Linking plasmid-based beta-lactamases to their bacterial hosts using single-cell fusion PCR
Figures
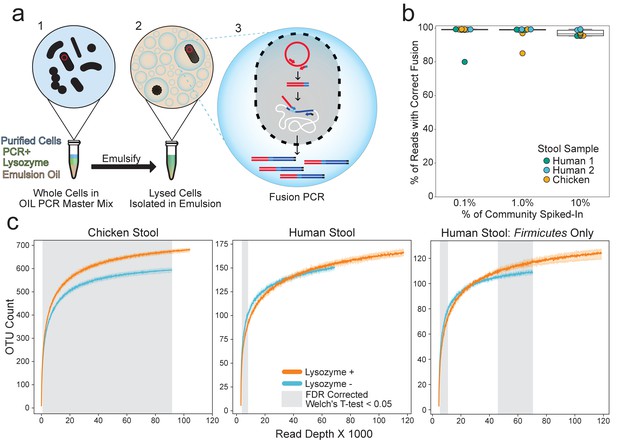
OIL-PCR can specifically link plasmid-encoded genes with their hosts.
(a) Depiction of the OIL-PCR method. (1) Nycodenz-purified cells are mixed with PCR master mix, lysozyme, and emulsion oil and shaken to create an emulsion. (2) Cells are lysed within the emulsion. (3) Fusion PCR is performed in droplets containing cells harboring the targeted gene. Fused amplicons between the gene of interest and the 16S rRNA gene are the product. (b) A boxplot showing the percent of Illumina reads containing correct fusion products, namely the fusion of plasmid-borne cmR and the 16S rRNA gene of E. coli MG1655. OIL-PCR was performed on two individuals’ and one chicken’s gut microbiome sample in triplicate, spiked with varying concentrations of E. coli. (c) Rarefaction analysis of chicken (left) or human gut microbiome sample (middle) with (orange) and without (blue) lysozyme treatment. At right is the rarefaction analysis performed on Firmicutes only in the human stool sample. Grayed regions in the plot represent areas where the curves, each composed of four technical replicates, are significantly different (p<0.05) from one another, according to an FDR-corrected Welch’s t-test.
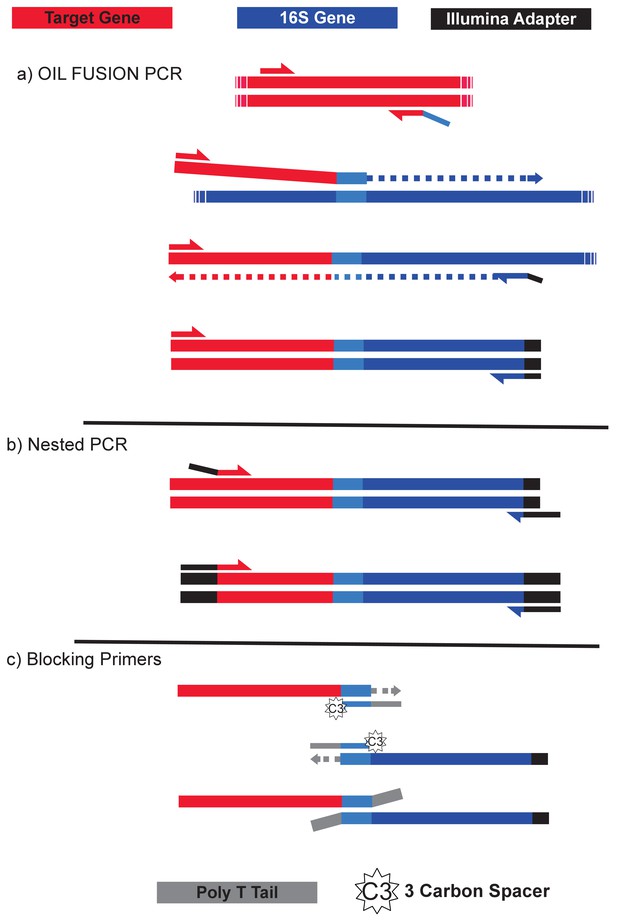
Depiction of the fusion PCR.
(a) PCR is initialized with primers to a target gene (red). The reverse primer contains a 5’ overhang complimentary to the universal V4 16S primer 519F (light blue). The product of this first amplification step can act as a forward primer for the 16S rRNA gene (blue). After extension of the full fusion product, the forward target primer can pair with the universal 16S reverse primer 786R (with a portion of an Illumina TruSeq adapter sequence) to amplify the fully fused PCR product. (b) Nested PCR is performed on the fused PCR products from (a) in order to filter out non-specific priming from the fusion PCR. The forward primer anneals within the target gene and has a TruSeq adapter at the 5’ end. The reverse primer also has the Illumina adapter sequence at it is 5’; end and anneals to the non-degenerate portion of 786R and the partial Illumina adapter sequence appended in (a). (c) Two blocking primers, both complementary to the 519F priming region, are included in the nested PCR to prevent unfused PCR products from annealing during the nested reaction. Blocking primers have a three-carbon spacer on the 3’ end to prevent extension and a poly-T tail that appends 10 As to the 3’ end of any unfused products, thus inactivating them from annealing or extension.
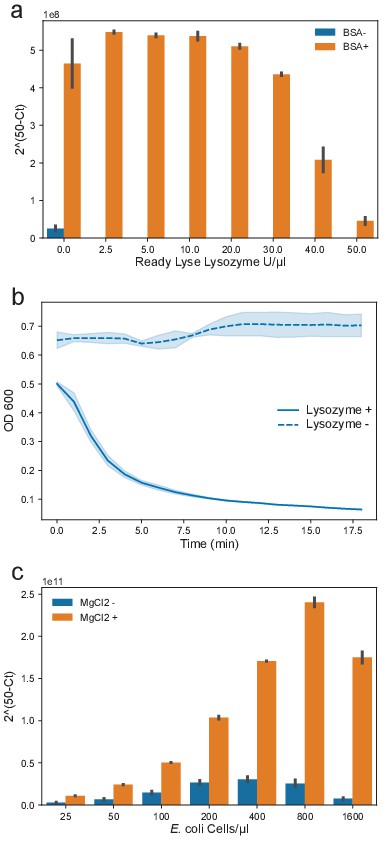
BSA and excess MgCl2 improve the efficiency of OIL-PCR and Ready Lyse Lysozyme remains active in OIL-PCR master mix.
(a) Sybr-based qPCR was performed on the cmR gene carried on pBAD33 with varying concentrations of lysozyme in the presence (orange) or absence (blue) of BSA. Higher 2(50-Ct) values represent greater amplification. (b) Lysozyme activity against B. subtilis suspended in the OIL-PCR optimized reaction mix with (solid line) and without (dashed line) lysozyme. (c) Sybr-based qPCR was performed on the cmR gene carried on the pBAD33 plasmid in E. coli MG1655 cells at increasing cell concentrations with (orange) and without (blue) additional MgCl2. Higher 2(50-Ct) values represent greater amplification.
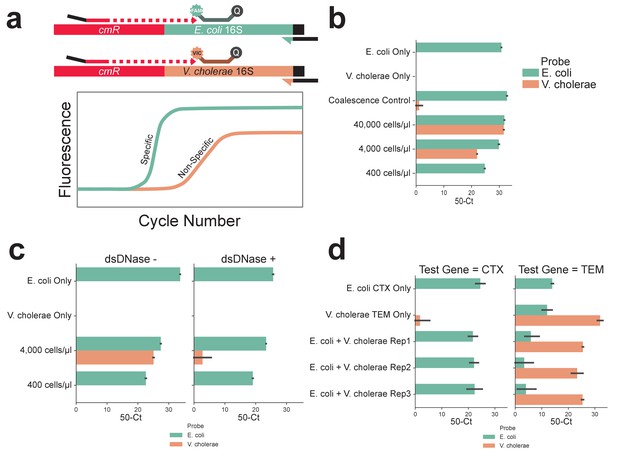
Cell concentration of 400 cells/μl, DNase treatment, and multiplexing PCR result in accurate OIL-PCR results.
(a) Diagram of the Taqman assay used to monitor OIL-PCR results. Briefly, Taqman probes were designed to be complementary for the 16S rRNA genes in either E. coli or V. cholerae, each with its own fluorophore. OIL-PCR was performed on E. coli carrying the cmR gene on the pBAD33 plasmid but not present in V. cholerae. Fusion PCR products were recovered and nested probe-based qPCR was performed. Upon amplification of the gene, the probe is cleaved by Taq polymerase releasing the fluorophore from the quencher. Specific amplification of the designated region is measured by fluorescence of the expected fusion product vs the non-specific product. (b) OIL-PCR with primers targeting a plasmid-borne cmR gene was performed with a 1:1 mix of cmR positive E. coli and cmR negative V. cholerae cell suspensions with. A gradient of cell concentrations was tested (400–40,000 cells/μl), in addition to E. coli and V. cholerae suspensions alone as positive and negative controls. Control emulsions were mixed 1:1 after emulsification to test for droplet coalescence. (c) OIL-PCR with primers targeting a plasmid-borne cmR was performed after pretreating cells with (right) and without (left) dsDNase at two different 1:1 E. coli to V. cholerae cell suspension concentrations as well as on the individual bacterial strains for controls. (d) Multiplexed OIL-PCR was performed with primer sets targeting a genomic blaCTX-M gene in E. coli and a plasmid-borne blaTEM gene in V. cholerae. Experiments were performed in triplicate and on each of the organisms separately. Results are shown for the blaCTX-M (left) and blaTEM (right).
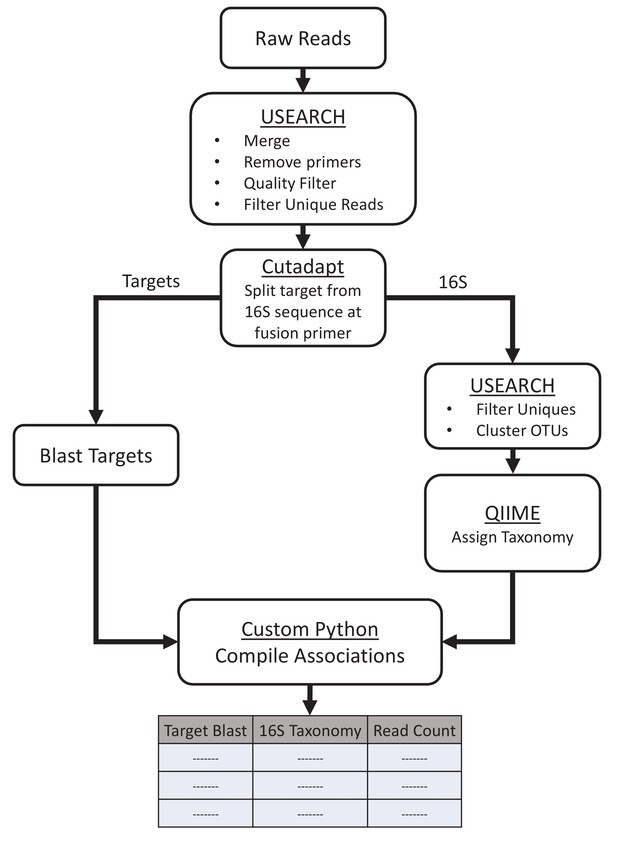
Computational workflow.
Paired-end reads were merged, quality filtered, and filtered for unique reads before splitting them at the fusion primer junction with Cutadapt. The target portion of each read was blasted to confirm the sequence while the 16S portion was clustered into OTUs and assigned taxonomy with the QIIME RDP classifier and Silva database. The target and 16S assignments were then compiled together in python for analysis.
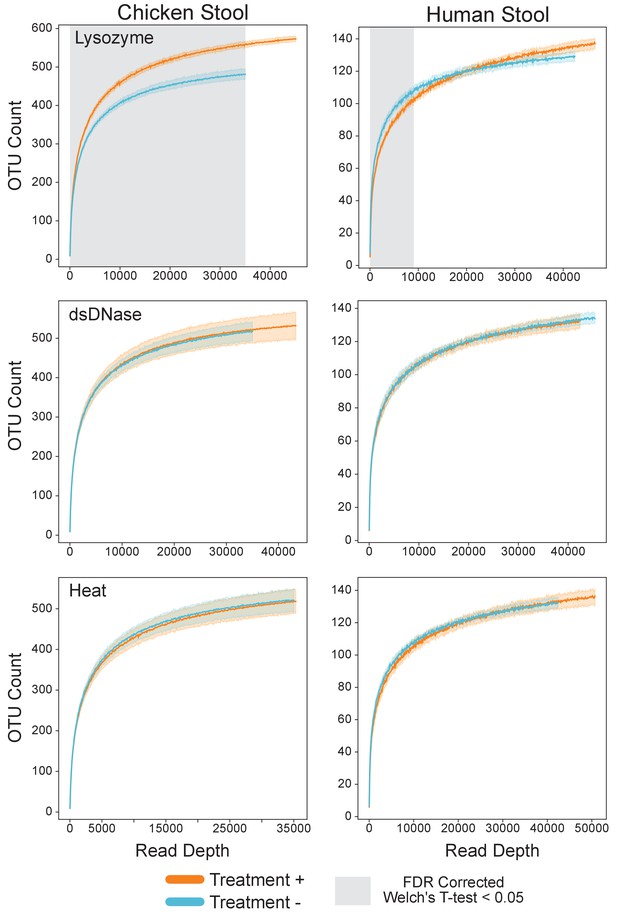
Lysozyme alone improved recovery of species.
Rarefaction analysis of chicken (left) or human (right) gut microbiome samples with (orange) and without (blue) lysozyme (top), dsDNA treatment (middle) and heat (bottom). Grayed regions in the plot represent areas where the curves are significantly different from one another (p<0.05), according to an FDR-corrected Welch’s t-test.
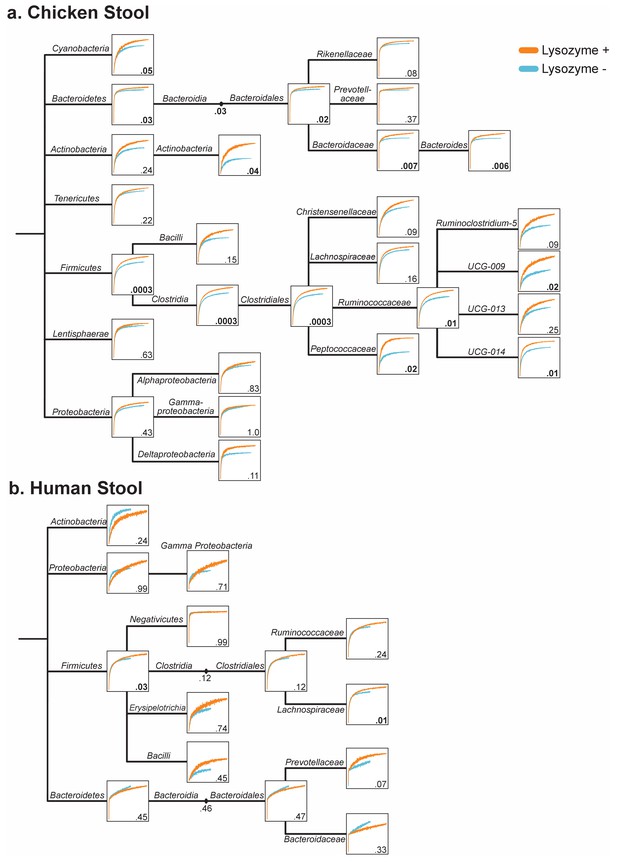
Lysozyme treatment improves OTU capture of most taxonomic groups in chicken and human stool.
Rarefaction trees for (a) chicken, and (b) human stool generated for samples with and without lysozyme treatment. Rarefaction curves were generated at each taxonomic level containing 10 or more OTUs from Phylum to Genus and organized on the phylogenetic tree. Numbers within each box indicate the FDR-corrected Welch’s t-test score at the greatest sampling depth for each curve. Three redundant curves were left out for clarity.
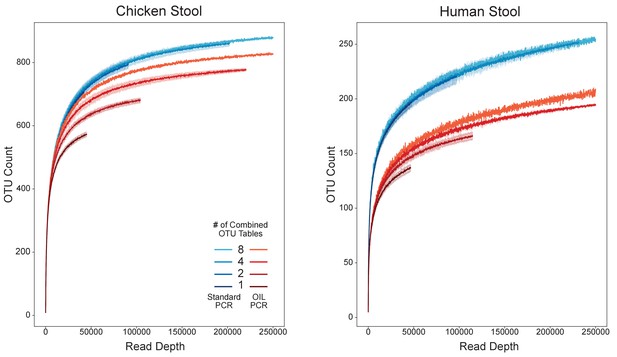
Combining replicates for increased depth improved recovery of species and reduced stochastic sampling bias.
Rarefaction analysis of chicken (left) or human (right) gut microbiome samples performed with OIL-PCR (red) or standard DNA extraction and library prep (blue). OTU tables were repeatedly subsampled in groups of two, four, or eight replicates from eight total replicate 16S libraries.
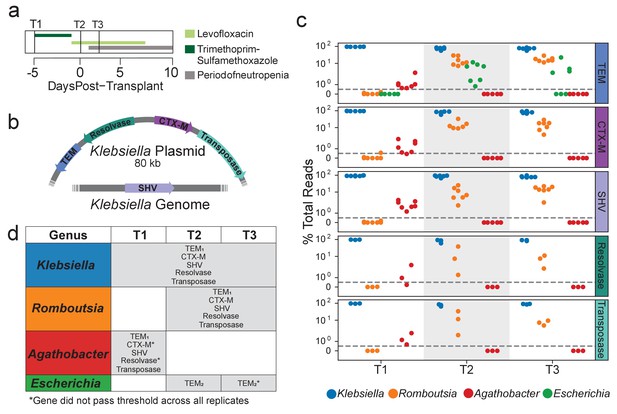
Extended spectrum beta-lactamase genes are associated with both pathogenic and commensal species.
(a) Summary of treatment and sample time points for patient B335. (b) Depiction of an 80 kb plasmid carried by K. pneumoniae harboring the blaCTX-M, blaTEM, Tn3 transposase and resolvase genes. The blaSHV gene is presumed to be carried within the K. pneumoniae genome. Placement of these genes was inferred from metagenomic assemblies of patient B335’s gut microbiome sample. (c) OIL-PCR results for each of the genes depicted in (a) patient B335 at three time points. For all gene-taxa associations, the percent of total OIL-PCR reads for that gene-time point is plotted. All species passing our detection threshold of 0.5% (dotted line) at any of the three time points is included in this plot. (d) A table summarizing the results in (b). All gene-taxa associations for each time point passing our detection thresholds are listed. Two SNP variants of TEM were detected and denoted with subscript numbering. Gene-taxa associations which did not consistently pass our detection threshold across all technical replicates are noted (*).
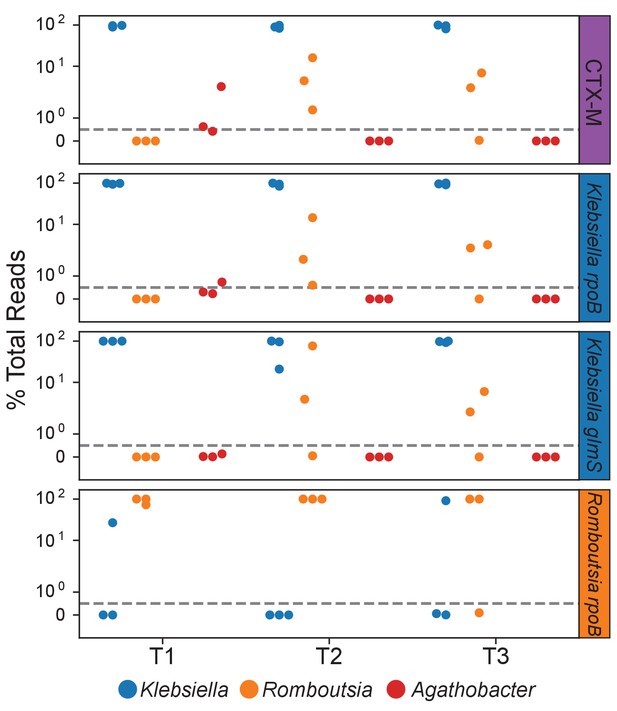
Species targeted fusion primers reveal the physical association of Klebsiella with Romboutsia.
OIL-PCR was performed with fusion primers targeting species-specific marker genes in Klebsiella pneumoniae and Romboutsia timonensis. Klebsiella sequences were found fused to Romboutsia 16S in time points T2 and T3, mirroring results for blaCTX-M.
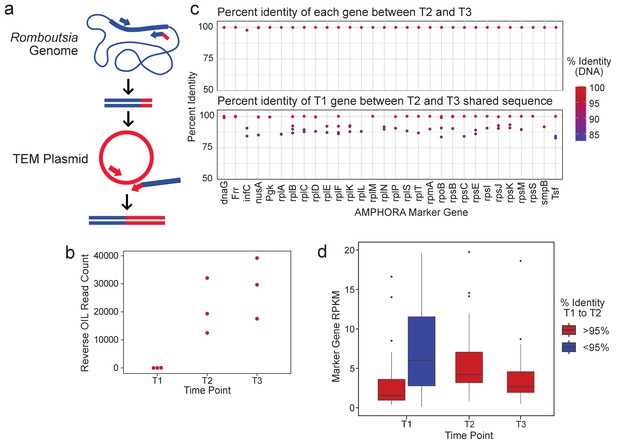
R.timonensis strains associated with the three beta-lactamase genes appear over the patient’s time course.
(a) Depiction of the reverse OIL-PCR in which Romboutsia-specific 16S rRNA sequences (blue) are fused with the blaTEM sequences (red). (b) OIL-PCR read counts of the reaction shown in (a) are plotted. (c) The percent sequence identity of assembled R. timonensis marker genes between genes identified in timepoints 2 and 3 (top) and between timepoint 1 and between sequences shared at timepoints 2 and 3 (bottom). (d) RPKM-normalized abundance-values for the assembled marker genes for each strain assembled in time point one and the major strain present in timepoints 2 and 3.
Additional files
-
Supplementary file 1
Strains and plasmids used in this study.
- https://cdn.elifesciences.org/articles/66834/elife-66834-supp1-v2.xlsx
-
Supplementary file 2
Primers used in this study.
- https://cdn.elifesciences.org/articles/66834/elife-66834-supp2-v2.xlsx
-
Supplementary file 3
OIL-PCR primer set table.
- https://cdn.elifesciences.org/articles/66834/elife-66834-supp3-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/66834/elife-66834-transrepform-v2.docx





