Kinetic analysis of ASIC1a delineates conformational signaling from proton-sensing domains to the channel gate
Figures
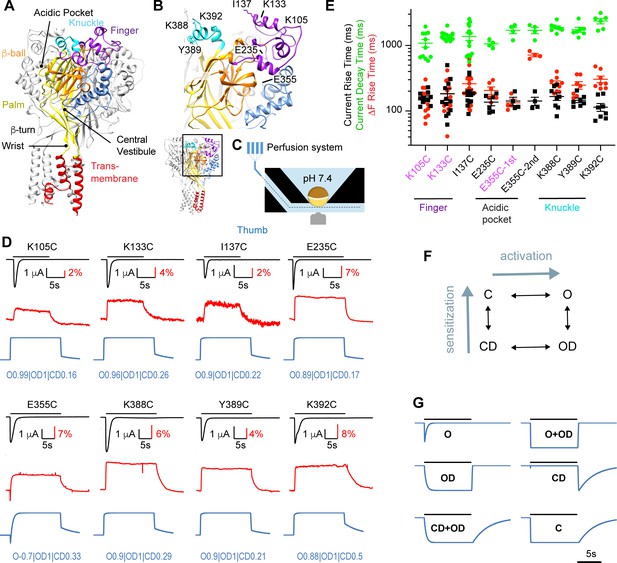
Rapid ΔF signals of mutants distant from the pore.
(A) Structural image of a trimeric ASIC1a channel with the domains colored and named, based on the Mit Toxin-opened structure (PDB identifier 4NTW, Baconguis et al., 2014); the acidic pocket, central vestibule, β-turn, and wrist are indicated. (B) Detailed structural view with indication of residues studied in Figure 1. (C) Schematic view of the oocyte chamber used for the kinetic measurements of currents and fluorescence signals (ΔF). (D) Current traces (black), fluorescence signals (red) and simulated fluorescence traces (blue) are shown. The conditioning pH between stimulations was pH7.4; the stimulation pH6 was applied as indicated by the horizontal black bars. Simulated traces were generated by the kinetic model (Materials and methods), using the parameters indicated in blue (Supplementary file 2). (E) Kinetics of current appearance and decay, and of ΔF onset, measured as rise time (RT) or decay time for an activation by pH6.0 (n = 4–14). An analysis was carried out to determine if the ΔF onset kinetics were correlated with current appearance or decay (Materials and methods). In mutants labeled in purple, the ΔF onset kinetics were correlated with current appearance. For E355C, ΔF onset kinetics are reported for both components (‘1st’ and ‘2nd’). (F) Representation of the kinetic model used for the simulation of fluorescence traces; C, closed; O, open; OD, open-desensitized; CD, closed-desensitized. (G) Model-generated fluorescence with proportionality factor = -1 for the state(s) indicated with the traces; for C, this factor was +1. Conditioning pH was 7.4; it was changed to pH6.0 for a duration of 10 s. The fall time of the pH change (speed of solution change) was set to 200 ms. Source data are provided in the file Figure 1—source data 1.
-
Figure 1—source data 1
Mutants distant from the pore.
- https://cdn.elifesciences.org/articles/66488/elife-66488-fig1-data1-v2.xlsx
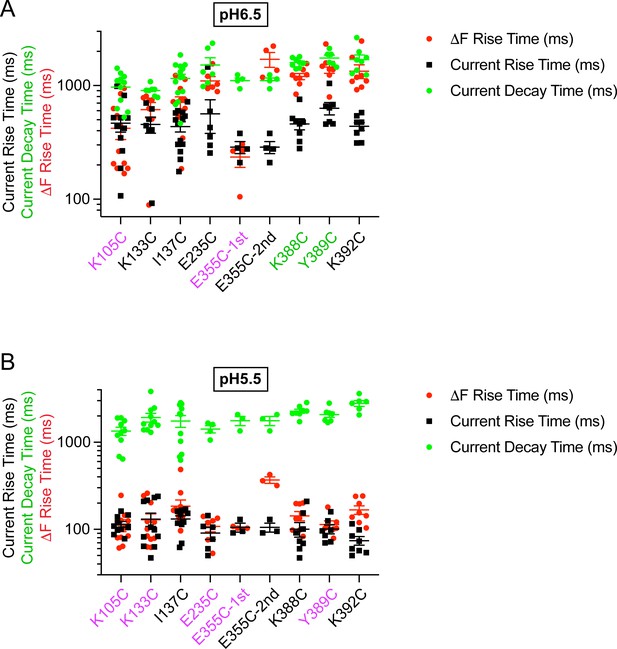
Kinetics of mutants with rapid ΔF located outside the wrist at pH6.5 and 5.5.
Current RT and decay time and ΔF RT obtained at pH6.5 (A) or 5.5 (B), as indicated, n = 3–14. The color of the labels of the mutants indicates that the ΔF onset kinetics are correlated with current appearance (purple) or with current desensitization (green; Materials and methods).
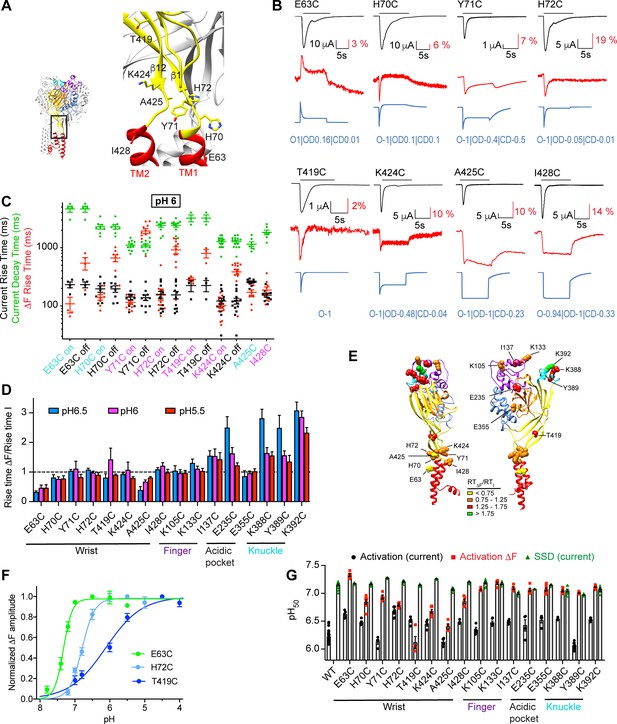
Transient, rapid ΔF signals in the wrist.
(A) Structural image indicating the positions of the tested mutants. (B) Representative current and fluorescence traces of the Cys mutants in the wrist. The conditioning pH between stimulations was pH7.4; the stimulation pH6 was applied as indicated by the horizontal black bars. The blue traces were generated by the kinetic model as described in the legend to Figure 1G. (C) The kinetics of current appearance and decay, and of ΔF onset, measured as rise time (RT) or decay time for an activation by pH6.0, are plotted (n = 3–14). Since the ΔF signal was transient in many mutants, the kinetics of the ΔF onset (‘on’) and decay (‘off’) are indicated. The color of the labels of the mutants indicates that the ΔF kinetics are correlated with current appearance (purple) or are faster than current appearance (cyan). (D) The RTΔF/RTI ratio is plotted for stimulation pH6.5, 6.0, and 5.5 (n = 3–14) of wrist and distant mutants. (E) Structural image of ASIC1a (seen from two different sides) indicating the position of the mutants studied in Figures 1 and 2. The color of each residue corresponds to a range of RTΔF/RTI ratio at pH6, as indicated in the figure. (F) Experimentally determined pH dependence of the ΔF amplitude for the indicated mutants; n = 4–5. (G) Summary of pH dependencies of current activation (black), current SSD (green) and ΔF amplitude (red), n = 4–30. In the experiments for ΔF measurements, the conditioning pH was 8.0. Source data are provided in the file Figure 2—source data 1.
-
Figure 2—source data 1
Wrist mutants.
- https://cdn.elifesciences.org/articles/66488/elife-66488-fig2-data1-v2.xlsx
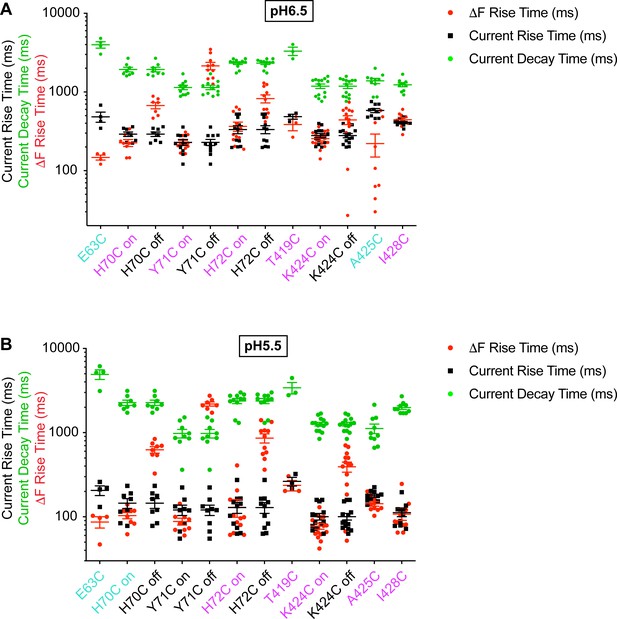
Rapid ΔF kinetics of wrist mutants at pH5.5 and 6.5.
Current RT and decay time and ΔF RT obtained at pH6.5 (A) or 5.5 (B), as indicated, n = 3–14. The color of the labels of the mutants indicates that the ΔF onset kinetics are correlated with current appearance (purple) or are faster than current appearance (cyan; Materials and methods).
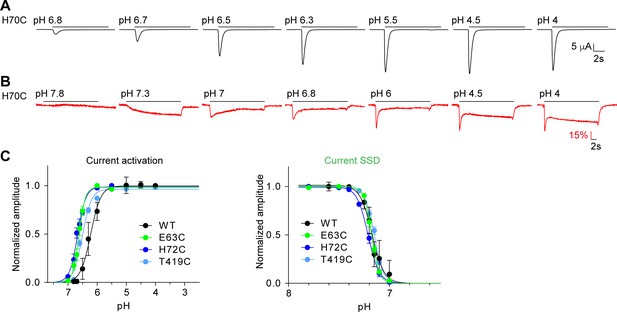
pH dependence of mutants involved in rapid conformational changes.
(A) and (B) Representative traces of experiments measuring the pH dependence of the current and of ΔF, respectively of the mutant H70C. (C) Activation curves for the current and steady-state desensitization (SSD) curve of the indicated mutants. The pH protocols were as illustrated in (A) and (B) and described in Materials and methods. For the analysis of the SSD pH dependence, the protocol was as follows: After exposure during 50 s to the conditioning pH, the channels were exposed during 10 s to pH4.5. This protocol was repeated at different conditioning pH values, and the normalized current is plotted as a function of the conditioning pH.
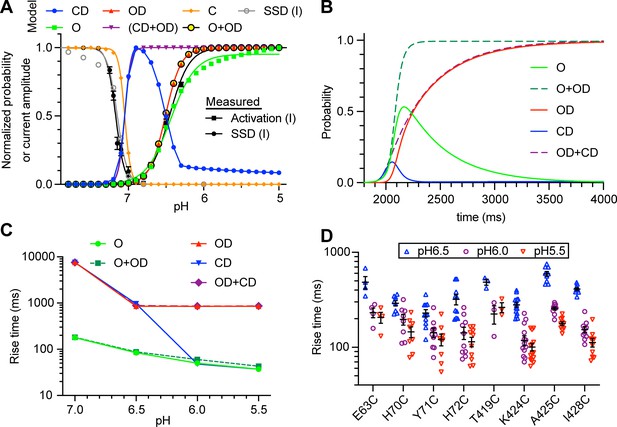
Analysis of pH dependence and kinetics of ΔF signals indicate that rapid ΔF signals are linked to channel opening.
(A) Model-generated normalized probability of occupancy of the indicated states (colored traces). The conditions of the simulations were as in Figure 1G, except that the duration of the acidic pulse was 20 s. The gray symbols represent the current SSD calculated with the model by applying the same protocol as applied in the experiments (Figure 2—figure supplement 2). The black symbols show the experimental pH dependence of current activation and SSD as determined in Figure 2—figure supplement 2. (B) Model-generated time dependence of state occupation upon a pH change from 7.4 to 6.0. Conditioning pH was 7.4; it was changed to pH6.0 at 2 s for a duration of 10 s. The fall time of the pH change was set to 200 ms. (C) Model-generated rise time of the occupancy of the indicated states upon acidification from pH7.4 to the indicated values; model parameters as in B. (D) Experimentally determined rise time of the ΔF signal measured for the wrist mutants at the three indicated pH values. Source data are provided in the file Figure 3—source data 1.
-
Figure 3—source data 1
Fluorescence kinetics of wrist mutants at pH 6.5, 6.0 and 5.5.
- https://cdn.elifesciences.org/articles/66488/elife-66488-fig3-data1-v2.xlsx
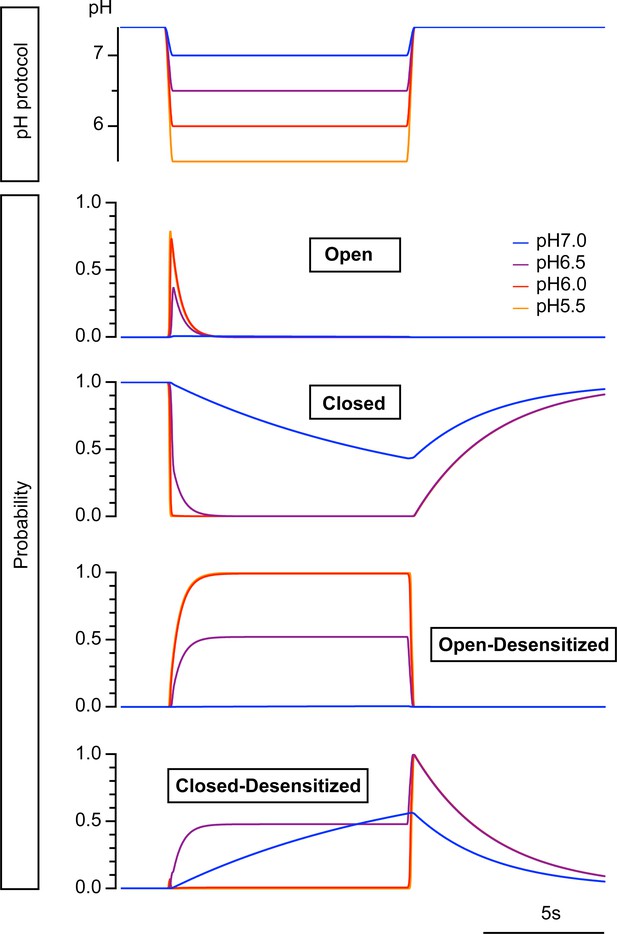
Modeled state probability at different pH conditions.
Time course of probability of the four model states for a pH change from 7.4 to 7.0, 6.5, 6.0, and 5.5. The conditioning pH7.4 was changed to the acidic pH at 2 s for a duration of 10 s. The fall time of the pH change was set to 200 ms.
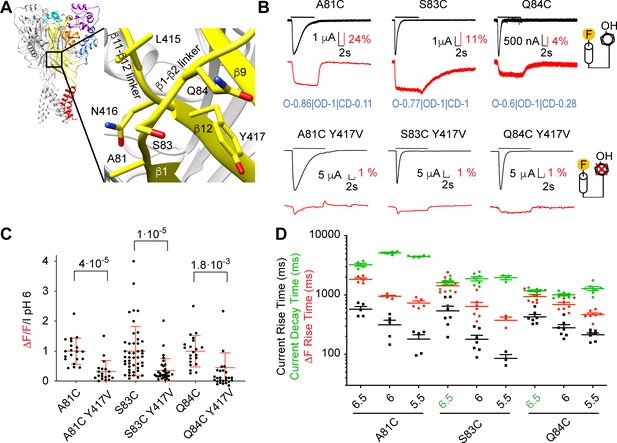
Removal of a fluorescence quenching group strongly reduces β1-β2 linker ΔF amplitude.
(A) Structural image of ASIC1a (left) with zoom on the lower palm domain (right). (B) Representative current and fluorescence traces of single (top) and double (bottom) palm mutants. Conditioning pH7.4 was used, and mutants were stimulated by pH6 for the duration indicated by the horizontal bars. On the right, cartoons of a fluorophore and a quenching Tyr residue illustrate that in the double mutants the quencher was removed. Proportionality factors of the corresponding simulated traces are indicated in blue. (C) Normalized ratio of the fluorescence change / total fluorescence, divided by the pH6-induced current amplitude (ΔF/F/IpH6) for single and double mutants; Kruskal-Wallis and Dunn's post-test, n = 19–45. Corresponding single and double mutants were measured on the same days, and for each experimental day, the ΔF/F/IpH6 ratio of each cell of a given single/double mutant pair was divided by the average ΔF/F/IpH6 ratio of the single mutant of that day. (D) Current RT and decay time and ΔF RT of single mutants obtained at pH6.5, 6.0 or 5.5 (n = 3–7). A green pH indication indicates that the ΔF onset kinetics are correlated with current decay. Source data are provided in the file Figure 4—source data 1.
-
Figure 4—source data 1
Palm mutants.
- https://cdn.elifesciences.org/articles/66488/elife-66488-fig4-data1-v2.xlsx
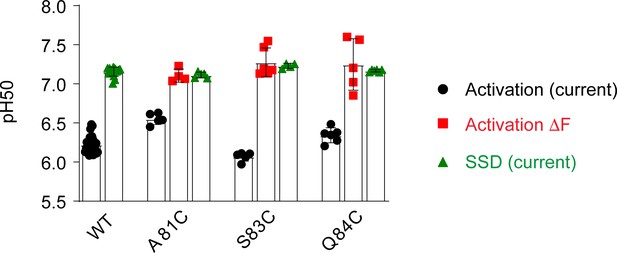
pH dependence of palm mutants.
pH50 values of current activation and SSD, and of ΔF are indicated.
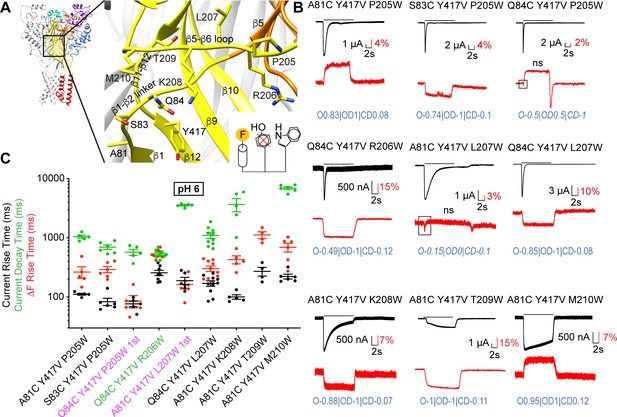
Slow structural rearrangements in the palm domain.
(A) Structural image of ASIC1a (left) with a zoom on the palm (right), showing the position of mutations studied in this figure. Bottom, cartoon illustrating the approach used for these triple mutants, with the fluorophore (‘F’, on the left), the original quenching group Tyr417 (center) mutated, and a new quenching Trp (right) introduced at a different position. (B) Representative current and fluorescence traces of the triple mutants. Conditioning pH7.4 was used, and mutants were stimulated by pH6 for the duration indicated by the horizontal bars. The black frames in some ΔF traces highlight the first ΔF component. ‘ns’ indicates that this ΔF component was potentially non-specific (Supplementary file 3). Proportionality factors of the corresponding simulated traces are indicated in blue. (C) Current RT and decay time and ΔF RT obtained at pH6, n = 4–10. The color of the labels of the mutants indicates that the ΔF onset kinetics are correlated with current appearance (purple) or decay (green; Materials and methods). Source data are provided in the file Figure 5—source data 1.
-
Figure 5—source data 1
Intrasubunit triple mutations palm.
- https://cdn.elifesciences.org/articles/66488/elife-66488-fig5-data1-v2.xlsx
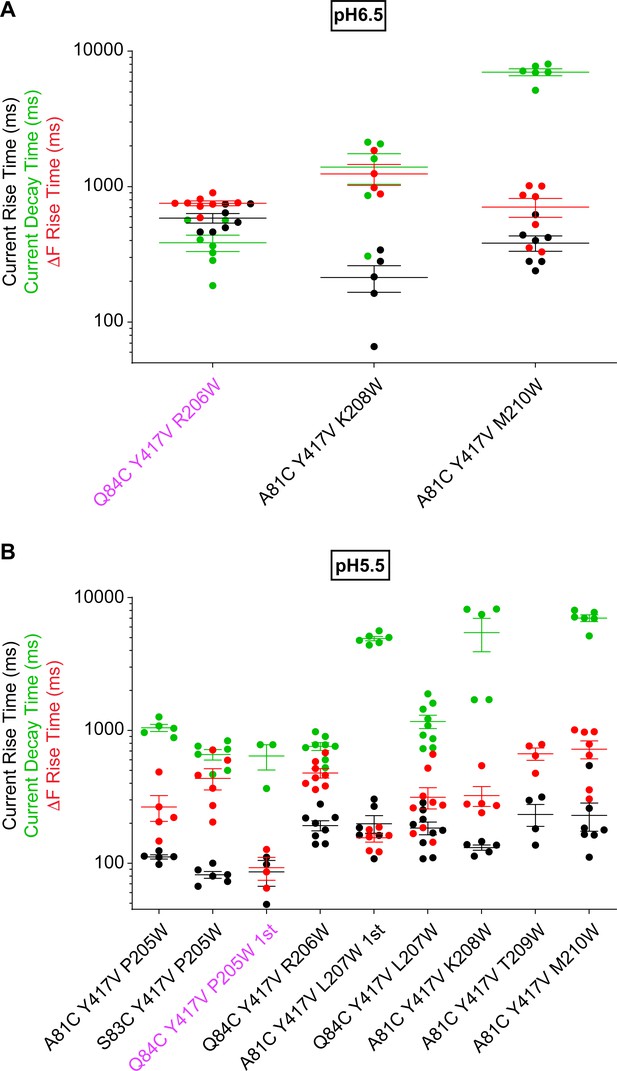
Slow ΔF kinetics of intrasubunit palm triple mutants at pH6.5 and 5.5.
Current RT and decay time and ΔF RT obtained at pH6.5 (A) or 5.5 (B), as indicated, n = 3–9. The color of the labels of the mutants indicates that the ΔF onset kinetics are correlated with current appearance (purple; Materials and methods).
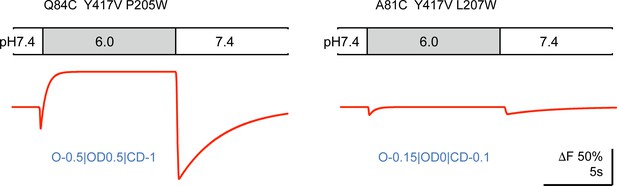
Model-generated ΔF patterns for two mutations of Figure 5.
The proportionality factors used for each pattern is indicated below the traces.
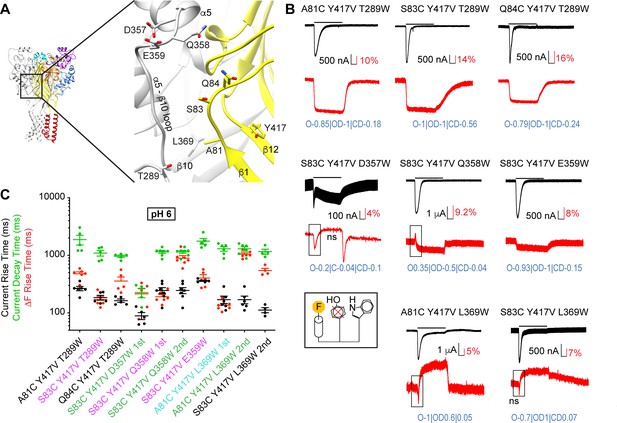
Fast structural rearrangements in the palm-thumb loops.
(A) Structural image of ASIC1a (left) with a zoom on the palm (right), showing the position of mutations studied in this figure. (B) Representative current and fluorescence traces of the triple mutants. Conditioning pH7.4 was used, and mutants were stimulated by pH6 for the duration indicated by the horizontal bars. The black frames in some ΔF traces highlight the first ΔF component. ‘ns’ indicates that this ΔF component was potentially non-specific (Supplementary file 3). Proportionality factors of the corresponding simulated traces are indicated in blue. (C) Current RT and decay time and ΔF RT obtained at pH6, n = 4–7. The color of the labels of the mutants indicates that the ΔF onset kinetics are correlated with current appearance (purple) or decay (green) or are faster than current appearance (cyan; Materials and methods). Source data are provided in the file Figure 6—source data 1.
-
Figure 6—source data 1
Intersubunit triple mutations palm.
- https://cdn.elifesciences.org/articles/66488/elife-66488-fig6-data1-v2.xlsx
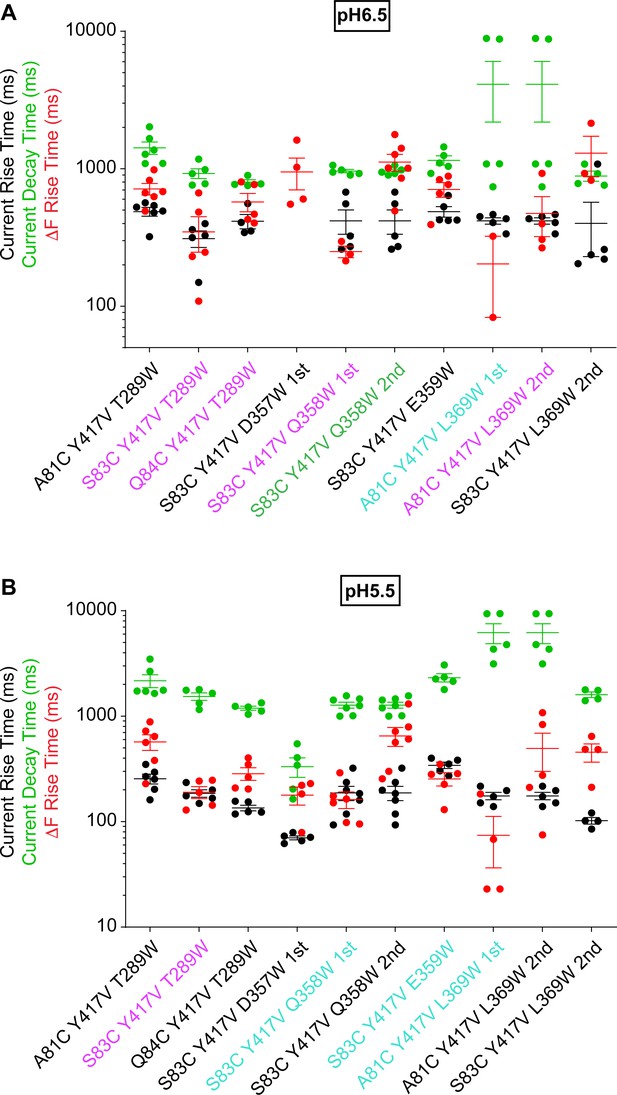
Fast ΔF kinetics of intersubunit palm triple mutants at pH6.5 and 5.5.
Current RT and decay time and ΔF RT obtained at pH5.5 (A) or 6.5 (B), as indicated, n = 2–7. The color of the labels of the mutants indicates that the ΔF onset kinetics are correlated with current appearance (purple) or decay (green; Materials and methods) or are faster than current appearance (cyan). At pH6.5, the current of the mutant S83C/Y417V/D357W was too small for fitting of the kinetics, therefore only the kinetics of the ΔF signal are shown.
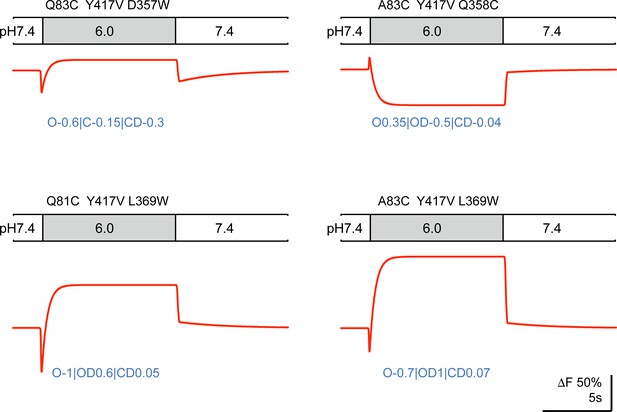
Model-generated ΔF patterns for four mutants of Figure 6.
The proportionality factors used for each pattern are indicated below the traces.
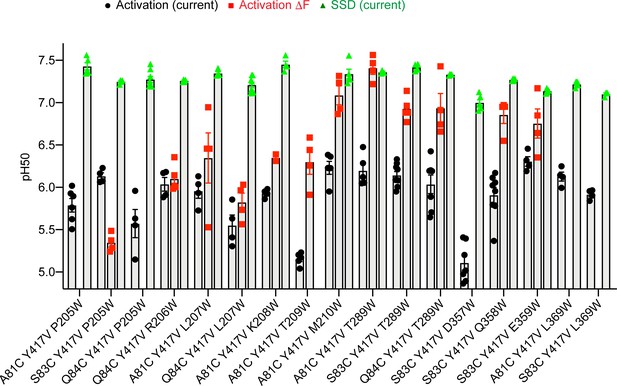
pH dependence of palm mutants.
pH50 values of current activation (black), current SSD (green) and ΔF amplitude (red) are indicated, n = 2–9. Note that ΔF pH50 values are not indicated for all mutants.
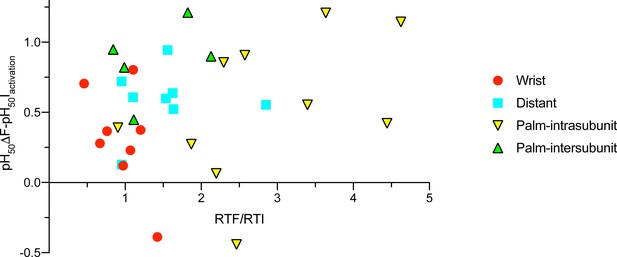
Correlation between the ΔF - current pH50 difference and ΔF kinetics.
The mean value of the pH50(ΔF)-pH50(current activation) difference is plotted as a function of the ratio of the rise time of ΔF / rise time of current activation, for the mutants analyzed in this study in different areas of ASIC1a. ‘Distant’ refers to the mutants of the finger, acidic pocket and knuckle, presented in Figure 1.
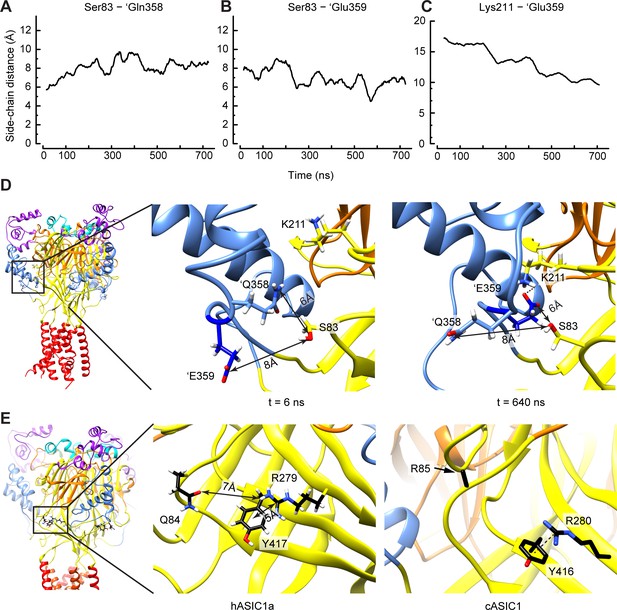
Structural interpretation of selected VCF-predicted distance changes.
(A–D) Data and structural images based on unbiased MD simulations starting from a closed structural model of hASIC1a, in which the protonation of residues mimicked pH5. (A–C) Time series reporting the average (n = three subunits) of the distance between the side chains of Ser83 and 'Gln358 (A), Ser83 and 'Glu359 (B), and Lys211 and 'Glu359. (D) Snapshots taken at the start (t = 6 ns, left) and toward the end (t = 640 ns, right) of the simulation, highlighting the switch in side chain orientation of 'Gln358 and 'Glu359, with 'Glu359 facing the solvent at start and approaching Lys211 after a few hundreds of ns, while 'Gln358 is exposed to the solvent toward the end of the simulation. Note that the switch in side chain orientation occurred in all three subunits, while the formation of the stable salt bridge occurred in one subunit interface only. The arrows indicate the distance between the center of mass of the side chain heavy atoms (Glu: OE1 and OE2, Gln: OE1, NE2, Ser: OG). (E) The hASIC1a, but not the cASIC1a structure, favors the approaching between Gln84 and Tyr417 during the open-to-desensitized transition. The homologous residues in cASIC1a are Arg85 and Tyr416. Left, structural image of hASIC1a. The frame indicates the magnified area shown in the center panel. Center, molecular representation from an MD trajectory of hASIC1a showing that Tyr417 forms a cation-π interaction with Arg279. Right, in the corresponding area of the cASIC structure (PDB 4NTW), Tyr416 also forms a cation-π interaction with Arg280 (homologous to hASIC1a-Arg279). However, in this case, Arg85 is not likely to approach this positively charged area. Note that the side chain of Arg85, except Cβ, is absent from this open as well as from the desensitized structures (PDB code 4NYK). Electrostatic interactions are symbolized with black dashed lines. Source data are provided in the file Figure 7—source data 1.
-
Figure 7—source data 1
Molecular Dynamics simulations.
- https://cdn.elifesciences.org/articles/66488/elife-66488-fig7-data1-v2.xlsx
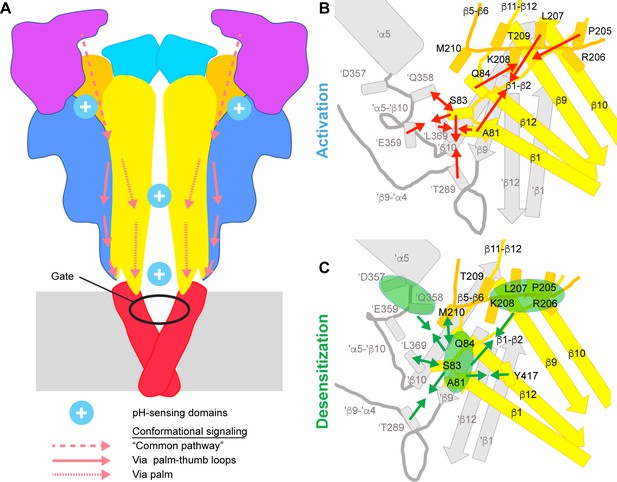
Conformational changes leading to activation and desensitization.
(A) Activation signaling pathways in ASIC1a. Cartoon of the ASIC structure, in which the predicted pH-sensing regions are highlighted (only two of the three acidic pockets are indicated) and hypothesized signaling pathways for channel activation from distal regions to the gate are indicated; dashed arrows, common pathway, dotted arrows; palm pathway; solid arrows, palm-thumb loop pathway, with the β-turn interacting with the upper end of the TM1. (B) and (C), Cartoons indicating the conformational changes in the palm and palm-thumb loops predicted by VCF to occur with ASIC activation (B) and desensitization (C). The lower palm domain of one subunit is shown in yellow, structural elements of a neighboring subunit are shown in gray. Arrows that point toward each other indicate an approaching between residues, while arrows pointing away from each other indicate an increase in distance. In C, the arrows indicate in some cases not the distance changes between individual residue pairs, but between groups of residues. The three groups indicated by green ellipses represent residues of the β1-β2 linker (Ala81, Ser83, Gln84), residues of the distal β5-β6 loop (Pro205, Arg206, Leu207, Lys208) and residues in proximity of the thumb α5 helix ('Asp357, 'Gln358). The individual distance changes during the closed-open and open-desensitized transitions are illustrated in the Videos 1 and 3, respectively.
Videos
Animation illustrating the VCF-predicted distance changes in the closed-open transition.
Animation based on a structural model of hASIC1a in the closed state (PDB 5WKU). Red solid lines indicate an approaching between the two residues of a given pair, while dashed lines indicate a distance increase between them. Note that the animation does not show the conformational changes; it just indicates where the distance changes are predicted to occur. The timing in the animation is not indicative of the timing of the distance changes in the represented pairs.
Animation showing a Molecular Dynamics simulation that highlights the flip in rotameric positions of 'Gln358 and 'Glu359.
The video shows the Molecular Dynamics trajectory, started from a closed state model of ASIC1a, analyzed in Figure 7A–D. The side chain and backbone atoms of residues Ser83, Lys211, 'Gln358 and 'Glu359 are shown, with the carbon, oxygen, and nitrogen atoms colored black, red and blue. Whenever a heavy atom of 'Gln358 or 'Glu359 is within a distance of ≤3 Å from a heavy atom of Ser83 or Lys211 during the simulation, the heavy atoms of the interacting residues are shown as spheres. The total MD simulation time covered by this video is 740 ns. In the presented subunit interface, the orientation exchange of 'Gln358 and 'Glu359 was followed by the subsequent formation of a salt bridge between Lys211 and ‘Glu359.
Animation illustrating the VCF-predicted distance changes in the open-desensitized transition.
Animation based on a structural model of hASIC1a in the open state (PDB 4NTW). Green solid lines indicate an approaching between the two residues of a given pair, while dashed lines indicate a distance increase between them. Note that the animation does not show the conformational changes; it just indicates where the distance changes are predicted to occur. The timing in the animation is not indicative of the timing of the distance changes in the represented pairs.
Tables
Summary of VCF analyses.
The polarity of the first or second ΔF signal components is indicated as negative (neg) or positive (pos), and the association of a ΔF signal component with either opening (o) or desensitization (d); ‘i’ stands for intermediate, that is between opening and desensitization. ‘neg’ and ‘pos’ are in regular font if no correlation with distance changes indicated by the structures is possible because the distance changes are too small (<1 Å), bold if they correlate with the structural prediction (‘o’ associated with closed-open distance change, ‘i’ and ‘d’ associated with open-desensitized distance change), and in italics if they contradict the structural predictions. *, these residue pairs were not used for structural interpretation; the pair A81-P205 because of the high distance (>20 Å, Table 2), and A81-T209 because the current of this mutant did not desensitize.
| Residue1 | Residue2 | VCF signal | ||
|---|---|---|---|---|
| Comp. | Signal | Association | ||
| Closed-to-open transition | ||||
| Q84 | P205 | 1st | neg | o |
| A81 | L207 | 1st | neg | o |
| S83 | T289 | neg | o | |
| S83 | Q358 | 1st | pos | o |
| S83 | E359 | neg | o | |
| A81 | L369 | 1st | neg | < o |
| Open-to-desensitized transition | ||||
| A81 | Y417 | neg | i | |
| S83 | Y417 | neg | i | |
| Q84 | Y417 | neg | i | |
| *A81 | P205 | pos | i | |
| S83 | P205 | neg | i | |
| Q84 | R206 | neg | d | |
| Q84 | L207 | neg | i | |
| A81 | K208 | neg | i | |
| *A81 | T209 | neg | i | |
| A81 | M210 | pos | i | |
| A81 | T289 | neg | i | |
| Q84 | T289 | neg | i | |
| S83 | D357 | 1st | neg | d |
| S83 | Q358 | 2nd | neg | d |
| A81 | L369 | 2nd | pos | d |
| S83 | L369 | 2nd | pos | i |
Distances between Cys / quencher pairs.
The distances were calculated between the β-carbon residues of the indicated residues from structural models of the closed (Yoder et al., 2018 PDB code 5WKU), Mit-toxin-opened (Baconguis et al., 2014 4NTW) and desensitized state (Gonzales et al., 2009 4NYK) of hASIC1a in Chimera (Pettersen et al., 2004). The distances were measured in all three subunits, and the average was calculated. ‘Average distance’ is the average of the distance measured in the structural models corresponding to the three functional states. The differences of these distances between two functional states are presented in a way that negative values indicate a shorter distance in open relative to the closed, and desensitized relative to the open conformation.
| Residue1 | Residue2 | Distance (from structure) | |||
|---|---|---|---|---|---|
| Average | Difference | ||||
| open- | desensitized | ||||
| closed (Å) | -open (Å) | ||||
| A81 | Y417 | 8.4 | 0.3 | −1.5 | |
| S83 | Y417 | 10.9 | 0.0 | −2.7 | |
| Q84 | Y417 | 10.5 | 0.0 | −0.4 | |
| A81 | P205 | 23.5 | 0.1 | −0.5 | |
| S83 | P205 | 17.4 | −0.4 | −0.4 | |
| Q84 | P205 | 13.0 | −0.1 | −0.6 | |
| Q84 | R206 | 11.0 | −0.2 | 0.0 | |
| A81 | L207 | 19.6 | 0.4 | −2.1 | |
| Q84 | L207 | 10.6 | −0.2 | 0.7 | |
| A81 | K208 | 15.0 | 0.3 | −1.3 | |
| A81 | T209 | 13.1 | −0.2 | −3.1 | |
| A81 | M210 | 11.7 | −0.4 | −3.0 | |
| A81 | T289 | 8.5 | 0.2 | 2.2 | |
| S83 | T289 | 12.4 | −2.0 | −0.9 | |
| Q84 | T289 | 16.2 | −0.8 | −1.7 | |
| S83 | D357 | 11.5 | 3.6 | −1.7 | |
| S83 | Q358 | 7.1 | 4.5 | −0.5 | |
| S83 | E359 | 8.8 | 3.0 | −1.0 | |
| A81 | L369 | 10.9 | −2.4 | −2.1 | |
| S83 | L369 | 18.2 | −3.7 | −0.9 | |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Recombinant DNA reagent (DNA plasmid) | psD5-hASIC1a | PMID:16282326 | For in vitro transcription | |
| Recombinant DNA reagent (DNA plasmid) | psD5-hASIC1a-A81C | This paper | hASIC1a containing mutation A81C for in vitro transcription; all mutants used in this study were prepared with the same approach. | |
| Commercial assay or kit | KAPA HiFi HotStart PCR polymerase | Roche Diagnostics | KK2501 | PCR |
| Commercial assay or kit | mMESSAGE mMACHINE SP6 | ThermoFisher | AM1340 | in vitro transcription |
| Sequence-based reagent | Primers for mutagenesis | Microsynth | See Supplementary file 4 | |
| Chemical compound, drug | 3-Maleimidopropionic acid | Bachem | 4038126.0001 | Blocking of free cysteines on the oocyte |
| Chemical compound, drug | CF488 maleimide | Biotium | 92022 | Fluorophore used in VCF |
| Chemical compound, drug | AlexaFluor 488 Maleimide | ThermoFisher | A10254 | Fluorophore used in VCF |
| Software, algorithm | HEKA Chartmaster and Fitmaster | Harvard Bioscience | RRID:SCR_016233 | Electrophysiology software, https://www.heka.com |
| Software, algorithm | Graphpad Prism | Graphpad Software | RRID:SCR_002798 | Data and statistical analysis, preparation of graphs |
| Software, algorithm | Origin PRO software | OriginLab Corp, Northampton, USA | RRID:SCR_002815 | Data analysis |
| Software, algorithm | UCSF Chimera | Resource for Visualization and Informatics | RRID:SCR_004097 | Structure analysis, distances measurements |
| Software, algorithm | GROMACS | Resource for MD simulations http://www.gromacs.org | RRID:SCR_014565 2018.6 | Simulations |
| Software, algorithm | VMD | Resource for MD Visualization and calculations | RRID:SCR_001820 1.9.3 | Structure and trajectory analysis |
| Software, algorithm | R RStudio | https://www.r-project.org/ | RRID:SCR_001905 1.3. | Statistics and data analysis |
| Software, algorithm | CHARMM GUI | http://www.charmm-gui.org/ | RRID:SCR_014892 | Construction of the systems of simulations |
| Software, algorithm | SWISS-MODEL | https://swissmodel.expasy.org/ | RRID:SCR_018123 | Homology modeling |
Additional files
-
Supplementary file 1
Decay kinetics of the ΔF signal when switching back from pH6.0 to pH7.4.
The decay kinetics were determined as decay time, the time to pass from 90% to 10% of the maximal amplitude (RTFoff). Note that for mutants whose signal was not sustained, the RTFoff could not be determined.
- https://cdn.elifesciences.org/articles/66488/elife-66488-supp1-v2.docx
-
Supplementary file 2
Parameters of kinetic models for the simulation of ΔF traces.
The modeling of the ΔF traces is described in Materials and methods. Note that F(C) was always = 0, except for S83C Y417V D357W where it was set to −0.15. a, F(OD) calculated from Fsust/Fpeak ratio; b, parameters chosen to reproduce the shape of the trace; c, F(O) calculated from the fluorescence rise time (RTF); d, F(O) and F(OD) determined from the amplitude ratio of the two components of the ΔF signal; *, experimental traces not matched by model, off kinetics are too slow in the model; & , F(C)=−0.15. F(CD) was in all cases except for b calculated from the RTF when the pH was switched back to pH7.4.
- https://cdn.elifesciences.org/articles/66488/elife-66488-supp2-v2.docx
-
Supplementary file 3
Test for intrinsic pH dependence of fluorophores.
The ratio of the ΔF/F of the acidification from the desensitized state (i.e., conditioning pH6.7, stimulation pH6)/ ΔF/F of the acidification from the closed state (pH7.4 / pH6) is shown as a percentage (n = 3–10, mean ± SEM). This ratio is not indicated here for the mutants of Figure 1 that had previously been measured with a different perfusion system. For those mutants, the ratio was <20% (Bonifacio et al., 2014; Gwiazda et al., 2015). For most mutants, the ratio was measured using the two fluorophores Alexa Fluor 488 and CF488. The fluorophore yielding the lower ratio was subsequently used for the experiments and its ratio (as a percentage) is indicated in this table. Negative values indicate that the two protocols produced signals of opposite polarity. Mutants labeled by CF488 are marked with C. Alexa Fluor 488 was used for all the remaining mutants.
- https://cdn.elifesciences.org/articles/66488/elife-66488-supp3-v2.docx
-
Supplementary file 4
Sequences of oligonucleotides for mutagenesis.
This table indicates the sequences of the oligonucleotides used for mutagenesis.
- https://cdn.elifesciences.org/articles/66488/elife-66488-supp4-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/66488/elife-66488-transrepform-v2.docx





