the transcription of target genes, such as AXIN2, as a co-activator of TCF/LEF transcription factors. (B) Cartoon depicting exon 2 of the CTNNB1 locus, which contains the start codon, and the CTNNB1 protein before (top) and after (bottom) introduction of the SGFP2 by CRISPR/Cas9-mediated homology directed repair. (C) Schematic of the experimental workflow and timeline for generating HAP1SGFP2-CTNNB1 clones. Cas9, gRNA and repair templates are transfected as plasmids. The repair template contains the coding sequence of SGFP2 surrounded by 800 bp homology arms on either side and lacks the gRNA recognition site (see supplement 2 of this figure). A short puromycin selection step is included from 24 to 48 hr after transfection to enrich for transfected cells. Haploid, GFP-positive cells are sorted, and single cell clones are expanded for further analysis. (D–F) FACS plots illustrating control (D) and SGFP2-CTNNB1-tagged cells (E–F). (D) Cells transfected with Cas9 and gRNA in the absence of a repair template were used to set the gate for SGFP2-positive events. (E) A small population of cells expressing low levels of SGFP2 can be detected when cells are transfected with Cas9, gRNA, and repair template. (F) Treatment for 24 hr of cells similar to those depicted in (D) with 8 μM CHIR99021 does not change the amount of cells that are SGFP2 positive, but increases the SFP2 signal, most likely reflecting an increase in SGFP2-tagged beta catenin levels on a per cell basis and supporting the notion that the gated events indeed represent successfully tagged cells.
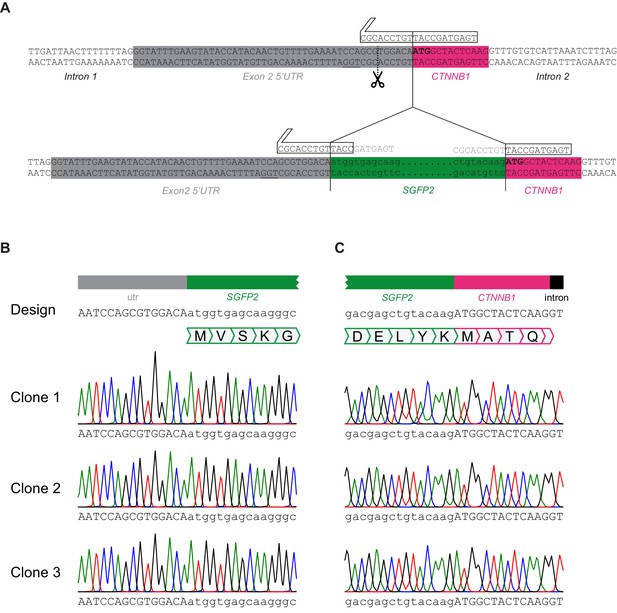
SGFP2-CTNNB1 locus.
(A) Detailed view of CTNNB1 exon two depicting gRNA design relative to the wildtype (top) and repaired (bottom) CTNNB1 allele. Note that the repair template contains the same sequence as the repaired allele depicted here. CTNNB1 sequences are shown in capital letters, SGFP2 sequences shown in lowercase. 5’ UTR, SGFP2 and CTNNB1 and intron regions are indicated below the colored boxes. The gRNA (white arrow box above sequence) overlaps the start codon (depicted in bold), resulting in a Cas9-mediated double-strand break in the 5’UTR (predicted cut site indicated by dotted line and scissor, PAM site underlined). After successful homologous recombination, most of the gRNA-binding site is destroyed, thus minimizing the chance of cutting the repair template or re-cutting the repaired allele. PCR-based screening confirmed that 22/23 single-cell sorted clones indeed showed an integration of the expected size at the CTNNB1 locus. Complete sequence coverage of the insertion site in exon 2 was obtained for 9/11 sequenced clones, of which eight showed the desired repair, and one clone showed an additional point mutation in the repaired locus. (B–C) Sequencing of three independent HAP1SGFP2-CTNNB1 clones on the 5’ (B) and the 3’ (C) end of SGFP2 integration in exon 2 of CTNNB1. Sanger sequencing of the endogenous CTNNB1 locus of clone 1, clone 2, and clone 3 shows an exact match to the design and thus correct homology directed repair.
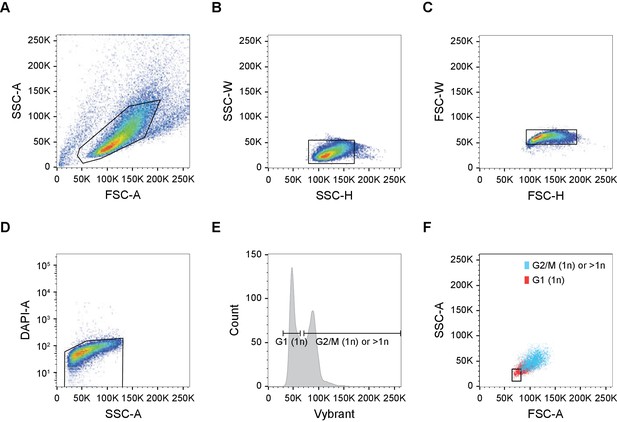
FACS Gating strategy for haploid HAP1 cells.
(A–C) Single-cell gating based on forward scatter (FSC) and side scatter (SSC). (D) Live cell gating based on DAPI exclusion. (E–F) Haploid cell sorting based on Vibrant live-cell DNA dye. (E) Haploid cell cycle profile. Only cells in G1 can be confidently identified as haploid (1 n). The second gate contains both G2/M haploid cells, as well as diploid (2 n) and polyploid events. Of note, the depicted HAP1WT population is mainly haploid. (F) Back-gating of the haploid G1 population from E onto the forward and side scatter plot. A stringent gate is set based on cell size to ensure only G1 (1 n) cells qualify for sorting.
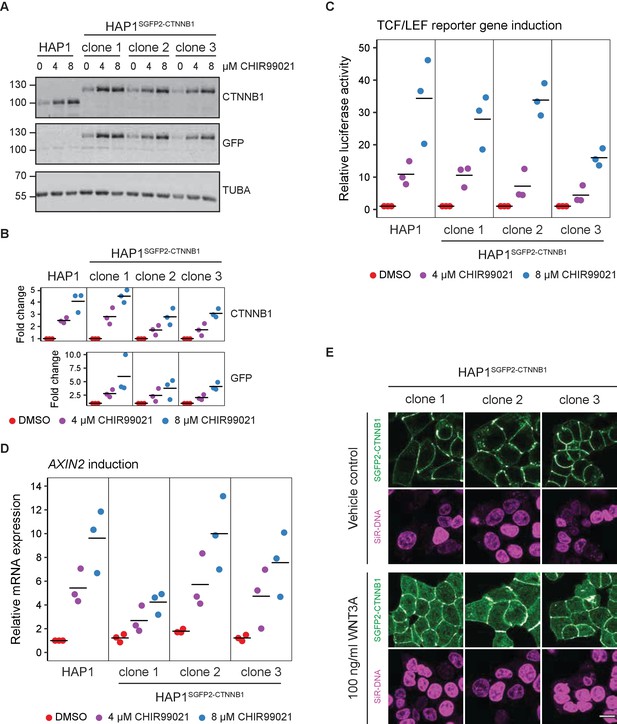
Functional validation of three independent HAP1SGFP2-CTNNB1 clones.
(A) Western blot, showing CTNNB1 (HAP1WT) and SGFP2-CTNNB1 (HAP1SGFP2-CTNNB1 clones 1, 2, and 3) accumulation in response to CHIR99021 treatment. All panels are from one blot that was cut at the 70 kDa mark and was stained with secondary antibodies with different fluorophores for detection. Top: HAP1WT cells express CTNNB1 at the expected wild-type size. Each of the three clonal HAP1SGFP2-CTNNB1 cell lines only express the larger, SGFP2-tagged form of CTNNB1, that runs at the expected height (~27 kDa above the wild-type CTNNB1). Middle: Only the tagged clones express the SGFP2-CTNNB1 fusion protein, as detected with an anti-GFP antibody at the same height. Bottom: alpha-Tubulin (TUBA) loading control. A representative image of n=3 independent experiments is shown. (B) Quantification of Western blots from n=three independent experiments, including the one in (A), confirming that the accumulation of CTNNB1 in response to WNT/CTNNB1 pathway activation is comparable between HAP1WT and HAP1SGFP2-CTNNB1 cells. Horizontal bar indicates the mean. (C) Graph depicting the results from a MegaTopflash dual luciferase reporter assay, showing comparable levels of TCF/LEF reporter gene activation for HAP1WT and HAP1SGFP2-CTNNB1 cells in response to CHIR99021 treatment. Data points from n=3 independent experiments are shown. Horizontal bar indicates the mean. Values are depicted relative to the DMSO control, which was set to one for each individual cell line. (D) Graph depicting AXIN2 mRNA induction in response to CHIR99021 treatment, demonstrating that induced expression of an endogenous target gene is comparable between HAP1WT and HAP1SGFP2-CTNNB1 cells. Data points represent n=3 independent experiments. Horizontal bar represents the mean. HPRT was used as a reference gene. Values are depicted relative to the HAP1WT DMSO control, which was set to 1. (E) Representative confocal microscopy images of the three HAP1 SGFP2-CTNNB1 clones after 4 hr vehicle control or 100 ng/ml WNT3A treatment from n=1 biological experiment, revealing intracellular accumulation of SGFP2-CTNNB1 (green). Nuclei were counterstained with SiR-DNA dye (magenta). Scale bar is 10 μm.
-
Figure 2—source data 1
Numerical data for Figure 2B,C,D and Figure 2—figure supplement 1A,C,D.
- https://cdn.elifesciences.org/articles/66440/elife-66440-fig2-data1-v2.xlsx
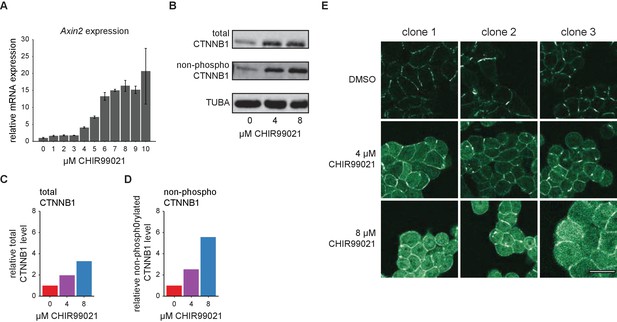
Verification of the WNT/CTNNB1 responsiveness of HAP1 cells.
(A) Graph depicting AXIN2 qRT-PCR results from HAP1WT cells treated with the indicated range of CHIR99021 (1–10 µM) or DMSO vehicle control (0 µM) for 24 hr. HPRT was used as a reference gene. Error bars represent standard deviation within technical triplicates from n=1 biological experiment. Based on this, we selected 4 μM and 8 µM as intermediate and high levels of WNT/CTNNB1 pathway induction for follow up experiments. (B) Western blot, showing the increase in total (top) and non-phosphorylated (i.e. active) CTNNB1 levels (middle) in response to pathway stimulation. HAP1WT cells were treated for 24 hr with 4 or 8 µM CHIR99021, or DMSO vehicle control (0 µM). Alpha-Tubulin (TUBA, bottom) serves as a loading control. (C–D) Quantification of the western blot from (B) depicting the relative fold change of total CTNNB1 (C) or non-phosphorylated CTNNB1 (D) to DMSO control corrected for Tubulin loading. (E) Representative confocal microscopy images of three independent HAP1SGFP2-CTNNB1 clones, treated for 24 hr with 4 or 8 µM CHIR99021, or DMSO vehicle control. Scalebar is 10 µm.
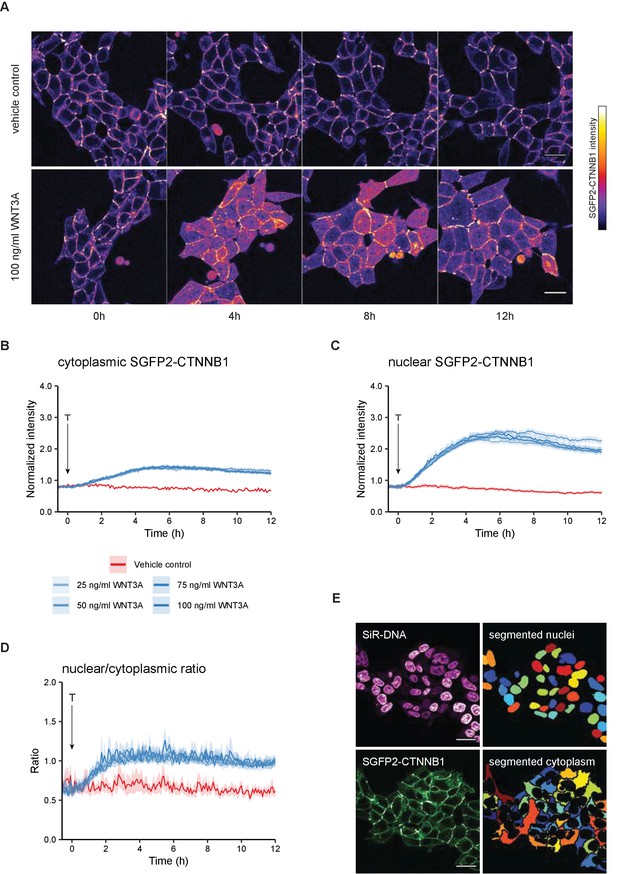
Live imaging of HAP1SGFP2-CTNNB1.
(A) Representative stills from confocal time-lapse experiments corresponding to Videos 1–2, showing an increase of SGFP2-CTNNB1 after treatment with 100 ng/ml WNT3A (bottom) relative to a vehicle control (BSA)-treated sample (top). Scale bar = 20 μm. (B–D) Quantification of time-lapse microscopy series, using the segmentation pipeline shown in (E). Arrow indicates the moment of starting the different treatments (T, see legend in B for details). (B–C) Graph depicting the normalized intensity of SGFP2-CTNNB1 in the cytoplasm (B) or nucleus (C) over time. Solid lines represent the mean normalized fluorescence intensity and shading indicates the 95% confidence interval. n=155–393 cells for each condition and time point, pooled data from n=three independent biological experiments. (D) Graph depicting the nuclear/cytoplasmic ratio of SGFP2-CTNNB1 over time, calculated from raw intensity values underlying (B) and (C). (E) Segmentation of nuclei (top) and cytoplasm (bottom) based on the SiR-DNA signal and SGFP2-CTNNB1 signal. Scale bar = 20 μm.
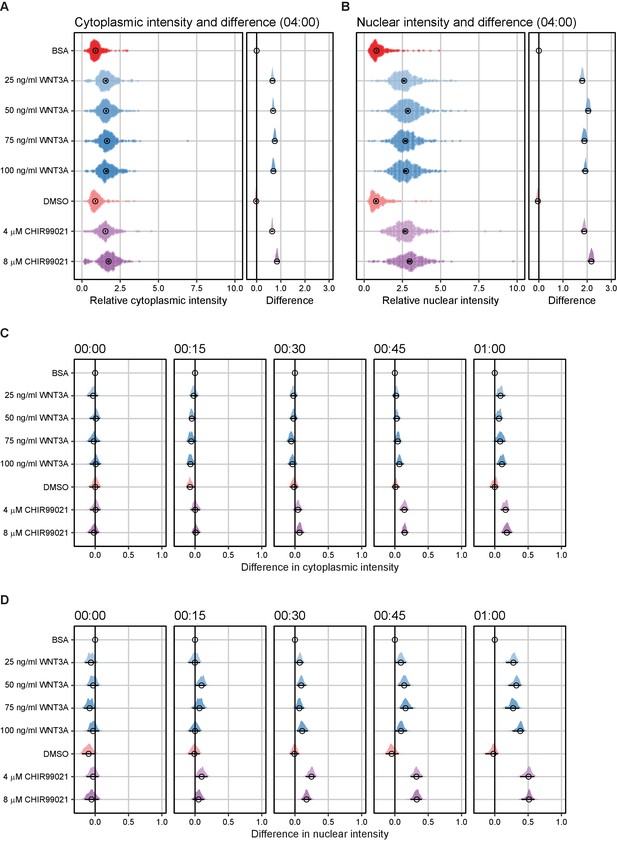
Difference analysis of SGFP2-CTNNB1 fluorescence.
(A–B) Plots depicting the relative intensity (left) and the difference in relative intensity to BSA treated cells (right) in the cytoplasm (A) and nucleus (B) after 4 hr of treatment. Circles indicate the median value and bars indicate the 95% CI. In the relative intensity plot (left), the distribution is built from individual data points in a violin-type fashion to faithfully represent the distribution of data. In the difference plot (right) the distribution of differences is represented in a half violin plot. If the 95% CI in the difference plot does not overlap the zero line, which indicates no difference, the sample is significantly different from BSA control condition. (C–D) Plots depicting the difference in relative intensity in the cytoplasm (C) and nucleus (D) between the moment of addition and 1 hr of treatment. Titles indicate the time (hh:mm). The distribution of differences is represented in a half violin plot. Circles indicate the median value and bars indicate the 95% CI. If the 95% CI does not overlap the zero line, which indicates no difference, the sample is significantly different from the BSA control condition.
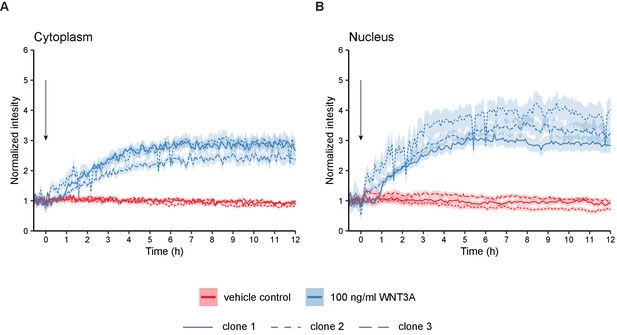
Graphs showing quantification of time-lapse microscopy experiments with three independent HAP1SGFP2-CTNNB1 clones.
Stills of this experiment are shown in Figure 3C. Segmentation was performed as described in Figure 4. Arrow indicates the moment of starting the different treatments (BSA in red or 100 ng/ml WNT3A in blue). Solid lines represent the mean normalized intensity and shading the 95% confidence interval in the cytoplasm (A) or nucleus (B). Line pattern indicates the three different clones. n=13–158 cells for each condition and time point for n=one biological experiment.
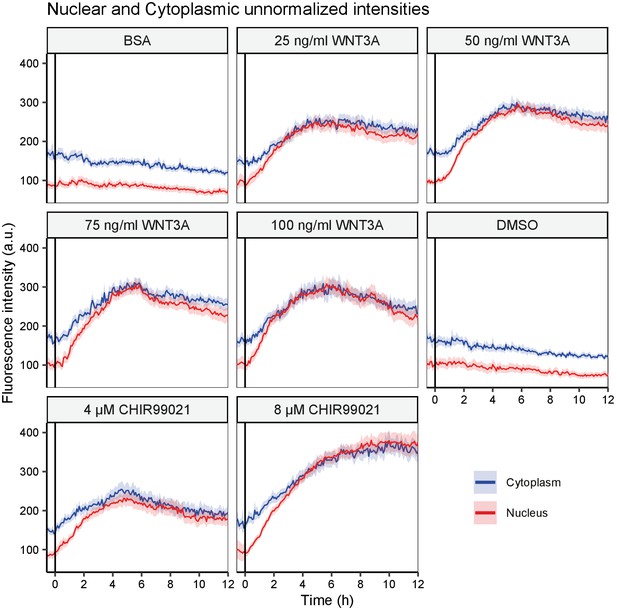
Unnormalized nuclear and cytoplasmic intensity measurements.
Graphs showing the unnormalized fluorescence intensities quantified for a single biological replicate shown in Figure 3. The vertical black lines indicate the moment of starting the different treatments (as indicated on top of each graph). Solid lines represent the mean unnormalized intensity and shading the 95% confidence interval in the cytoplasm (blue) or nucleus (red). n=64–148 cells for each condition and time point.
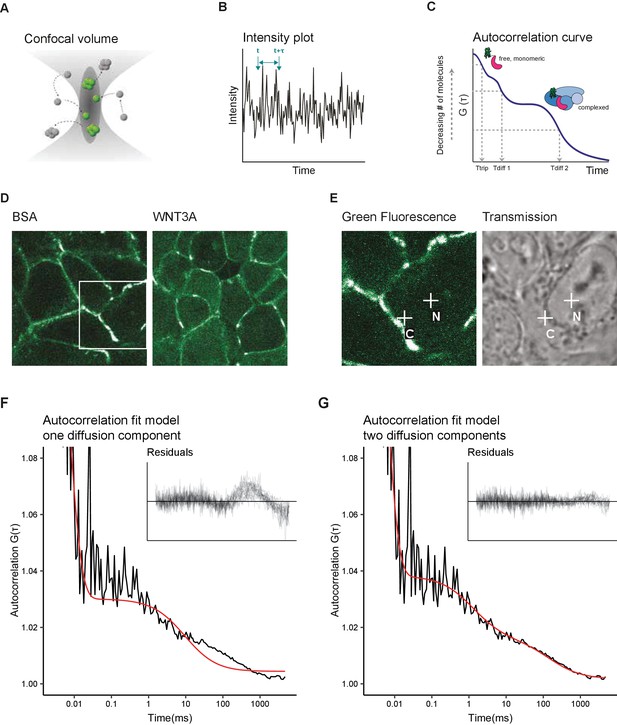
Two diffusion-component fit-model for SGFP2-CTNNB1 FCS measurements.
(A) Schematic representation of the point FCS technique, depicting the confocal volume with fluorescent particles diffusing in and out. Particles in FCS are defined by their coherent movement; therefore, a particle can be made up of monomers or multimers in isolation or complexed to unlabeled molecules. (B) Schematic representation of intensity fluctuations over time as measured in the confocal volume. Fluctuations are the result of both photo-physics (e.g. blinking of the fluorophore), diffusion and the number of particles in the confocal volume. (C) Graphical representation of the two diffusion-component fitting model used for our autocorrelation curves. Ttrip describes the blinking of the SGFP2 fluorophore and the after-pulsing artefact. Tdiff1 and Tdiff2 describe the monomeric and complexed form of SGFP2-CTNNB1, respectively. Details of all fitting parameters are described in Materials and methods. (D) Representative confocal images of HAP1SGFP2-CTNNB1 cells treated for 4 hr with BSA (left) or 100 ng/ml WNT3A (right). (E) Zoom in of the white rectangle in (D), with representative locations of FCS measurement points for cytoplasm (C) and nucleus (N) indicated with white crosses in the SGFP2-CTNNB1 channel and transmission channel. (F–G) Fitting of a representative autocorrelation curve with one unfixed diffusion-component (F) or a two diffusion-component model (G), where the first diffusion component was fixed to the speed of free monomeric SGFP2-CTNNB1 (14.9 µm2/s) and the second diffusion component was unfixed. The black line represents the autocorrelation curve generated from the FCS measurement; the red line represents the fitted model. The residuals after fitting of 25 individual curves are shown in the upper right corner of the graphs.
-
Figure 4—source data 1
Numerical data for Figure 4F,G and Figure 4—figure supplement 1.
- https://cdn.elifesciences.org/articles/66440/elife-66440-fig4-data1-v2.xlsx
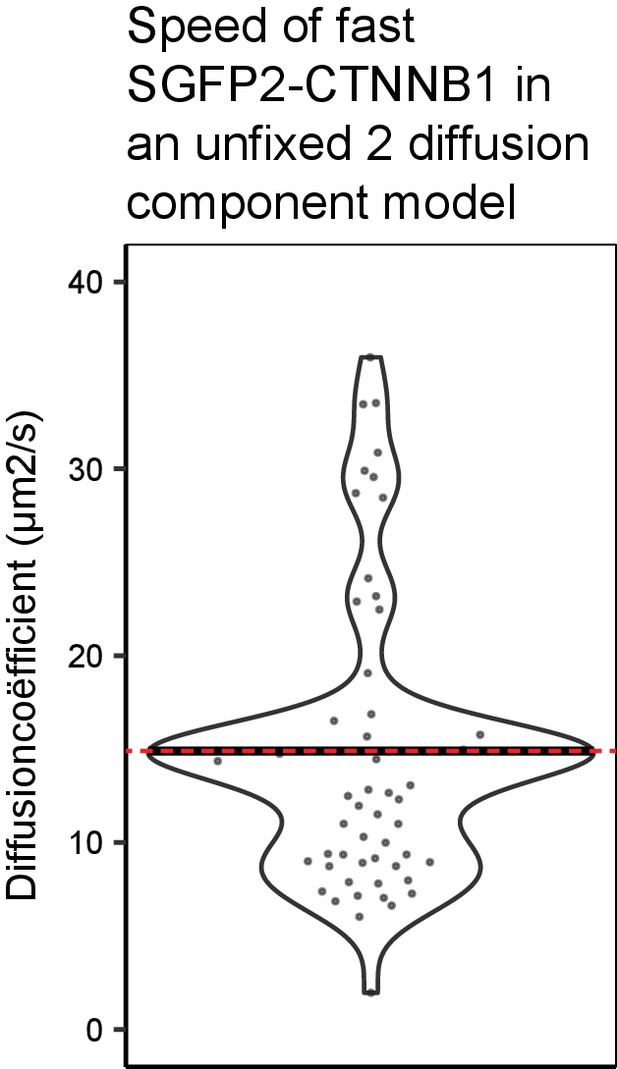
Quantification of SGFP2-CTNNB1 fast component using an unfixed two diffusion component model.
Graph depicting the first (i.e. fast) diffusion coefficient in SGFP2-CTNNB1 cells (treated with BSA or WNT3A, data as in Figures 4 and 5) in a two diffusion component model where both the first and second diffusion time were unfixed (see Materials and methods for details). The red dotted line shows the theoretically established diffusion coefficient (14.9 μm2/s) overlapping with the median (black line, 14.8 μm2/s) of the unfixed first diffusion coefficient found in this unfixed model.
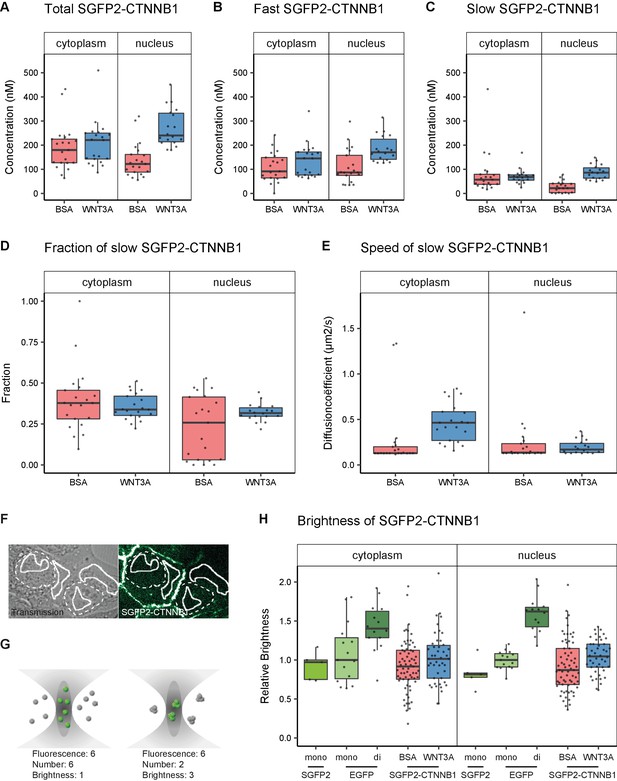
Abundance and mobility of SGFP2-CTNNB1 molecules in living cells after 4 hr WNT3A treatment or control.
Details on sample size and statistics can be found in Supplementary file 1. (A) Graph depicting the total concentration of SGFP2-CTNNB1 particles (monomeric plus complexed) as measured with FCS. (B) Graph depicting the concentration of SGFP2-CTNNB1 particles with the fast diffusion component (i.e. free monomeric). (C) Graph depicting the concentration of SGFP2-CTNNB1 containing particles with the slow diffusion component (i.e. complex associated). (D–E) Graphs depicting the fraction (D) and speed (E) of the second diffusion component (i.e. SGFP2-CTNNB1 containing complex) measured by FCS. (F) Example of typical regions of interest in two cells used in N and B analysis. Solid line represents the analysis ROI, dashed line, marks the outline of the nuclear envelope. (G) Schematic representation of a confocal volume with different brightness species. On the left are six monomers with a brightness of 1, on the right two trimers with a brightness of 3, both result in a fluorescence of 6. N and B analysis is able to extract the number and the brightness of such samples, for more detail see supplement 1 of this figure. (H) Graph depicting the molecular brightness of SGFP2-CTNNB1 in the cytoplasm and nucleus relative to controls as measured with N and B in the same subcellular compartments. EGFP monomer was used for normalization and EGFP dimer as a control for N and B measurements.
-
Figure 5—source data 1
Numerical data for Figure 5 (all graphs) and Figure 5—figure supplement 1 (all panels).
- https://cdn.elifesciences.org/articles/66440/elife-66440-fig5-data1-v2.xlsx
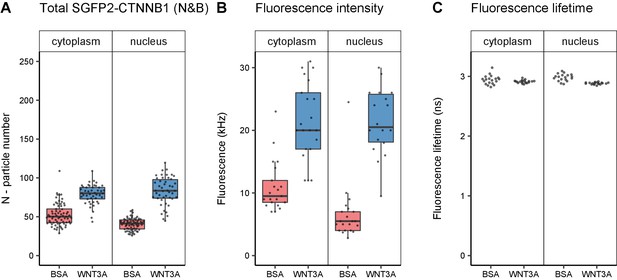
Quantification of SGFP2-CTNNB1 particles, fluorescence and fluorescence lifetime.
Details on sample size and statistics can be found in Supplementary file 1. (A) Graph depicting the total number of SGFP2-CTNNB1 particles as measured with N and B. (B) Graph depicting the average fluorescence intensity at the start of the FCS measurement. The increase in SGFP2-CTNNB1 fluorescence in the cytoplasm (2.1-fold) exceeds the increase in the SGFP2-CTNNB1 concentration (1.1-fold, Figure 5A), but does correspond to the relative increase measured by time-lapse imaging (1.7-fold, Figure 4C). (C) Graph depicting fluorescence lifetimes calculated from the FCS measurements. The Fluorescence lifetime of SGFP2-CTNNB1 is independent of the subcellular compartment and treatment. Therefore, photophysical effects are not the cause for the difference between the fold-change in fluorescence and concentrations of the FCS measurements as described in (B).
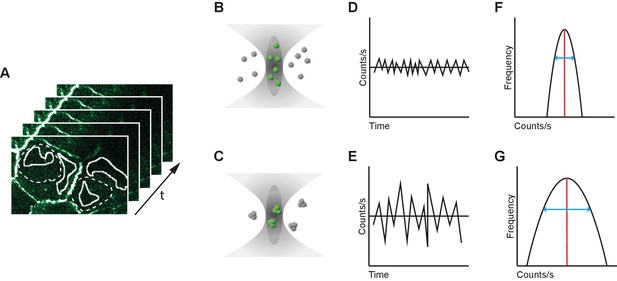
Number and Brightness analysis.
(A) A series of images are acquired over time (t) (B-C) Representation of the confocal volume in a single pixel of the image in (A) representing monomers with a brightness of 1 (B) and trimers with a brightness of 3 (C) both confocal volumes emit a fluorescence of 6. (D–E) Graph depicting the variation in fluorescence, measured by an APD in counts/s, in a single pixel over time for the monomer (D) and trimer (E). With a higher brightness the variation around the average becomes larger. Note that the timescale in N and B is much larger than in FCS (Figure 4A–C) due to the use of images rather than a single point and therefore the variation is no longer dependent on diffusion kinetics. (F–G) Graph depicting the distribution of fluorescence observed in a single pixel. The average (red line) in our case is the same for six monomers (F) or two trimers (G), but the standard deviation (blue line) is much larger for the trimer. The average and standard deviation are used to calculate the number of molecules and their relative brightness.
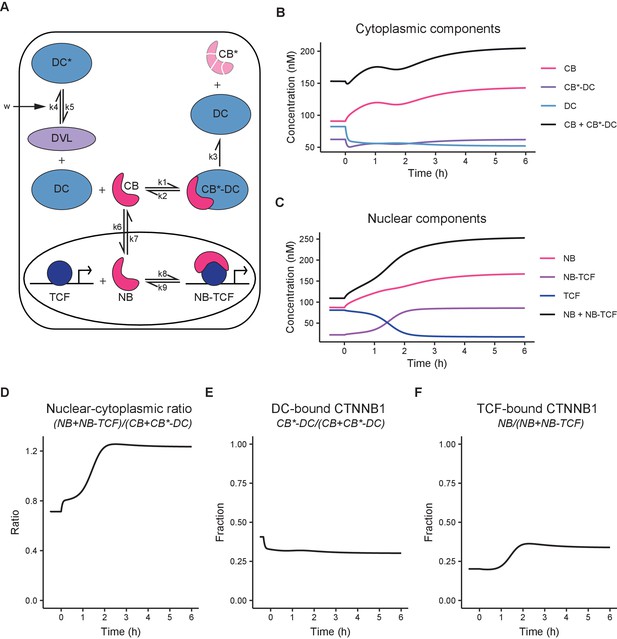
Computational model of WNT/CTNNB1 based on FCS concentrations for free and complexed CTNNB1 (Tables 1–2).
(A) Schematic overview of the model. DC=destruction complex, DC* = DVL-inactivated DC, CB=cytoplasmic CTNNB1, CB*=phosphorylated CB, NB=nuclear CTNNB1, TCF=TCF/LEF transcription factors, DVL=WNT-activated DVL. In WNT OFF, w=0, and therefore k5/k4 does not play a role and no inactivated destruction complex is formed. In WNT ON, w=1, and k5/k4 is put into action, resulting in an increase in DC* at the expense of DC. Under the assumption that k3 remains equal and given that CB*-DC was experimentally determined to be the same in WNT ON and WNT OFF, removal of DC, results in an increase in CB. Changes in k6/k7 and k9/8 further increase NB and NB-TCF in WNT ON. Note that CB* is degraded and therefore plays no role in the model. (B) Graph depicting the modeled concentrations of cytoplasmic components over time. The black line indicates total concentration of cytoplasmic CTNNB1, corresponding to Figure 3B. (C) Graph depicting the modeled concentrations of nuclear components over time. The black line indicates total concentration of nuclear CTNNB1, corresponding to Figure 3C. (D) Graph depicting the ratio of total nuclear and cytoplasmic CTNNB1 over time, corresponding to the measurements in Figure 3D. (E) Graph depicting the DC-bound CTNNB1 fraction ratio over time. (F) Graph depicting the TCF-bound CTNNB1 fraction ratio over time.
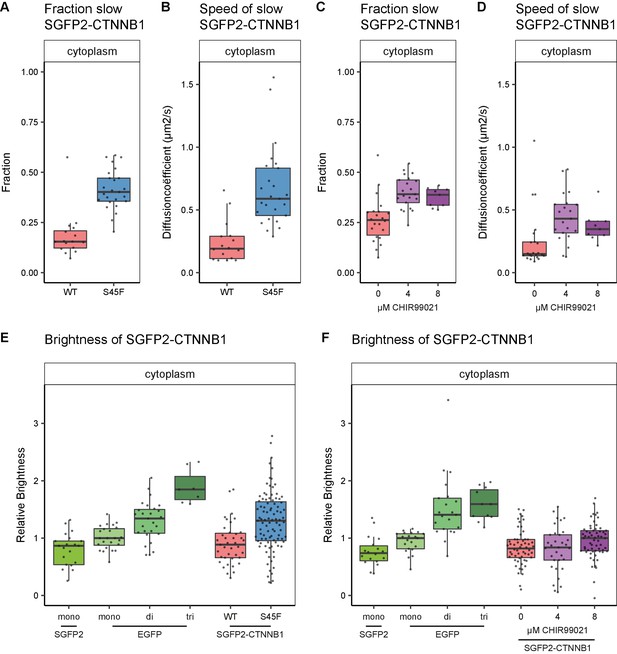
Cytoplasmic complex characteristics in absence of SGFP2-CTNNB1 N-terminal phosphorylation.
The S45F mutant was introduced using CRISPR (see Figure 7—figure supplement 1) and CHIR treated and control cells were measured after 24 hr. Details on sample size and statistics can be found in Supplementary file 1. (A) Graph depicting the fraction of particles with the second diffusion component (i.e. SGFP2-CTNNB1 containing complex) measured by FCS for S45F mutant (B) Graph depicting the speed of the second diffusion component (i.e. SGFP2-CTNNB1 containing complex) measured by FCS for S45F mutant. (C).Graph depicting the fraction of particles with the second diffusion component (i.e. SGFP2-CTNNB1 containing complex) measured by FCS after 24 hr treatment with CHIR99021 (C) Graphs depicting the speed of the second diffusion component (i.e. SGFP2-CTNNB1 containing complex) measured by FCS after 24 hr treatment with CHIR99021. (E–F) Graphs depicting the molecular brightness of SGFP2-CTNNB1 in the cytoplasm relative to controls as measured with N and B in the same subcellular compartments for S45F mutant CTNNB1 (E) or after 24 hr of CHIR99021 treatment (F). EGFP monomer was used for normalization and EGFP dimer and trimer as controls for N and B measurements.
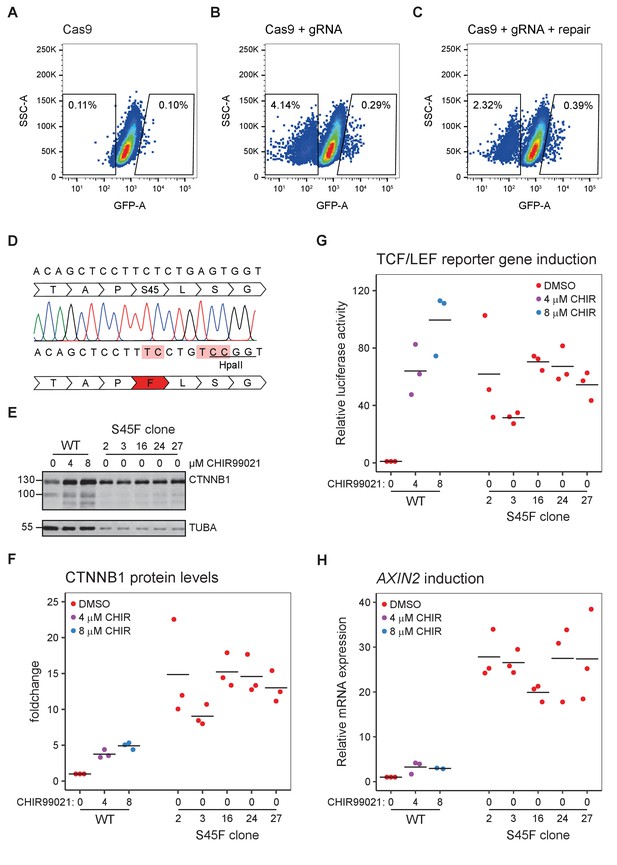
Generation and characterization of a S45F mutant cell line (HAP1SGFP2-CTNNB1(S45F)).
(A–C) FACS graphs showing the selection strategy for HAP1SGFP2-CTNNB1(S45F) cells. HAP1SGFP2-CTNNB1 clone one was transfected with CRISPR constructs similar to Figure 1C, see Materials and methods for details. Transfection with a guide RNA in the third exon of CTNNB1 resulted in a population with reduced SGFP2 fluorescence and a population with increased fluorescence (B) compared to a Cas9 only control (A). Co-transfection of a single-stranded oligo repair, led to a reduction in the population with decreased SGFP2 fluorescence (representing cells repaired with NHEJ that resulted in a loss of the SGFP2-CTNNB1 fusion protein) and an increase in the population with increased SGFP2 fluorescence (representing cells repaired with NHEJ that resulted in a stabilizing SGPF2-CTNNB1 truncation, mutation, insertion or deletion as well as cells with the correct HDR resulting in the stabilizing S45F mutation). Cells with increased fluorescence were sorted as single haploid cells (see Figure 1—figure supplement 2) and expanded for further analysis (D) Graphical representation of part of the 3rd exon of SGFP2-CTNNB1 before (top) and after (bottom) CRISPR/Cas9-mediated mutagenesis. Sanger sequencing is shown for one correctly targeted clone. The mutation changes the 45th serine to a phenylalanine (S45F) and additionally removes the gRNA-binding site and a silent mutation that introduces a HpaII restriction site for screening purposes. Mutated bases are indicated in light red. Fourteen of 32 single-cell clones showed the expected genotyping fragment size and 7/32 showed large insertions or deletions. Seven of 14 clones showed the expected genotyping band contained the HpaII restriction site, 3/14 did not. Sanger sequencing coverage for the exon3 locus was obtained for 6/7 clones and all showed correct targeting (as shown for one clone here). Five clones were further analyzed; Clone 2, 3, 16, and 24 are from one single transfection, clone 27 from an independent transfection. (E) Western blot, showing SGFP2-CTNNB1WT (WT) and SGFP2-CTNNB1S45F (S45F clone) protein levels. All panels are from one blot that was cut at the 70 kDa mark. Top: HAP1SGFP2-CTNNB1(WT)and HAP1SGFP2-CTNNB1(S45F) clones express the SGFP2-CTNNB1 protein at the expected height. Bottom: alpha-Tubulin (TUBA) loading control. A representative image of n=3 independent experiments is shown. (F) Quantification of Western blots from n=3 independent experiments, including the one in (E), normalized to the SGFP2-CTNNB1WT DMSO control, showing that the S45F clones express higher levels of SGFP2-CTNNB1 than their HAP1SGFP2-CTNNB1(WT) parental line, even after 24 hr treatment with 8 µM CHIR99021. Horizontal bar indicates the mean. (G) Graph depicting the results from a MegaTopflash dual luciferase reporter assay, showing increased levels of TCF/LEF reporter gene activation for and HAP1SGFP2-CTNNB1(S45F), comparable to HAP1SGFP2-CTNNB1(WT) cells treated for 24 hr with 8 µM CHIR99021 treatment. Data points from n=three independent experiments are shown. Horizontal bar indicates the mean. Values are depicted relative to the HAP1SGFP2-CTNNB1(WT) DMSO control, which was set to 1. (H) Graph depicting AXIN2 mRNA induction activation for and HAP1SGFP2-CTNNB1(S45F) and HAP1SGFP2-CTNNB1(WT), showing increased levels of AXIN2 expression for HAP1SGFP2-CTNNB1(S45F), exceeding those in HAP1SGFP2-CTNNB1(WT) cells treated for 24 hr with 8 µM CHIR99021 treatment. Data points from n=3 independent experiments are shown. Horizontal bar indicates the mean. Values are depicted relative to the HAP1SGFP2-CTNNB1(WT) DMSO control, which was set to 1.
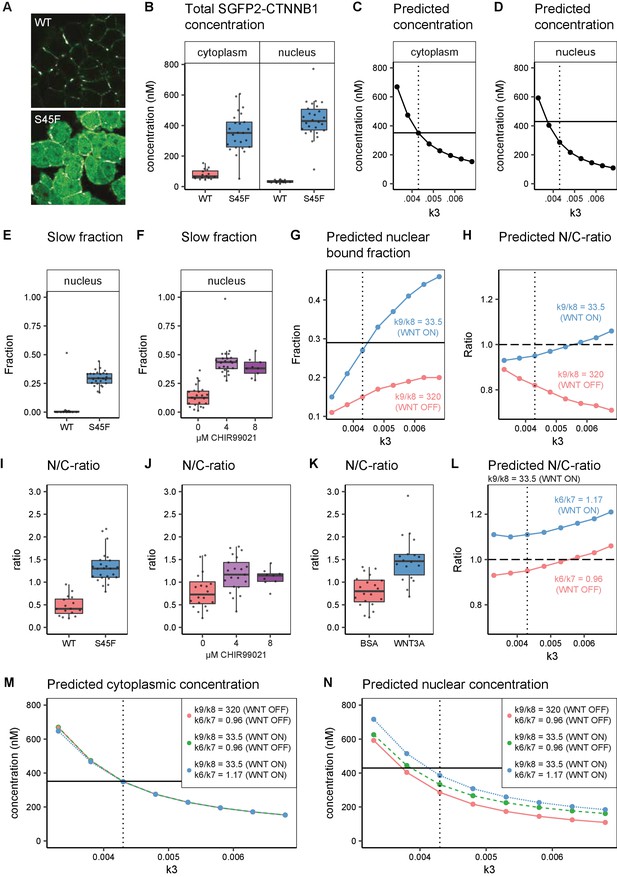
In silico and experimental perturbation of WNT signaling.
Details on experimental sample size and statistics can be found in Supplementary file 1. (A) Representative confocal images of HAP1-SGFP2WT (WT, top) and HAP1-SGFP2S45F (S45F, bottom) cells acquired with the same image settings. The S45F mutation leads to the accumulation and nuclear enrichment of CTNNB1 in the cell. (B) Graph depicting the total concentration of SGFP2-CTNNB1 particles (monomeric plus complexed) as measured with FCS. (C–D) Inhibition of CTNNB1 degradation is modelled as a reduction in the value of k3. (C) Graph depicting the predicted total cytoplasmic CTNNB1 concentration as a function of k3. A reduction in k3 from 0.0068 (Table 5, WNT ON and WNT OFF conditions) to ~0.0043 (dotted line) corresponds to the cytoplasmic concentration observed (solid line). (D) Graph depicting the predicted total nuclear CTNNB1 concentration as a function of k3. The solid horizontal line indicates the concentration measured for the S45F mutant by FCS. Note that the value of k3 that matches the observed cytoplasm concentration (dotted line) does not match the experimentally determined concentration in the nucleus (solid line). (E–F) Graphs depicting the fraction of particles with the second diffusion component (i.e. SGFP2-CTNNB1 containing complex) measured by FCS for wild-type and S45F mutant (E) and after 24 hr CHIR99021 treatment (F). The increase in the bound fraction in the oncogenic mutant or after GSK3 inhibition we find, is comparable to what we observed in WNT3A stimulated cells (Figure 5D). (G) Graph showing the predicted nuclear bound fraction of CTNNB1as a function of k3 with the TCF/CTNNB1 binding affinity of the model (Table 4) for WNT OFF (k9/k8 = 320, pink line) and for WNT ON (k9/k8=33.5, blue line). Note that for WNT ON, the value for the nuclear bound fraction approximates the experimentally determined slow fraction for the S45F mutant (solid line, panel E) at the value for k3 that matches the cytoplasmic concentration of CTNNB1 (dotted line). (H) Graph showing the predicted nuclear/cytoplasmic (N/C)-ratio as a function of k3 with TCF/CTNNB1 binding affinity of the model (Table 4) for WNT OFF (k9/k8=320, pink line) and WNT ON (k9/k8=33.5, blue line). Note that, although for WNT ON the value of the N/C-ratio increases with k3, there is still nuclear exclusion (N/C-ratio lower than 1, dashed line) at the value of k3 that matches the cytoplasmic CTNNB1-concentration (dotted line). (I–K) The N/C-ratio as measured by FCS for wild-type and S45F mutant (I), after 24 hr CHIR99021 treatment (J) and after 4 hr WNT3A treatment (K). Note that all perturbations lead to nuclear accumulation (N/C-value exceeding 1). (L) Graph showing the predicted N/C-ratio as a function of k3 with the WNT ON value for k9/k8 with the nuclear shuttling ratio of the model (k6/k7 Table 4),corresponding to WNT OFF (k6/k7=0.96, pink line) and WNT ON (k6/k7=1.17, blue line), respectively. Note that the WNT ON value of k6/k7 increases the N/C-ratio to nuclear accumulation at the value for k3 that matches the cytoplasmic concentration (dotted line). (M) Graph depicting the predicted total cytoplasmic CTNNB1 concentration as a function of k3 with WNT ON and WNT OFF values for k9/k8 and k6/k7. Note that modulation of k9/k8 and k6/k7 has virtually no effect on the predicted cytoplasmic concentration of CTNNB1, resulting in overlapping points and lines in the graph. The horizontal solid line is the experimentally determined cytoplasmic CTBNN1 concentration (cf. panel B); the vertical dotted line is at the value of k3 that best reproduces this experimental finding in the model. (N) Graph depicting the predicted total nuclear CTNNB1 concentration as a function of k3 for WNT ON and WNT OFF values for k9/k8 and k6/k7. Note that if both k9/k8 and k6/7 are changed from their WNT OFF values the predicted nuclear concentration of CTBNN1 better matches the experimentally determined concentration (horizontal solid line) at the value for k3 that matches the cytoplasm concentration (vertical dotted line).
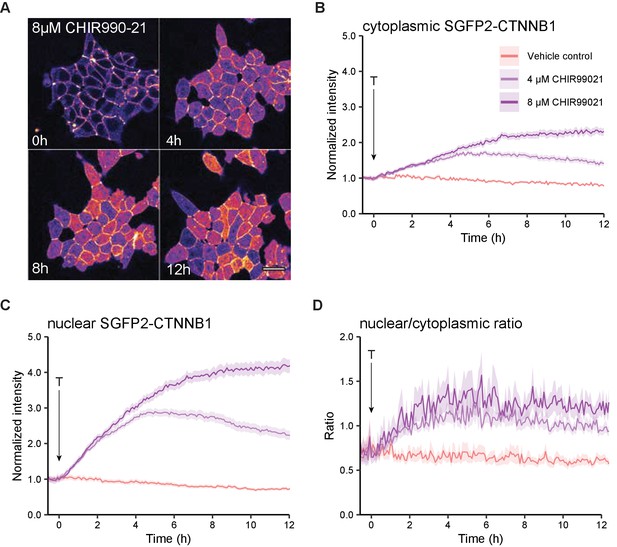
Live imaging of HAP1SGFP2-CTNNB1 upon CHIR99021 stimulation.
(A) Representative stills from confocal time-lapse experiments corresponding to Video 3, showing an increase of SGFP2-CTNNB1 after treatment with 8 µM CHIR99021. Scale bar = 20 μm. (B–D) Quantification of time-lapse microscopy series, using the segmentation pipeline shown in Figure 3E. Arrow indicates the moment of starting the different treatments (T, see legend in B for details). (B–C) Graph depicting the normalized intensity of SGFP2-CTNNB1 in the cytoplasm (B) or nucleus (C) over time. Solid lines represent the mean normalized fluorescence intensity and shading indicates the 95% confidence interval. n=166–400 cells for each condition and time point, pooled data from n=three independent biological experiments. (D) Graph depicting the nuclear/cytoplasmic ratio of SGFP2-CTNNB1 over time, calculated from raw intensity values underlying (B) and (C). This experiment was recorded simultaneously with the data shown in Figure 3. Additional data representation of this experiment is found in Figure 3—figure supplement 1 and Videos 4–6.
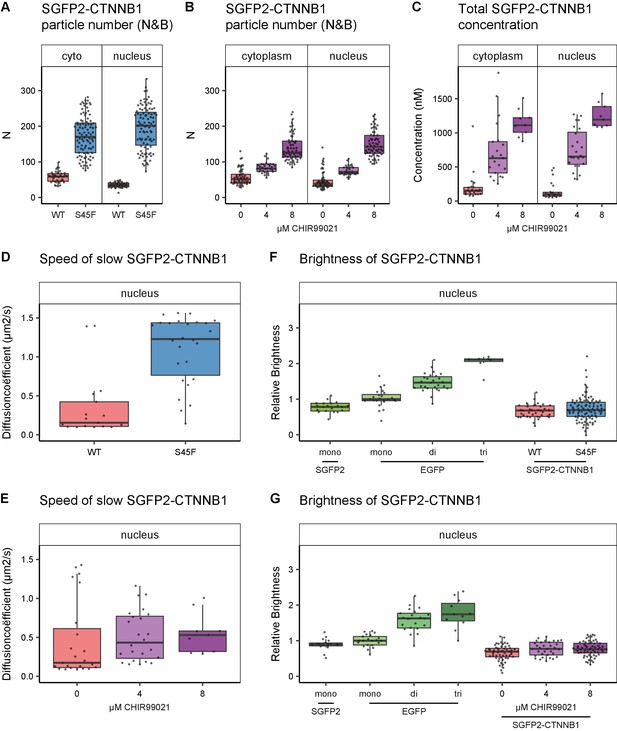
additional biophysical properties of SGFP2-CTNNB1S45F and SGFP2-CTNNB1 under CHIR99021 stimulation.
The S45F mutant was introduced using CRISPR (see Figure 7—figure supplement 1) and CHIR-treated and control cells were measured after 24 hr. Details on sample size and statistics can be found in Supplementary file 1. (A–B) Graph depicting the total number of SGFP2-CTNNB1 particles as measured with N and B upon S45F mutation (A) or CHIR99021 stimulation (B). (C) Graph depicting the total concentration of SGFP2-CTNNB1 particles (monomeric plus complexed) as measured with FCS upon CHIR99021 treatment. (D–E) Graphs depicting the speed of the second diffusion component (i.e. SGFP2-CTNNB1 containing complex) in the nucleus measured by FCS for S45F mutant (D) or 24 hr CHIR99021 treatment (E). (F-G) Graph depicting the molecular brightness of SGFP2-CTNNB1 in the nucleus relative to controls as measured with N and B for S45F mutant (F) or 24-hour CHIR99021 treatment (G). EGFP monomer was used for normalization and EGFP dimer and trimer as controls for N and B measurements.
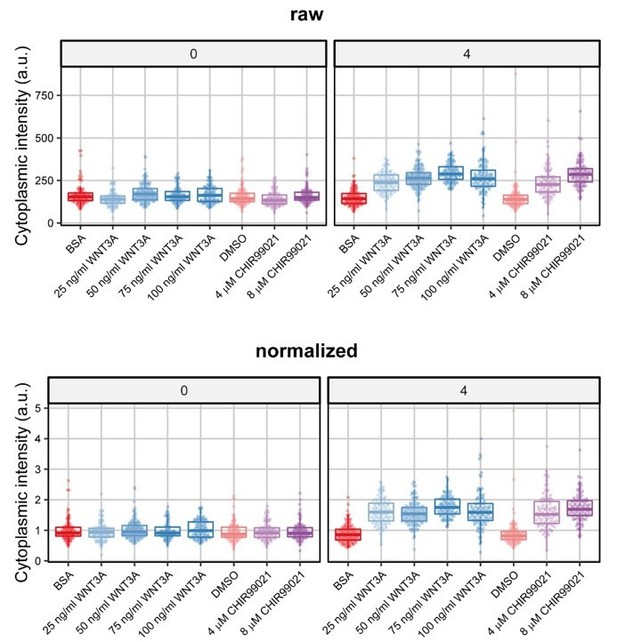
Boxplots and individual datapoints from a single biological experiment (that is part of Figure 3 previously Figure 4) at 0 and 4 hours after treatment.
The upper panels (raw) show the unnormalized intensity and the bottom panels (normalized) the normalized data. Note that although the axis is very different (0-750 compared to 0-5), the spread in the data is highly similar before and after normalization.
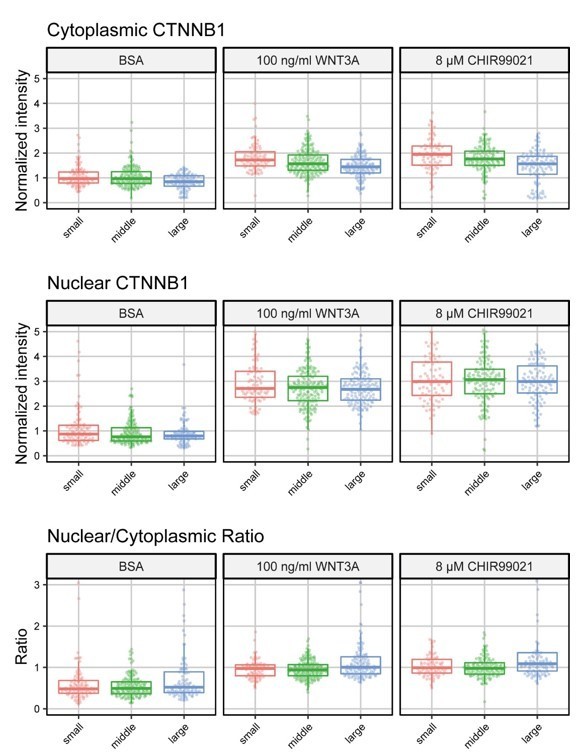
Cells were divided into 3 size categories (small: <500 pixels, middle: 500-1500 pixels, large: >1500 pixels).
Graphs show the normalized cytoplasmic intensity (top), normalized nuclear intensity (middle) and the nuclear/cytoplasmic ratio (bottom) for cells treated for 4 hours with BSA (control) 100 ng/ml WNT3A or 8 µM CHIR99021.
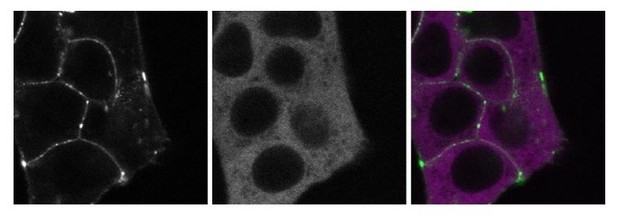
These images show a double-tagged line that has endogenously tagged SGFP2-CTNNB1 (left panel, green) and endogenously tagged mScI-GSK3B (middle panel, magenta).
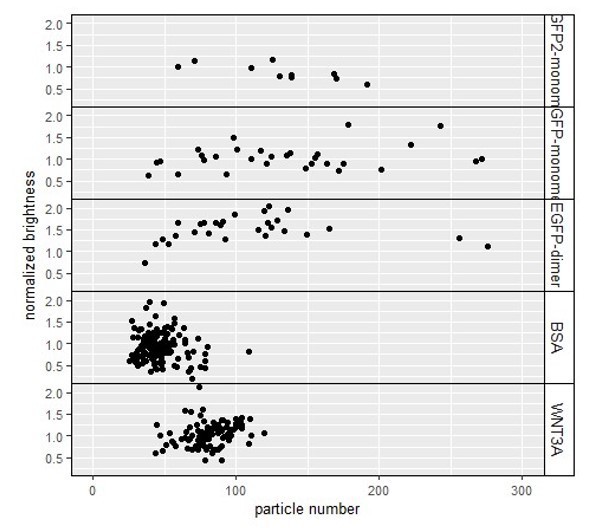
Figure displaying the relation between the particle number (x-axis) and the normalized brightness (y-axis).
The number of particles is up to 3fold higher than on average in our endogenously tagged SGFP2CTNNB1 cell lines. However, we see no apparent effect of these higher particle numbers on the normalized brightness.
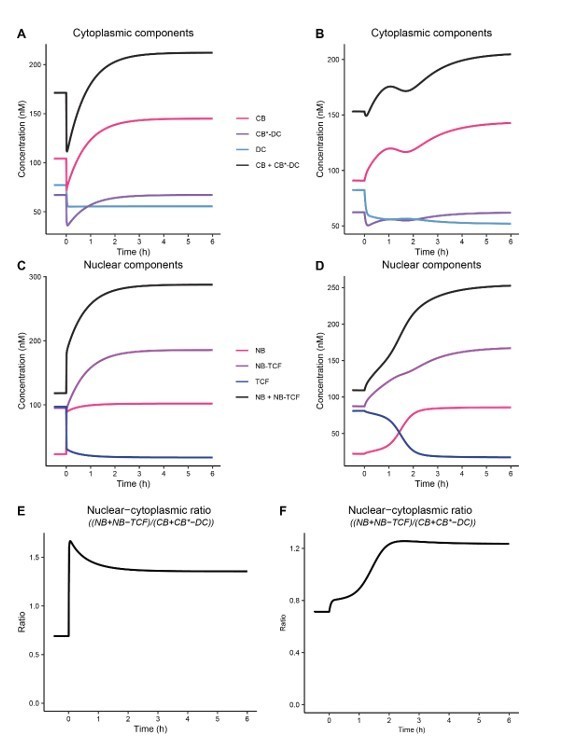
Comparison of old model without steep (A,C,E) and new model with steep (B,D,F).
In the old model the change in k6/k7 and k9/k8 is immediate at the addition of WNT3A. This results in a sudden loss of cytoplasmic CTNNB1 (A) and gain in the nucleus (C) as is also apparent from the nuclear-cytoplasmic ratio (E). Our live imaging experiment shows that the changes. This excluded the slow adjustment of the k6/k7 and k8/k9 (called steep in the newest version of the model). Without this adjustment the parameters in the nucleus adjust immediately resulting in an immediate change in the nuclear-cytoplasmic ratio (C) and the TCF-bound fraction (E). This also leads to a dip in the cytoplasmic concentrations of CTNNB1 (B) and immediate increase in cytoplasmic concentrations (C).
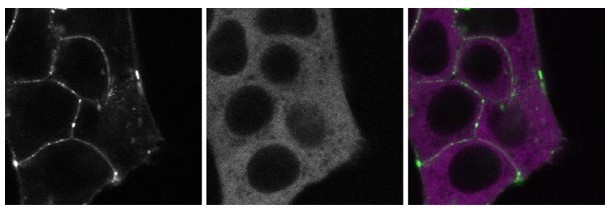
These images show a double-tagged line that has endogenously tagged SGFP2CTNNB1 (left panel, green) and endogenously tagged mScI-GSK3B (middle panel, magenta).
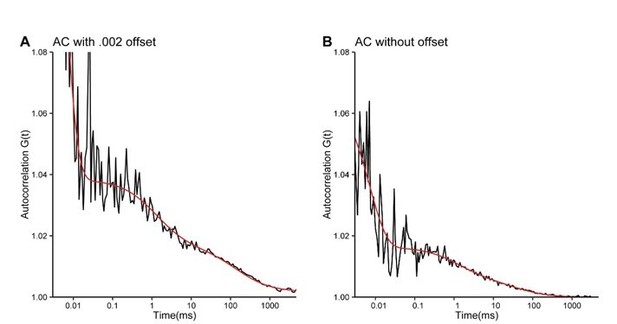
Autocorrelation curves from the same experiment with a small offset of 0.
002 (A) as original example in Figure 5G (now Figure 4G) or without offset (B).
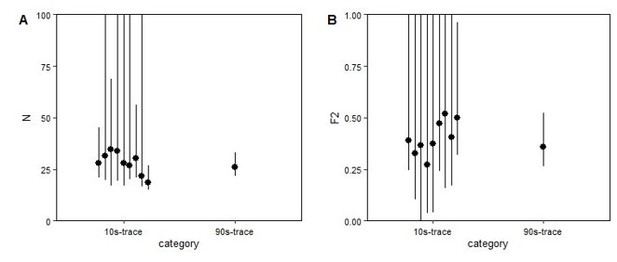
Comparison of analysis of short and long FCS traces.
Autocorrelation curves were made from nine 10s portion of the measurement (trace), or a single 90s trace from the same FCS measurement of SGFP2-CTNNB1 and subsequently fitted with the same fitting model. The graphs depict the fitted values (dots) and confidence intervals determined by the goodness of fit in the FCS analysis software (lines) for the particle number (N, panel A) and the fraction of slow component (F2, panel B) of the 10s and 90s.

SGFP2-CTNNB1 parameters fitted by a two-diffusion component model where T1 is free (A-C) or fixed at 14.9 um2/s (D-F).
(A and D) Concentration of SGFP2-CTNNB1 (B and E) Fraction of slow SGFP2-CTNNB1 (C and F) Speed of slow SGFP2-CTNNB1.

Differences between EGFP and SGFP2.
(A) protein alignment of EGFP (top) and SGFP2 (bottom) showing the 4 amino acid difference. (B) Independent measurements in an additional mammalian cell line of monomeric EGFP and SGFP2 brightness. The same control constructs, measured in HAP1 cells, were already included in figures 5 and 7. The molecular brightness of sGFP2 is slightly lower, but well within the 95% confidence interval of the biological measurements. Any differences between the proteins are thus too small to be significant.
Videos
Representative video of confocal time-lapse experiments, showing SGFP2-CTNNB1 (left, green), SiR-DNA staining (middle, magenta), and transmission image (right, gray) after treatment with vehicle control (BSA).
Time of addition is at 00:00:00 (indicated at the top left). Scale bar in the lower right represents 20μm.
Representative video of confocal time-lapse experiments, showing SGFP2-CTNNB1 (left, green), SiR-DNA staining (middle, magenta), and transmission image (right, gray) after treatment with 100 ng/ml WNT3A.
Time of addition is at 00:00:00 (indicated at the top left). Scale bar in the lower right represents 20μm.
Representative video of confocal time-lapse experiments, showing SGFP2-CTNNB1 (left, green), SiR-DNA staining (middle, magenta), and transmission image (right, gray) after treatment 8 µM CHIR99021.
Time of addition is at 00:00:00 (indicated at the top left). Scale bar in the lower right represents 20μm.
Video showing the quantification of the normalized intensity of SGFP2-CTNNB1 in the cytoplasm of time-lapse microscopy series (from Figure 4 and Videos 1–3) at each time point showing all individual cells from three biological experiments.
Time of addition of the indicated substances is at 00:00:00 (indicated at the top left). The left graph represents the raw data (colored dots, each dot is one cell, n=155–400 cells for each condition and time point), the median (black circle) and the 95% CI of the median (black bar). The right graph represents the median difference (black circle) from the treatments to the control (BSA). When the 95% CI (black bar) does not overlap 0, the difference between the two conditions is significant. Significant changes in intensity can first be observed after 40 min of 8 µM CHIR99021, and after 70–80 min of 4 µM CHIR99021 or 25–100 ng/ml WNT3A treatment.
Video showing the quantification of of the normalized intensity of SGFP2-CTNNB1 in the nucleus of time-lapse microscopy series (from Figure 4 and Videos 1–3) at each time point showing all individual cells from three biological experiments.
Time of addition of the indicated substances is at 00:00:00 (indicated at the top left). The left graph represents the raw data (colored dots, each dot is one cell, n=155–400 cells for each condition and time point), the median (black circle) and the 95% CI of the median (black bar). The right graph represents the median difference (black circle) from the treatments to the control (BSA). When the 95% CI (black bar) does not overlap 0, the difference between the two conditions is significant. Significant changes in intensity can be observed for all treatments (but not controls) after 20–50 min.
Video showing the quantification of the nuclear-cytoplasmic ratio of SGFP2-CTNNB1, calculated from raw intensity values underlying Videos 4 and 5.
At each time point showing all individual cells from three biological experiments. Time of addition of the indicated substances is at 00:00:00 (indicated at the top left). The left graph represents the raw data (colored dots, each dot is one cell, n=155–400 cells for each condition and time point), the median (black circle) and the 95% CI of the median (black bar). The right graph represents the median difference (black circle) from the treatments to the control (BSA). When the 95% CI (black bar) does not overlap 0, the difference between the two conditions is significant. Significant changes in the nuclear-cytoplasmic ratio can be observed for all treatments (but not controls) after 20–50 min.
Tables
Total number of SGFP2-CTNNB1 molecules in the confocal volume and corresponding calculated concentrations obtained from FCS measurements in n=3 independent experiments.
The concentration is calculated from the number of molecules in the confocal volume and the calibrated confocal volume (see Materials and methods). The number of molecules is consistent with those measured with N and B analysis (Figure 5—figure supplement 1A, Supplementary file 1).
| Number of molecules | Concentration (nM) | |||||
|---|---|---|---|---|---|---|
| Compartment | Treatment | N | Median | 95% CI | Median | 95% CI |
| Cytoplasm | BSA | 21 | 80 | 70–116 | 180 | 127–218 |
| WNT3A | 21 | 95 | 85–122 | 221 | 144–250 | |
| Nucleus | BSA | 21 | 63 | 53–72 | 122 | 91–158 |
| WNT3A | 18 | 135 | 127–150 | 240 | 217–325 | |
Number of SGFP2-CTNNB1 molecules in the confocal volume and corresponding calculated concentration of SGFP2-CTNNB1 molecules with the fast or slow diffusion coefficient obtained from FCS measurements in n=3 independent experiments.
The concentration is calculated from the number of molecules and the calibrated confocal volume (see Materials and methods).
| Fast SGFP2-CTNNB1 | Slow SGFP2-CTNNB1 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Number of molecules | Concentration (nM) | Number of molecules | Concentration (nM) | |||||||
| Compartment | Treatment | n | Median | 95% CI | Median | 95% CI | Median | 95% CI | Median | 95% CI |
| Cytoplasm | BSA | 21 | 51 | 40–63 | 91 | 66–139 | 29 | 20–37 | 57 | 38–76 |
| WNT3A | 21 | 60 | 47–80 | 145 | 76–168 | 35 | 30–41 | 68 | 57–76 | |
| Nucleus | BSA | 21 | 48 | 41–66 | 87 | 78–119 | 13 | 2–22 | 22 | 4–40 |
| WNT3A | 18 | 96 | 81–101 | 170 | 147–214 | 47 | 37–49 | 86 | 64–104 | |
Brightness of SGFP2-compared relative to EGFP-monomer and -dimer controls in n=2 independent experiments. N is the number of analyzed cells.
p-Values were calculated using PlotsOfDifferences that uses a randomization test (Goedhart, 2019).Note that only the EGFP-dimer is significantly different to the EGFP-monomer control, while SGFP2-CTNNB1 is not.
| Fluorophore | Compartment | Treatment | N | Median | 95 CI median | p-value to matched control (EGFP monomer in the nucleus or cytoplasm) |
|---|---|---|---|---|---|---|
| EGFP-monomer | Cytoplasm | NA | 15 | 1 | 0.79–1.34 | 1.000 |
| EGFP-dimer | Cytoplasm | NA | 14 | 1.4 | 1.29–1.60 | 0.011* |
| SGFP2-CTNNB1 | Cytoplasm | BSA | 69 | 0.92 | 0.83–1.00 | 0.738 |
| SGFP2-CTNNB1 | Cytoplasm | 100 ng/ml WNT3A | 46 | 1.01 | 0.93–1.11 | 0.919 |
| EGFP-monomer | Nucleus | NA | 15 | 1 | 0.91–1.07 | 1.000 |
| EGFP-dimer | Nucleus | NA | 14 | 1.62 | 1.44–1.69 | <0.001* |
| SGFP2-CTNNB1 | Nucleus | BSA | 69 | 0.87 | 0.78–0.96 | 0.192 |
| SGFP2-CTNNB1 | Nucleus | 100 ng/ml WNT3A | 46 | 1.05 | 0.95–1.15 | 0.578 |
Variables Minimal Model of WNT signaling.
| Model name | Variable | Compound | Values obtained from | WNT OFF (nM) | WNT ON (nM) |
|---|---|---|---|---|---|
| CB | Free cytoplasmic CTNNB1 | FCS data this report | 91 | 145 | |
| DC | Free destruction complex | Model equations | 82.4 | 52 | |
| CB*-DC | DC-bound phosphorylated CTNNB1 | FCS data this report* | 62.5* | 62.5* | |
| DC* | Inactivated destruction complex | Model equations | 0** | 30.5** | |
| NB | Free nuclear CTNNB1 | FCS data this report | 87 | 170 | |
| TCF | Free TCF | Model equations | 81 | 17 | |
| NB-TCF | TCF-bound nuclear CTNNB1 | FCS data this report | 22.2 | 86 | |
| TCF0 | Total TCF | and Tan et al., 2012 - Figure 11 | 103 | 103 |
-
*Under the assumption that k3 does not change, the levels of CB*-DC remain equal. Since there was no significant difference between the concentration of slow SGFP2-CTNNB1 in the absence or presence of WNT3A (57 nM versus 68 nM, not significant, Table 2) the average of both medians (62.5 nM) was used. ** In WNT OFF, w=0, and no inactivated destruction complex is formed. In WNT ON, w=1, which induces the formation of inactivated destruction complex at the expense of free destruction complex (see Equations 7a and 9 in the model description in Materials and methods).
Equilibrium conditions for the Minimal Model of WNT signaling.
All rates are multiplied with factor R=20, so that the equilibrium is reached at 4.5 hr according to Figure 4C–D.
| Rate constant | Biological process | Values based on | Wnt off | Wnt on | |
|---|---|---|---|---|---|
| b | nMmin−1 | CTNNB1 synthesis | from Lee | 0.423 | 0.423 |
| nM | Binding to and phosphorylation by the destruction complex of cytoplasmic CTNNB1 | from Lee | 120 | 120 | |
| min−1 | Dissociation and degradation of phosphorylated CTNNB1 from the destruction complex | Deduced from and | 0.0068 | 0.0068 | |
| nM | Inactivation of the destruction complex by activated DVL | Fitted to and | N.A.* | 1.7 | |
| Ratio between nuclear import and export of CTNNB1 | Deduced from and | 0.96 | 1.17 | ||
| nM | Dissociation of nuclear CTNNB1 from TCF | Deduced from , , | 320 | 33.6 |
-
*In WNT OFF, w=0, and no inactive destruction complex is formed. Only in WNT ON, w=1, which induces the formation of inactivated destruction complex at the expense of free destruction complex (see Equations 7a and 9 in the model description in Materials and methods).
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Homo sapiens) | HAP1 | Whitehead Institute | Cellosaurus: CVCL_Y019 | kind gift from Thijn Brummelkamp (NKI) |
| Cell line (Homo sapiens) | HAP1SGFP2-CTNNB1 | This paper | ||
| Cell line (Homo sapiens) | HAP1SGFP2-CTNNB1(S45F) | This paper | ||
| Transfected construct (Homo sapiens) | pSpCas9(BB)−2A-Puro (PX459) V2.0 | Ran et al., 2013 | RRID:Addgene_62988 | |
| Transfected construct (Homo sapiens) | MegaTopflash | Hu et al., 2007 | kind gift from Christophe Fuerer and Roel Nusse, Stanford University | |
| Transfected construct (Homo sapiens) | CMV Renilla | Promega | E2261 | |
| Transfected construct (Homo sapiens) | pSGFP2-C1 | Kremers et al., 2007 | RRID:Addgene_22881 | kind gift from Dorus Gadella |
| Transfected construct (Homo sapiens) | pmScarlet-i_C1 | Bindels et al., 2017 | RRID:Addgene_85044 | kind gift from Dorus Gadella |
| Transfected construct (Homo sapiens) | pSYFP2-C1 | Kremers et al., 2006 | RRID:Addgene_22878 | kind gift from Dorus Gadella |
| Transfected construct (Homo sapiens) | mTurquoise2-C1 | Goedhart et al., 2012 | RRID:Addgene_54842 | kind gift from Dorus Gadella |
| Transfected construct (Homo sapiens) | pEGFP | Clontech | ||
| Transfected construct (Homo sapiens) | pEGFP2 | Pack et al., 2006 | kind gift from Masataka Kinjo | |
| Transfected construct (Homo sapiens) | pEGFP3 | Pack et al., 2006 | kind gift from Masataka Kinjo | |
| Transfected construct (Homo sapiens) | pBluescript II KS(+) | Stratagene | ||
| Transfected construct (Homo sapiens) | pX459-CTNNB1-ATG | This paper | RRID:Addgene_153429 | |
| Transfected construct (Homo sapiens) | pX459-CTNNB1-S45 | This paper | RRID:Addgene_164587 | |
| Transfected construct (Homo sapiens) | pRepair-SGFP2-CTNNB1 | This paper | RRID:Addgene_153430 | |
| Recombinant DNA reagent | pRepair-mScI-CTNNB1 | This paper | RRID:Addgene_153431 | |
| Recombinant DNA reagent | pRepair-SYFP2-CTNNB1 | This paper | RRID:Addgene_153432 | |
| Recombinant DNA reagent | pRepair-mTq2-CTNNB1 | This paper | RRID:Addgene_153433 | |
| Chemical compound, drug | CHIR99021 | Biovision | 1677–5 | 6 mM stock in DMSO |
| Peptide, recombinant protein | Recombinant Mouse Wnt-3a | R and D systems | 1324-WN-002 | 10 µg/ml stock solution in 0.1% BSA in PBS |
| Chemical compound, drug | Dapi | Invitrogen | D1306 | |
| Chemical compound, drug | Vybrant DyeCycle Violet Stain | Invitrogen | V35003 | |
| Chemical compound, drug | Vybrant DyeCycle Ruby Stain | Invitrogen | V10273 | |
| Antibody | Non-phosphorylated (Active) β-catenin clone D13A1 (Rabbit monoclonal) | Cell Signaling | 8814S RRID:AB_11127203 | WB (1:1000) |
| Antibody | Total β-catenin clone 14 (mouse monoclonal) | BD | 610153 RRID:AB_397554 | WB (1:2000) |
| Antibody | α-Tubulin clone DM1A (mouse monoclonal) | Sigma-Aldrich | T9026 RRID:AB_477593 | WB (1:1000) |
| Antibody | GFP Antibody (Rabbit polyclonal) | Invitrogen | A-6455 RRID:AB_221570 | WB (1:1000) |
| Antibody | IRDye 680LT Goat anti-Rabbit IgG | LI-COR | 926–68021 RRID:AB_10706309 | WB (1:20,000) |
| Antibody | IRDye 800CW Donkey anti-Mouse IgG | LI-COR | 926–32212 RRID:AB_621847 | WB (1:20,000) |
| Chemical compound, drug | SiR-DNA | Spirochrome | SC007 | |
| Chemical compound, drug | Alexa Fluor 488 NHS Ester | Molecular probes | A20000 | |
| Software, algorithm | FlowJo | |||
| Software, algorithm | CellProfiler pipeline | This paper | available at https://osf.io/6pmwf/ | |
| Software, algorithm | FIJI/ImageJ | |||
| Software, algorithm | FFS Dataprocessor version 2.3 | SSTC | ||
| Software, algorithm | ptu converter | Crosby et al., 2013 | ||
| Software, algorithm | ImageJ macro script | modified from Crosby et al., 2013 | available at https://osf.io/ys5qw/ | |
| Software, algorithm | PlotsOfDifferences | Goedhart, 2019 | https://huygens.science.uva.nl/PlotsOfDifferences/ | |
| Software, algorithm | RStudio | |||
| Software, algorithm | R script | This paper | available at https://osf.io/sxakf/ | |
| Software, algorithm | R shiny app | This paper | WNT_minimal_model_v2.4.R | app available at https://wntlab.shinyapps.io/WNT_minimal_model/, source script available at https://osf.io/27ya6/ |
primers/oligonucleotides used in this study.
| primer | sequence |
|---|---|
| RVA24 | CAAGTTTGTTGTAGGATATGCCC |
| RVA25 | CGATGTCAATAGGACTCCAGA |
| RVA124 | AGTGTGAGGTCCACGGAAA |
| RVA125 | CCGTCATGGACATGGAAT |
| RVA555 | GCCAAACGCTGGACATTAGT |
| RVA558 | AGACCATGAGGTCTGCGTTT |
| RVA561 | CACCGTTGCCTTTACCACTCAGAGA |
| RVA562 | AAACTCTCTGAGTGGTAAAGGCAAC |
| RVA567 | CACCGTGAGTAGCCATTGTCCACGC |
| RVA568 | AAACGCGTGGACAATGGCTACTCAC |
| RVA581 | tgctcaccatggtggGATTTTCAAAACAGTTGTATGGTATACTTC |
| RVA582 | actgttttgaaaatcCCACCATGGTGAGCAAGGGC |
| RVA583 | agtagccattgtccaCTTGTACAGCTCGTCCATGCCG |
| RVA584 | gacgagctgtacaagTGGACAATGGCTACTCAAGGTTTG |
| RVA618 | atacgactcactatagggcgaattggagctGATGCAGTTTTTTTCAATATTGC |
| RVA619 | ttctagagcggccgccaccgcggtggagctCTCTCTTTTCTTCACCACAACATTTTATTTAAAC |
| RVA622 | AAGAGAGAGCTCCACCGCGGTGGCGGCCG |
| RVA623 | TGCATCAGCTCCAATTCGCCCTATAGTGAGTCG |
| RVA1616 | tgtccacgctgGATTTTCAAAACAGTTGTATGG |
| RVA1617 | atacaactgttttgaaaatccagcgtggacaATGGTGAGCAAGGGCGAG |
| RVA1618 | cacaaaccttgagtagccatCTTGTACAGCTCGTCCATGC |
| RVA1619 | ATGGCTACTCAAGGTTTGTGTCATTAAATC |
| RVA2540 | CTTACCTGGACTCTGGAATCCATTCTGGTGCCACTACCACAGCTCCTTTCCTGTCCGGTAAAGGCAATCCTGAGGAAGAGGATGTGGATACCTCCCAAGT |
This table describes different scenarios of how different brightness species would impact the average apparent brightness.
| Scenario 1 | actual brightness | actual fraction (%) | apparant fraction (%) | average apparent brightness |
|---|---|---|---|---|
| monomer SGFP2-CTNNB1 | 1 | 50 | 50 | - |
| complex containing 1 SGFP2-CTNNB1 | 1 | 50 | 50 | - |
| 1 | ||||
| Scenario 2 | actual brightness | actual fraction (%) | apparant fraction (%) | average apparent brightness |
| monomer SGFP2-CTNNB1 | 1 | 50 | 20 | - |
| complex containing 2 SGFP2-CTNNB1 | 2 | 50 | 80 | - |
| 1.67 | ||||
| Scenario 3 | actual brightness | actual fraction (%) | apparant fraction (%) | average apparent brightness |
| monomer SGFP2-CTNNB1 | 1 | 80 | 50 | - |
| complex containing 2 SGFP2-CTNNB1 | 2 | 20 | 50 | - |
| 1.33 | ||||
| Scenario 4 | actual brightness | actual fraction (%) | apparant fraction (%) | average apparent brightness |
| monomer SGFP2-CTNNB1 | 1 | 50 | 10 | - |
| complex containing 3 SGFP2-CTNNB1 | 3 | 50 | 90 | - |
| 2.5 | ||||
| Scenario 5 | actual brightness | actual fraction (%) | apparant fraction (%) | average apparent brightness |
| monomer SGFP2-CTNNB1 | 1 | 90 | 50 | - |
| complex containing 3 SGFP2-CTNNB1 | 3 | 10 | 50 | - |
| 1.5 | ||||
| Scenario 6 | actual brightness | actual fraction (%) | apparant fraction (%) | average apparent brightness |
| monomer SGFP2-CTNNB1 | 1 | 99 | 50 | - |
| complex containing 10 SGFP2-CTNNB1 | 10 | 1 | 50 | - |
| 1.83 |
Additional files
-
Source data 1
Numerical data for Figure 3B,C,D, Figure 3—figure supplement 2, Figure 3—figure supplement 3 and Figure 8—figure supplement 1B,C,D.
- https://cdn.elifesciences.org/articles/66440/elife-66440-data1-v2.xlsx
-
Supplementary file 1
Tables of all summary statistics (mean, median, 95% confidence intervals, differences, p-values) of the FCS and N and B parameters show in Figures 5, 7 and 8 and accompanying supplements.
- https://cdn.elifesciences.org/articles/66440/elife-66440-supp1-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/66440/elife-66440-transrepform-v2.docx