Altered temporal sequence of transcriptional regulators in the generation of human cerebellar granule cells
Figures
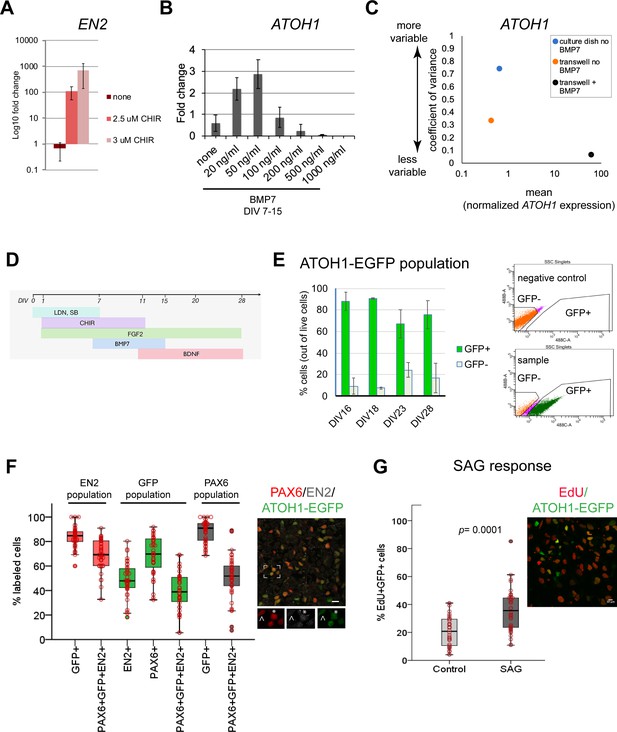
Derivation of the human ATOH1 lineage.
(A) EN2 expression (log10 fold change of no CHIR99021) in dual SMAD+FGF2-treated human pluripotent stem cells (hPSCs) in the absence and presence of CHIR99021 by RT-qPCR at day in vitro (DIV) 11. (B) ATOH1 expression (fold change of no BMP7) at DIV16 in response to a BMP7 concentration series added at DIV7–15. (C) Dot plot showing the coefficient of variance of mean ATOH1 expression detected by RT-qPCR at DIV16 in cultures grown on regular tissue culture dishes (-BMP7, blue) versus on transwell membranes (-BMP7, orange; +BMP7, black). (D) Schematic of the protocol for derivation of the ATOH1 lineage. (E) Left: the percentage of EGFP+ (green) and EGFP- (gray) cells at DIV16, 18, 23, and 28 of differentiation of the ATOH1-EGFP line by fluorescence-activated cell (FAC)-sorting (the change in EGFP+ population across DIVs compared by ANOVA: p=0.053). Right: representative FACS charts showing separation of ATOH1-EGFP+/EGFP- cells. (F) Left: box plot showing the percentages of EGFP, EN2, PAX6 single- and triple-positive cells by immunocytochemistry, within the EN2, EGPF (ATOH1), and PAX6 populations at DIV28–30. Right: representative merged image of the immunocytochemistry labeling. Boxed area is magnified at the bottom with individual channels displayed. Note asterisk highlighting a triple-positive cell, while cell above the arrowhead is EN2-;PAX6+;EGFP+. (G) Left: box plot showing the percentage of EdU+;EGFP+ double-positive cells per Dapi nuclei ± SAG treatment after 48 hr (DIV28–30). Right: a representative merged image of the labeling. N = 3 independent experiments except in (B), which shows technical replicates. Bar graphs show mean ± 1 SD. Scale bars: 10 μm.
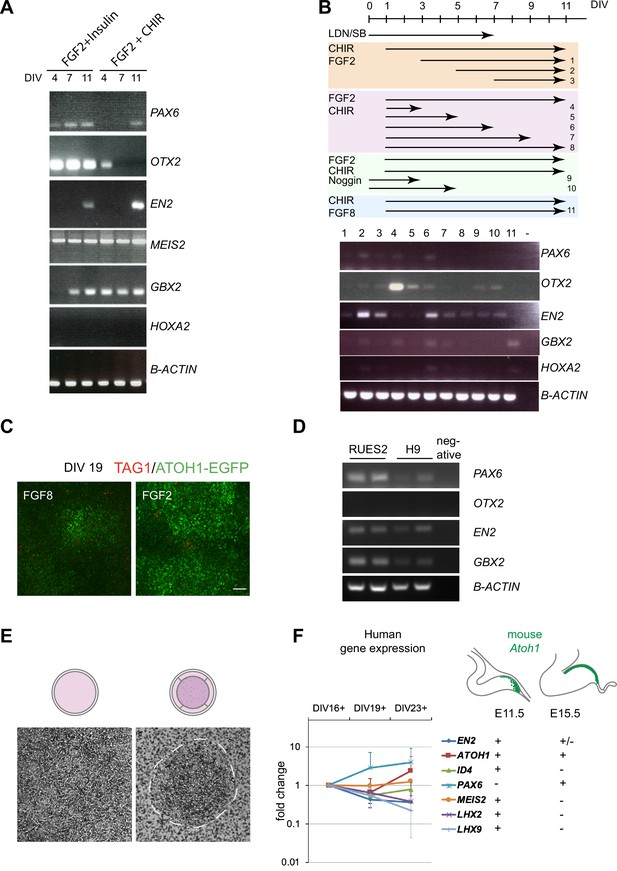
Derivation and characterization of the human ATOH1 lineage.
(A) Gene expression by RT-PCR at days in vitro (DIV) 4, 7, and 11 comparing dual SMAD inhibition plus FGF2/insulin versus FGF2/CHIR99021 treatment. (B) Top: schematic showing 11 different time intervals of FGF2, CHIR99021, and Noggin treatment in the presence of dual SMAD inhibition at DIV0–7 versus FGF8/CHIR99021. Bottom: RT-PCR at DIV11 showing the resulting expression of various markers in the 11 different conditions. (C) Representative examples of cultures at DIV19, treated with either FGF8 or FGF2 at DIV1–11, showing ATOH1-EGFP and TAG1 labeling as an indication of neuronal survival. (D) RT-PCR of granule cell progenitor markers at DIV16 in RUES2 and H9 human embryonic stem cell (hESC) lines upon dual SMAD inhibition (DIV0–7) plus FGF2/CHIR99021 (DIV1–11) treatment. (E) Phase-contrast images comparing differentiating cells on regular culture plates (left) versus transwell plates (right). Dashed lines demarcate the edge of a colony. (F) Right: schematic showing how gene expression in the mouse Atoh1 lineage changes from embryonic day (E) 11.5, prior to external granule cell layer (EGL) establishment, to E15.5, after EGL establishment. + indicates presence of expression, – indicates absence of expression. Left: RT-qPCR detection of gene expression of listed genes in ATOH1-EGFP fluorescence-activated cell (FAC)-sorted cells at DIV16, 19, and 23 of culture. All genes were detected at DIV16, and fold change is relative to levels at DIV16. N = 3 independent experiments. LDN, LDN193189; SB, SB431542. Scale bar: 50 μm.
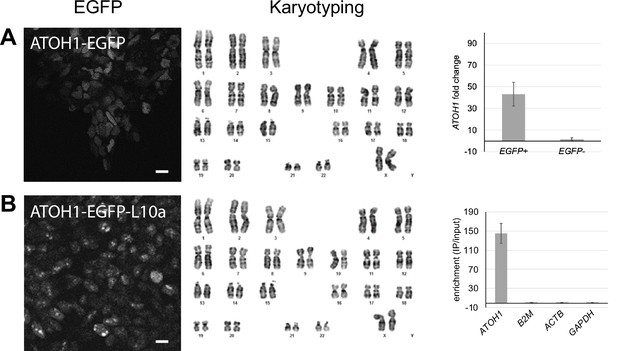
Characterization of ATOH1-EGFP and ATOH1-EGFP-L10a transgenic lines.
(A) Left: representative image of EGFP expression upon differentiation of the ATOH1-EGFP lines at day in vitro (DIV)16. Middle: normal karyotype detected. Right: bar chart showing the mean ± SD of normalized ATOH1 expression levels in fluorescence-activated cell (FAC)-sorted EGFP+ cells as fold change of expression in EGFP- cells at DIV28 by RT-qPCR. (B) Left: representative image of EGFP-L10a expression upon differentiation of the ATOH1-EGFP-L10a lines at DIV16. Note the marked difference in the localization of EGFP compared to (A). Middle: normal karyotype detected. Right: bar chart showing the mean ± SD of the level of enrichment of ATOH1 compared to three housekeeping genes in ATOH1-EGFP-L10a TRAP IPs versus input at DIV28 by RNA-seq.
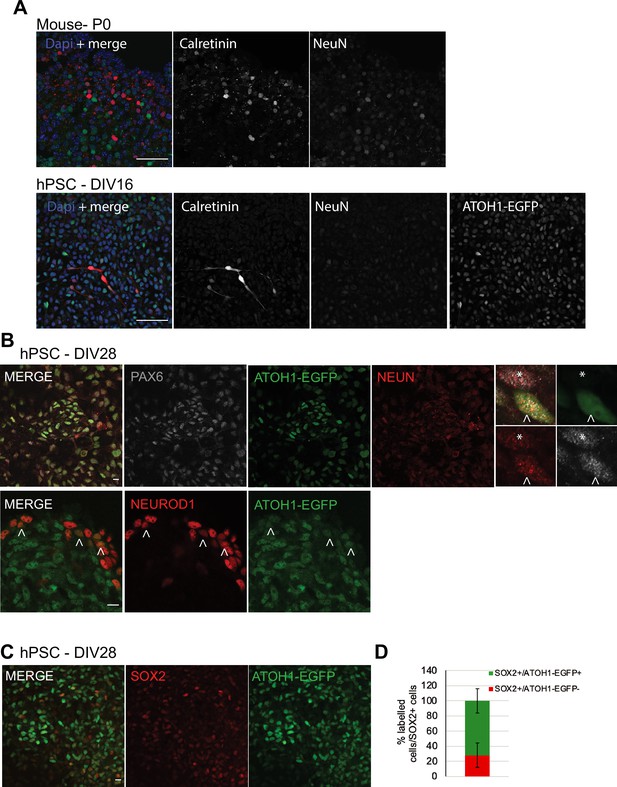
Characterization of ATOH1-EGFP cells (until day in vitro [DIV] 28).
(A) Top panel: Calretinin and NeuN expression in the cerebellar nuclei at P0 in mice. Lower panel: Calretinin, NeuN, and ATOH1-EGFP expression in human pluripotent stem cell (hPSC) cultures at DIV16. (B) Top panel: ATOH1-EGFP, NeuN, and PAX6 expression at DIV28. Far-right panel: high magnification showing cells that are PAX6+;NeuN+; ATOH1-EGFP- (asterisk) or triple positive (arrowhead). Note that NeuN expression increases with time in culture. Lower panel: NEUROD1 and ATOH1-EGFP expression at DIV28. Arrowheads highlight examples of double-positive cells. (C) SOX2 expression at DIV28. (D) Bar chart showing the proportion of ATOH1-EGFP+/- cells per SOX2+ cells. N = 2–3 independent cultures/time point, two mouse cerebella. Scale bars: 50 μm in (A), 10 μm in (B, C).
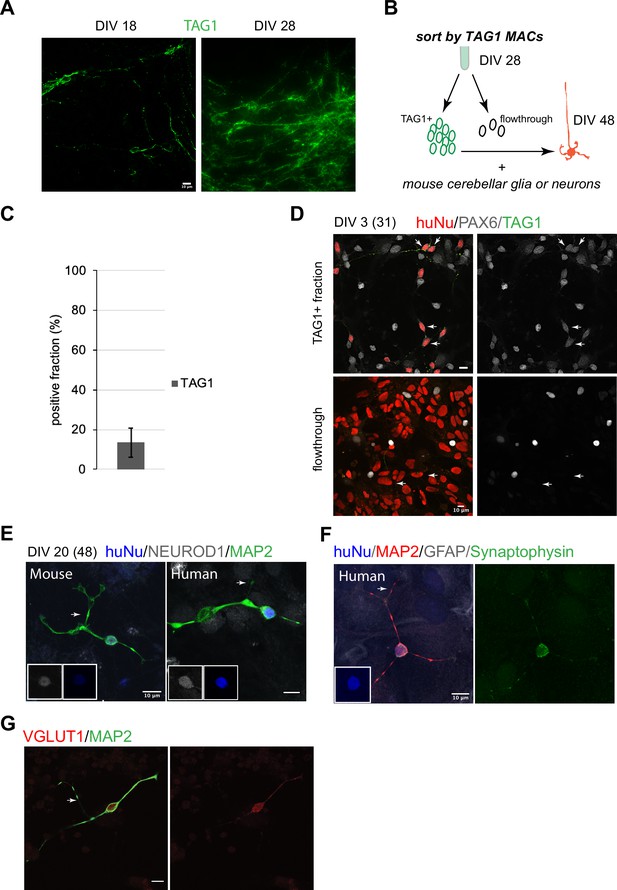
Human granule cell (GC) differentiation from human pluripotent stem cells (hPSCs).
(A) TAG1 expression at day in vitro [DIV]18 (left) and DIV28 (right). (B) Schematic of the sorting strategy of TAG1+ cells by magnetic-activated cell sorting (MACS) at DIV28 and co-culture with mouse cerebellar neurons or glia until DIV48. (C) Bar chart (mean ± 1 SD) of TAG1+ cells/total cells at DIV28, N = 7 independent experiments. (D) Top: TAG1+ cells (green) in co-culture with mouse cerebellar neurons and glia for 3 days express PAX6 (red + white, arrows). Bottom: TAG1- cells (flowthrough) in co-culture with mouse neurons and glia have larger nuclei and are PAX6- (arrows). (E) TAG1+ cells in co-culture with mouse neurons and glia for 20 days (DIV48 total) display small round nuclei (inset, blue), bifurcated neuronal extensions, and are NEUROD1+;MAP2+. A mouse GC cultured in the same dish for the same period of time, shown for comparison. (F) TAG1+ cell in co-culture with glia only for 20 days (DIV48 total) expresses synaptophysin. (G) TAG1+ cell in co-culture with mouse neurons and glia (DIV48 total) expresses VGLUT1.
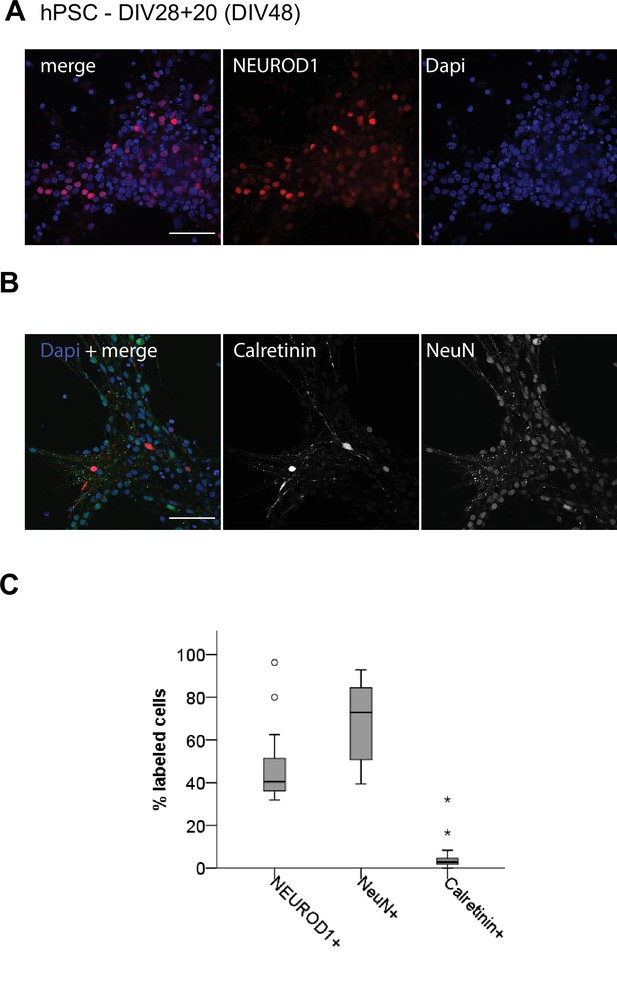
Characterization of TAG1-negative fraction at day in vitro (DIV)28 + 20.
(A) NEUROD1 expression and (B) NeuN and Calretinin expression in the TAG1-negative fraction after magnetic-activated cell sorting at DIV28 followed by differentiation of cells for another 20 days (DIV28 + 20). (C) Box plot showing the percentages of cells expressing each marker (per total cells). N = 3 independent experiments. Scale bars: 50 μm.
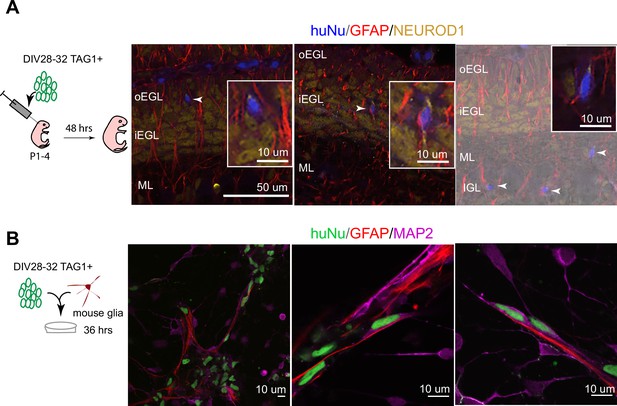
Human postmitotic granule cells (GCs) undergo glial-guided neuronal migration and integrate into the mouse cerebellum upon transplantation.
(A) Left: schematic outlining transplantation of MACsorted (day in vitro [DIV]28–32) TAG1+ human cells into the early postnatal mouse cerebellum. Images show three representative coronal sections of mouse cerebella (N = 5 mice), 48 hr post transplantation. Far-right image is overlaid on a DIC image. Arrowheads highlight human cells integrated in the mouse internal granule cell layer (IGL). Boxed images are higher magnifications of migrating cells. (B) Day 28–32 TAG1+ human cells in co-culture with mouse glia after 36 hr, showing examples of migrating neurons with elongated nuclear morphologies along glia. Far-left image shows a lower-magnification view containing migrating neurons on glia as well as non-migrating neurons with rounded morphologies. HuNu, human nuclear antigen; oEGL, outer external granule cell layer; iEGL, inner external granule cell layer; ML, molecular layer. Scale bars as indicated on images.
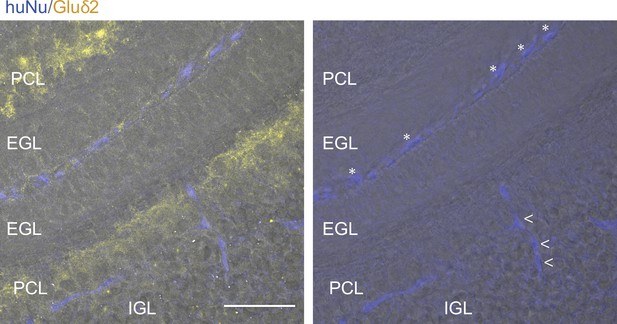
Integration of human pluripotent stem cell-granule cells (hPSC-GCs) into the mouse cerebellum upon transplantation.
Example of a coronal section of a mouse cerebellum showing human cells (blue), some of which integrated into the internal granule cell layer (IGL) (arrowheads) below the Purkinje cell layer marked by Gluδ2 expression. Note that the majority of the human cells are still on the pial surface of the mouse cerebellum (asterisks) at 48 hr post transplantation. Scale bar: 50 μm.
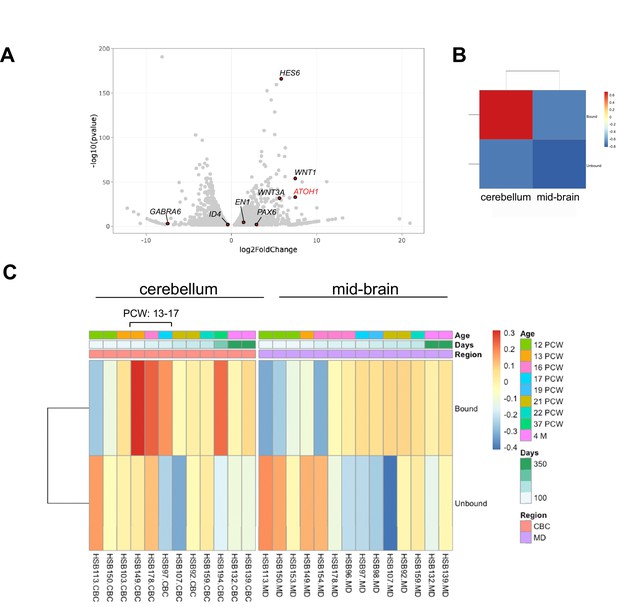
The human pluripotent stem cell (hPSC)-derived ATOH1 lineage resembles the human cerebellum in the second trimester by translational profiling.
(A) Volcano plot of log2 fold change global gene expression in ATOH1-TRAP IPs versus input. Key granule cell (GC) genes are highlighted by red dots (Figure 4—source data 1). The fully differentiated GC marker GABRA6 is depleted while progenitor genes are enriched. (B) Heatmap showing Gene Set Enrichment Analysis (GSEA) of log2 fold-enriched genes in day in vitro (DIV) 28 ATOH1-TRAP versus the PsychEncode dataset for the developing human cerebellum and midbrain from 12 post coitus week (PCW) until 4 months of age (combined). (C) Heatmap of data in (B) but divided by timeline with columns representing our data (bound [IP] and unbound [input]) compared to individuals from the PsychEncode project (identifiers depicted at the bottom, Figure 4—source data 2). CBC, cerebellum; MB, midbrain.
-
Figure 4—source data 1
DESeq2 analysis of ATOH1-EGFP-L10a TRAP IP versus input.
- https://cdn.elifesciences.org/articles/67074/elife-67074-fig4-data1-v2.xlsx
-
Figure 4—source data 2
Comparison of ATOH1-EGFP-L10a TRAP IP to human developmental data from PsychEncode.
- https://cdn.elifesciences.org/articles/67074/elife-67074-fig4-data2-v2.xlsx
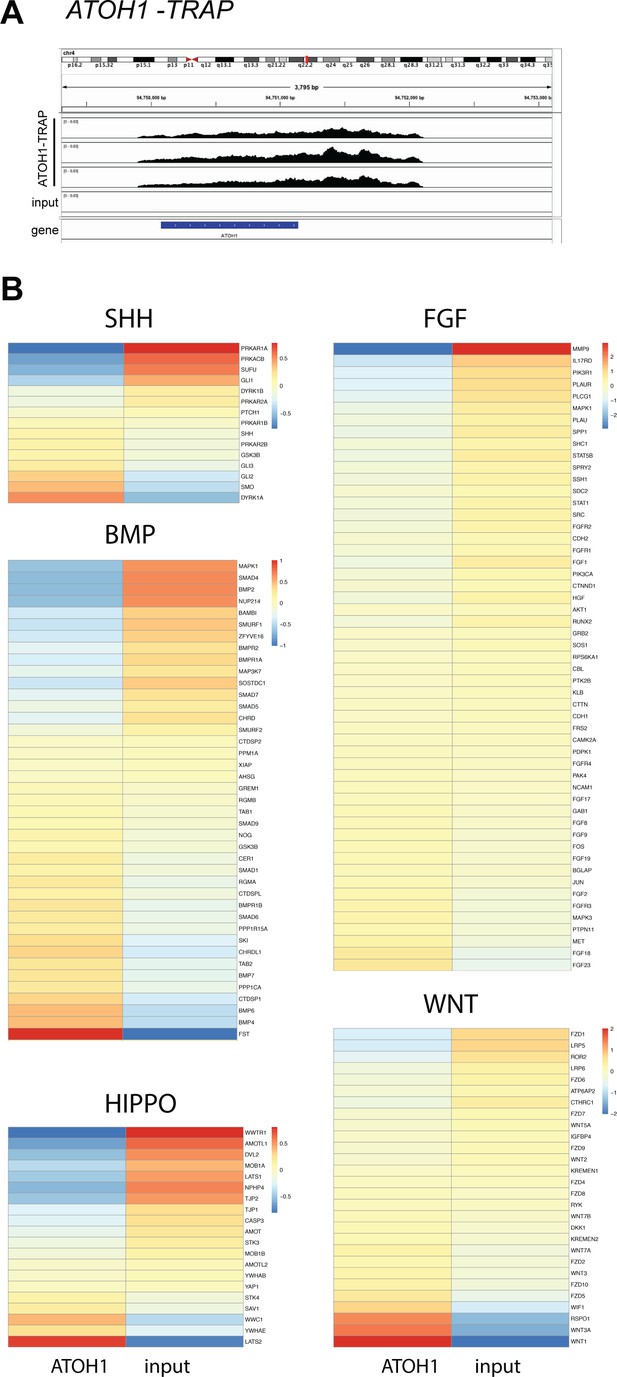
Heatmaps of key developmental signaling pathways in the hPSC-ATOH1 lineage.
(A) RNA sequencing of immunoprecipitated mRNAs from an ATOH1-EGFP-L10a hPSC TRAP line differentiated until day in vitro [DIV]28 shows enrichment of ATOH1 reads in the ATOH1-TRAP IPs compared to the input, depicted by Integrative Genomics Viewer. (B) Heatmaps showing the enrichment of genes in the ATOH1-EGFP-L10a TRAP versus input. Genes have been organized according to the developmental pathways they are associated with including the SHH, BMP, HIPPO, FGF, and WNT pathways.
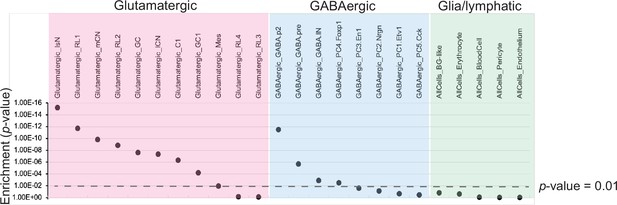
Comparison of ATOH1-EGFP-L10a TRAP against single-cell RNA-seq data from the developing mouse cerebellum.
Dot plot representation of the cumulative enrichment (by p-value) of the ATOH1-EGFP-L10a data against glutamatergic, GABAergic, and non-neuronal cell clusters identified by Wizeman et al., 2019.
-
Figure 4—figure supplement 2—source data 1
Comparison of ATOH1-EGFP-L10a TRAP IP to scRNA-seq from Wizeman et al., 2019, See Figure 4—figure supplement 2.
- https://cdn.elifesciences.org/articles/67074/elife-67074-fig4-figsupp2-data1-v2.xlsx
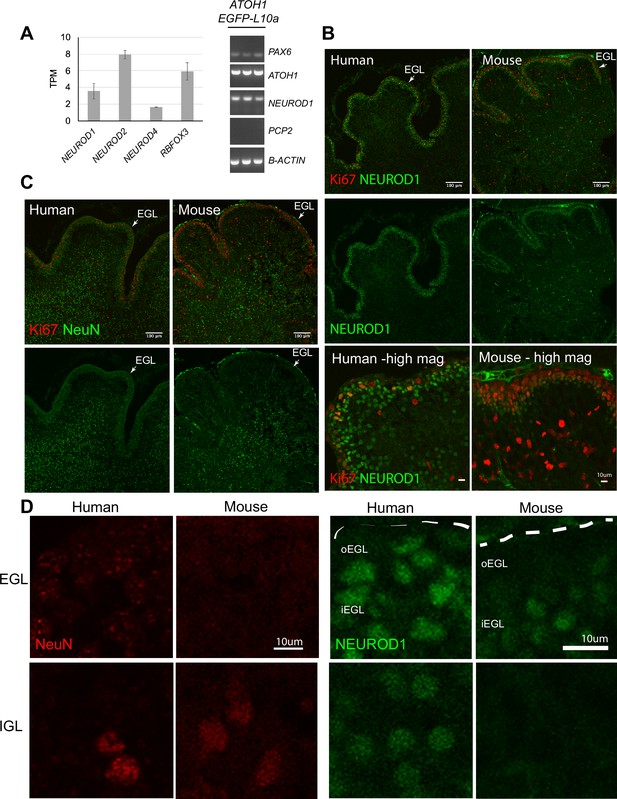
Temporal shift in the expression of transcriptional regulators in the human external granule cell layer (EGL) compared to mouse.
(A) Left: bar chart showing the mean ± 1 SD normalized expression of transcriptional regulators in ATOH1-TRAP IPs at day in vitro (DIV) 28 by RNA-seq. Right: the expression of PAX6 (GCP marker), and coexpression of ATOH1 and NEUROD1, but not PCP2 (Purkinje cells marker) in ATOH1-TRAP IPs by RT-PCR. (B) Sagittal sections though the vermis showing NEUROD1 and Ki67 expression by immunohistochemistry in the human cerebellum at 17 post coitus week (PCW) and mouse at P0. Note the similarities in foliation depth and pattern. Bottom: higher magnifications (scale bars: 10 μm) of a lobule in human and mouse. (C) Mid-sagittal sections showing NeuN (RBFOX3) and Ki67 in the human (17 PCW) and mouse (P0). (D) Higher magnification of NeuN and NEUROD1 labeling in the human versus mouse EGL and internal granule cell layer (IGL) (scale bars: 10 μm). Left panel: note the punctate NeuN labeling in the human but not in the mouse EGL. Right panel: NEUROD1. Dashed lines demarcate the pial surface. N = 2 cerebella/species. o, outer; , inner; TPM, transcripts per million reads. Scale bars: 100 μm unless stated otherwise.
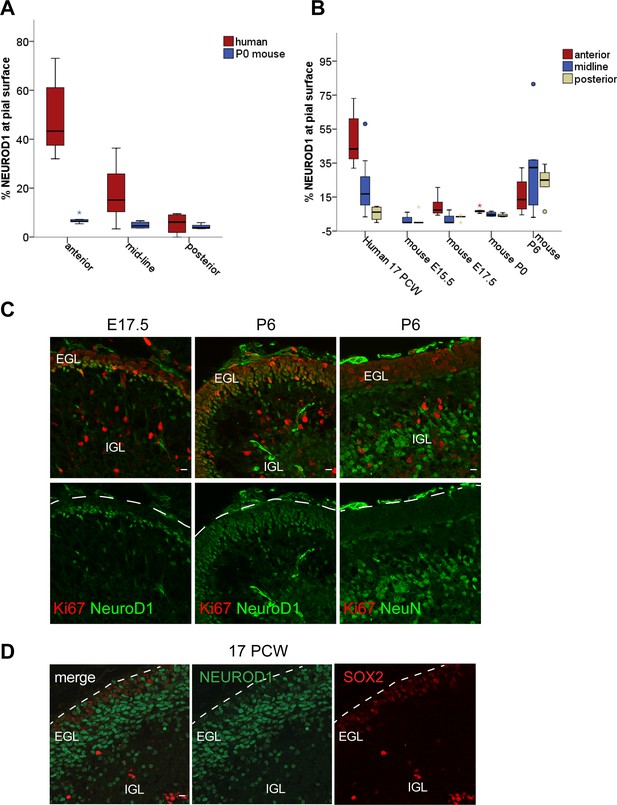
Marker expression in the developing human cerebellum versus mouse.
(A) Box plot showing the percentages of NEUROD1+ nuclei per total nuclei (Dapi) along the length of the pial surface in anterior, midline, and posterior regions of the developing cerebellum comparing mouse (P0) to human (17 post coitus week [PCW]). Note that the percentage of NEUROD1+ cells at the pia is low across the anterior-posterior axis of the mouse cerebellum, in contrast to the human where the anterior-posterior axis differs (Human n. cells at the pia counted = 613, Human NEUROD1+ fraction anterior: 48.4 ± 6.5 SE, midline: 21.1 ± 4.3 SE, posterior: 5.4 ± 2.2 SE. P0 Mouse n. cells at the pia counted = 369, P0 Mouse NEUROD1+ fraction anterior: 7.1 ± 0.8 SE, midline: 4.8 ± 0.7 SE, posterior: 4.3 ± 0.8 SE). (B) Box plot showing the percentages of NEUROD1+ nuclei per total nuclei (Dapi) along the length of the pial surface in anterior, midline, and posterior regions of the developing human cerebellum at 17 PCW compared to additional stages of development in mouse (E15.5, E17.5, P0, and P6). (C) NEUROD1, NeuN, and Ki67 expression on sagittal sections through the mouse cerebellum at embryonic day (E)17.5 and P6. (D) SOX2 expression in the human external granule layer (EGL) at 17 PCW. Scale bars: 10 μm. IGL, internal granule layer.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (human) | RUES2 | Human embryonic stem cell line | NIH registration number: NIHhESC-09-0013 | |
| Cell line (human) | H9 (WA09) | Human embryonic stem cell line | NIH registration number: NIHhESC-10-0062 | |
| Cell line (human) | ATOH1-EGFP | Human embryonic stem cell line | This study (RUES2 line) | |
| Cell line (human) | ATOH1-EGFP-L10a | Human embryonic stem cell line | This study (RUES2) | |
| Recombinant DNA reagent | pPS-EF1α-GFP-RFP | System Biosciences | LV603PA-1 | Lentiviral vector |
| Recombinant DNA reagent | J2XnGFP | Dr. Jane Johnson | GFP plasmid | |
| Recombinant DNA reagent | pSIN-hATOH1 enhancer- hβ-globin-nlsEGFP-bGH polyA-hPGK-Puromycin | This study | ATOH1-EGFP lentiviral construct | |
| Recombinant DNA reagent | pSIN-hATOH1 enhancer- hβ-globin-nlsEGFP-bGH polyA-hPGK-Puromycin | This study | ATOH1-EGFP- L10a lentiviral construct | |
| Biological sample (human) | Human fetal cerebellum (17 PCW) | Human Developmental Biology Resource | http://www.hdbr.org/ | |
| Biological sample (mouse) | C57Bl/6J mice | Jackson Laboratory | Embryos and pups obtained from times matings | |
| Antibody | Calretinin (rabbit polyclonal) | Swant | 7699/4 | (1:1000) |
| Antibody | EN2 (C19) (goat polyclonal) | Santa Cruz | SC-8111 | (1:50) |
| Antibody | GFAP (chicken polyclonal) | EnCor | CPCA-GFAP | (1:1500) |
| Antibody | GFP (rabbit polyclonal) | Invitrogen | A-111122 | (1:500) |
| Antibody | GFP (chicken polyclonal) | Aves labs | GFP-1020 | (1:1000) |
| Antibody | HuNu (anti-human nuclei) (mouse monoclonal) | Millipore | MAB1281 | (1:100) |
| Antibody | Ki67 (rabbit monoclonal) | Vector Laboratories | VP-RM04 | (1:100) |
| Antibody | Ki67 (rabbit polyclonal) | EnCor | RPCA-Ki67 | (1:1000) |
| Antibody | Map2 (chicken polyclonal) | Abcam | ab5392 | (1:1000) |
| Antibody | NeuN (mouse monoclonal) | Millipore | MAB377 | (1:100) |
| Antibody | NeuroD1 (mouse monoclonal) | BD Pharmingen | 563000 | (1:300) |
| Antibody | Pax6 (rabbit polyclonal) | BioLegend | 901301 | (1:300) |
| Antibody | TAG1 (mouse monoclonal) | Tom Jessell | (1:2) | |
| Antibody | Synaptophysin (mouse monoclonal) | Millipore | MAB329 | (1:500) |
| Antibody | VGLuT1 (mouse monoclonal) | Millipore | MAB5502 | (1:100) |
| Antibody | SOX2 (rabbit monoclonal) | Cell Signaling | 3579 | (1:200) |
| Antibody | GluR-d2 (goat polyclonal) | Santa Cruz | Sc-26118 | (1:100) |
| Antibody | Anti-goat Alexa Fluor 633 (donkey polyclonal) | Invitrogen | A21082 | (1:300) |
| Antibody | Anti-rabbit Alexa Fluor 555 (donkey polyclonal) | Invitrogen | A-31572 | (1:300) |
| Antibody | Anti-mouse IgM Alexa Fluor 488 (goat polyclonal) | Invitrogen | A-21042 | (1:300) |
| Antibody | Anti-mouse Alexa Fluor 555 (donkey polyclonal) | Invitrogen | A-31570 | (1:300) |
| Antibody | Anti-chicken IgY 488 (donkey polyclonal) | Jackson ImmunoResearch | 703-545-155 | (1:300) |
| Antibody | Anti-chicken IgY Cy3 (donkey polyclonal) | Jackson ImmunoResearch | 703-165-155 | (1:300) |
| Antibody | Anti-rabbit Alexa Fluor 647 (donkey polyclonal) | Invitrogen | A-31573 | (1:300) |
| Antibody | Anti-mouse Alexa Fluor 647 (donkey polyclonal) | Invitrogen | A-31571 | (1:300) |
| Antibody | Anti-mouse Alexa Fluor 488 (donkey polyclonal) | Invitrogen | A-21202 | (1:300) |
| Antibody | Anti-mouse Fluor 405 (donkey polyclonal) | Abcam | Ab175658 | (1:300) |
| Sequence-based reagent | ATOH1 | This paper | PCR primer | Forward 5′-GCGCA AAAGAATTT GTCTCC-3′ |
| Sequence-based reagent | ATOH1 | This paper | PCR primer | Reverse 5′-GCG AAGTTTTGCTG TTTTCC-3′ |
| Sequence-based reagent | ID4 | This paper | PCR primer | Forward 5′-GC TCACTGCGCT CAACACC-3′ |
| Sequence-based reagent | ID4 | This paper | PCR primer | Reverse 5′-GAA TGCTGTCGCC CTGCTTG-3′ |
| Sequence-based reagent | EN2 | This paper | PCR primer | Forward 5′- GG CGTGGGTCTA CTGTACG-3′ |
| Sequence-based reagent | EN2 | This paper | PCR primer | Reverse 5′- TACCTGTTG GTCTGGAA CTCG-3′ |
| Sequence-based reagent | PAX6 | This paper | PCR primer | Forward 5′-TCA CCATGGCAA ATAACCTG-3′ |
| Sequence-based reagent | PAX6 | This paper | PCR primer | Reverse 5′-CA GCATGCAGG AGTATGAGG-3′ |
| Sequence-based reagent | NEUROD1 | This paper | PCR primer | Forward 5′-GGACGA GGAGCAC GAGGCAG ACAAGAA-3′ |
| Sequence-based reagent | NEUROD1 | This paper | PCR primer | Reverse 5′- TTCCTCA GTGAGTCC TCCTCTG CGTTCA-3′ |
| Sequence-based reagent | PCP2 | This paper | PCR primer | Forward 5′- GACC AGGAGGG CTTCTTCAATCT-3′ |
| Sequence-based reagent | PCP2 | This paper | PCR primer | Reverse 5′- CATG TCCATGA GGCTGT CCATCT-3′ |
| Sequence-based reagent | OTX2 | This paper | PCR primer | Forward 5′-ACAA GTGGC CAATTCA CTCC-3′ |
| Sequence-based reagent | OTX2 | This paper | PCR primer | Reverse 5′-GAGG TGGACAA GGGATCTGA-3′ |
| Sequence-based reagent | MEIS2 | This paper | PCR primer | Forward 5′-CCAG GGGACT ACGTTTCTCA-3′ |
| Sequence-based reagent | MEIS2 | This paper | PCR primer | Reverse 5′-TAA CATTGT GGGGC TCTGTG-3′ |
| Sequence-based reagent | GBX2 | This paper | PCR primer | Forward 5′-GTTCC CGCCG TCGCTGATGAT-3′ |
| Sequence-based reagent | GBX2 | This paper | PCR primer | Reverse 5′-GCC GGTGTA GACGAA ATGGCCG-3′ |
| Sequence-based reagent | HOXA2 | This paper | PCR primer | Forward 5-CGT CGCTC GCTGA GTGCCTG-3′ |
| Sequence-based reagent | HOXA2 | This paper | PCR primer | Reverse 5′-TGTC GAGTGTG AAAGCG TCGAGG-3′ |
| Sequence-based reagent | LHX2 | This paper | PCR primer | Forward 5′- GGTCCTC CAGGTCT GGTTC-3′ |
| Sequence-based reagent | LHX2 | This paper | PCR primer | Reverse 5′- TAAGAG GTTGCGC CTGAACT-3′ |
| Sequence-based reagent | LHX9 | This paper | PCR primer | Forward 5′- GCT GGGAGT GGACATCGTCA-3′ |
| Sequence-based reagent | LHX9 | This paper | PCR primer | Reverse 5′- CATG GTCCGGA GCTGGTGAT-3′ |
| Sequence-based reagent | β-ACTIN | This paper | PCR primer | Forward 5′-AAAC TGGAAC GGTGAAGG-3′ |
| Sequence-based reagent | β-ACTIN | This paper | PCR primer | Reverse 5′-AGA GAAGT GGGGTGGCTT-3′ |
| Sequence-based reagent | ATP5O | This paper | PCR primer | Forward 5′- cgcta tgccac agctcttta-3′ |
| Sequence-based reagent | ATP5O | This paper | PCR primer | Reverse 5′- atgg aacgcttc acataggg-3′ |
| Peptide, recombinant protein | Human bFGF | Invitrogen | Catalog # 13256-029 | |
| Peptide, recombinant protein | Human/mouse/ ratBDNF | PeproTech | Catalog # 450-02 | |
| Peptide, recombinant protein | Mouse BMP7 | R&D Systems | Catalog # 5666BP-010 | |
| Peptide, recombinant protein | Human recombinant insulin | Tocris | Catalog # 3435 | |
| Peptide, recombinant protein | Human BMP4 | R&D Systems | Catalog # 314BP-050 | |
| Peptide, recombinant protein | Human BMP6 | R&D Systems | Catalog # 507BP-020 | |
| Peptide, recombinant protein | Human/mouse FGF8b | R&D Systems | Catalog # 423-F8-025 | |
| Commercial assay or kit | RNeasy micro kit | QIAGEN | Catalog # 74004 | |
| Commercial assay or kit | RNeasy Plus mini kit | QIAGEN | Catalog # 74134 | |
| Commercial assay or kit | Transcription First Strand cDNA Synthesis Kit | Roche Life Sciences | Catalog # 04379012001 | |
| Commercial assay or kit | HotStarTaq PLUS DNA Polymerase kit | QIAGEN | Catalog # 203603 | |
| Commercial assay or kit | Click-iT EdU Cell Proliferation Kit for Imaging | Invitrogen | Catalog # C10338 | |
| Commercial assay or kit | SMART-Seq v4 Ultra Low Input RNA Kit | TaKaRa Bio | Catalog # 634888 | |
| Commercial assay or kit | Nextera XT DNA library preparation kit | Illumina | Catalog # FC-131-1024 | |
| Commercial assay or kit | RNA 6000 Pico Kit | Agilent | Catalog # 5067-1513 | |
| Commercial assay or kit | In-fusion HD cloning plus (Clontech) | TaKaRa Bio | Catalog # 638909 | |
| Commercial assay or kit | Anti-mouse IgM microbeads | Miltenyi Biotec | Catalog # 130-047-302 | |
| Commercial assay or kit | MS columns | Miltenyi Biotec | Catalog # 130-042-201 | |
| Chemical compound, drug | ROCK-inhibitor Y-27632 | Abcam | Catalog # ab 120129 | |
| Chemical compound, drug | SB431542 | Tocris | Catalog # 1614 | |
| Chemical compound, drug | LDN-193189 | Stemgent/ Tocris | Catalog # 6053 | |
| Chemical compound, drug | CHIR99021 | Stemgent/ REPROCELL | Catalog # 04-0004-02 | |
| Software, algorithm | Primer3 | Open source | Primer3, RRID:SCR_003139 | |
| Software, algorithm | Salmon quantification software (version 0.8.2) | Open source | Salmon, RRID:SCR_017036 (Patro et al., 2017) | |
| Software, algorithm | R statistical software | Open source | R Project for Statistical Computing, RRID:SCR_001905 | |
| Software, algorithm | Tximport (version 1.8.0). | Open source | tximport, RRID:SCR_016752 (Love et al., 2016) | |
| Software, algorithm | DESeq2 (version 1.20.0) | Open source | DESeq2, RRID:SCR_015687 (Love et al., 2018) | |
| Software, algoritham Software, algorithm | rtracklayer package (version 1.40.6) | Open source | rtracklayer, RRID:SCR_021325 | |
| Software, algorithm | GSVA (version 1.34.0) | Open source(Hänzelmann et al., 2013) | GSVA, RRID:SCR_021058 | |
| Software, algorithm | Pheatmap R package (version 1.0.10) | Open source | ncv | pheatmap, RRID:SCR_016418 |
| Software, algorithm | topGO Bioconductor package | Open source | topGO, RRID:SCR_014798 | |
| Software, algorithm | GOseq Bioconductor package | Open source (Young et al., 2010) | Goseq, RRID:SCR_017052 | |
| Software, algorithm | ImageJ (version 2.1.0/1.53c) | Open source, NIH | ImageJ, RRID:SCR_003070 | |
| Software, algorithm | SPSS software | IBM | ||
| Software, algorithm | BD FACSDiva 8.0.1 software | BD Biosciences | ||
| Software, algorithm | ZEN imaging software | Zeiss |
Table of primers.
| Gene | Forward primer | Reverse primer | °C |
|---|---|---|---|
| ATOH1 | 5′-GCGCAAAAGAATTTGTCTCC-3′ | 5′-GCGAAGTTTTGCTGTTTTCC-3′ | 60 |
| ID4 | 5′-GCTCACTGCGCTCAACACC-3′ | 5′-GAATGCTGTCGCCCTGCTTG-3′ | 60 |
| EN2 | 5′-GGCGTGGGTCTACTGTACG-3′ | 5′-TACCTGTTGGTCTGGAACTCG-3′ | 59 |
| PAX6 | 5′-TCACCATGGCAAATAACCTG-3′ | 5′-CAGCATGCAGGAGTATGAGG-3′ | 60 |
| NEUROD1 | 5′-GGACGAGGAGCACGAGGCAGACAAGAA-3′ | 5′-TTCCTCAGTGAGTCCTCCTCTGCGTTCA-3′ | 56 |
| PCP2 | 5′-GACCAGGAGGGCTTCTTCAATCT –3′ | 5′-CATGTCCATGAGGCTGTCCATCT-3′ | 56 |
| OTX2 | 5′-ACAAGTGGCCAATTCACTCC-3′ | 5′-GAGGTGGACAAGGGATCTGA-3′ | 60 |
| MEIS2 | 5′-CCAGGGGACTACGTTTCTCA-3′ | 5′-TAACATTGTGGGGCTCTGTG-3′ | 50 |
| GBX2 | 5′-GTTCCCGCCGTCGCTGATGAT-3′ | 5′-GCCGGTGTAGACGAAATGGCCG-3′ | 60 |
| HOXA2 | 5-CGTCGCTCGCTGAGTGCCTG-3′ | 5′-TGTCGAGTGTGAAAGCGTCGAGG-3′ | 60 |
| LHX2 | 5′- GGTCCTCCAGGTCTGGTTC-3′ | 5′-TAAGAGGTTGCGCCTGAACT-3′ | 60 |
| LHX9 | 5′- GCTGGGAGTGGACATCGTCA-3′ | 5′-CATGGTCCGGAGCTGGTGAT-3′ | 60 |
| β-ACTIN | 5′-AAACTGGAACGGTGAAGG-3′ | 5′-AGAGAAGTGGGGTGGCTT-3′ | 59 |
| ATP5O | 5′-cgctatgccacagctcttta-3′ | 5′-atggaacgcttcacataggg-3′ | 60 |
Table of antibodies.
| Antibody | Species | Source | Dilution | Catalog # |
|---|---|---|---|---|
| Calretinin | Rabbit | Swant | 1:1000 | 7699/4 |
| EN2 (C19) | Goat | Santa Cruz | 1:50 | SC-8111 |
| GFAP | Chicken | EnCor Biotechnology | 1:1500 | CPCA-GFAP |
| GFP | Rabbit | Invitrogen | 1:500 | A-111122 |
| GFP | Chicken | Aves labs | 1:1000 | GFP-1020 |
| HuNu (anti human nuclei) | Mouse | Millipore | 1:100 | MAB1281 |
| Ki67 | Rabbit | Vector Laboratories | 1:100 | VP-RM04 |
| Ki67 | Rabbit | EnCor | 1:1000 | RPCA-Ki67 |
| MAP2 | Chicken | Abcam | 1:1000 | ab5392 |
| NEUN | Mouse | Millipore | 1:100 | MAB377 |
| NEUROD1 | Mouse | BD Pharmingen | 1:300 | 563000 |
| PAX6 | Rabbit | BioLegend | 1:300 | 901301 |
| TAG1 | Mouse IgM | T.Jessell | 1:2 | N/A |
| Synaptophysin | Mouse IgM | Millipore | 1:500 | MAB329 |
| VGLUT1 | Mouse | Millipore | 1:100 | MAB5502 |
| SOX2 (D6D9) | Rabbit | Cell Signaling | 1:200 | 3579 |
| GluR-δ2 | Goat | Santa Cruz | 1:100 | Sc-26118 |
| Anti-goat Alexa Fluor 633 | Donkey | Invitrogen | 1:300 | A21082 |
| Anti-rabbit Alexa Fluor 555 | Donkey | Invitrogen | 1:300 | A-31572 |
| Anti-mouse IgM Alexa Fluor 488 | Goat | Invitrogen | 1:300 | A-21042 |
| Anti-mouse Alexa Fluor 555 | Donkey | Invitrogen | 1:300 | A-31570 |
| Anti-chicken IgY 488 | Donkey | Jackson ImmunoResearch | 1:300 | 703-545-155 |
| Anti-chicken IgY Cy3 | Jackson ImmunoResearch | 1:300 | 703-165-155 | |
| Anti-rabbit Alexa Fluor 647 | Donkey | Invitrogen | 1:300 | A-31573 |
| Anti-mouse 405 | Donkey | Abcam | 1:300 | Ab175658 |
| Anti-mouse Alexa Fluor 647 | Donkey | Invitrogen | 1:300 | A-31571 |
| Anti-mouse Alexa Fluor 488 | Donkey | Invitrogen | 1:300 | A-21202 |
Additional files
-
Supplementary file 1
GO terms for TRAP-seq data.
- https://cdn.elifesciences.org/articles/67074/elife-67074-supp1-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/67074/elife-67074-transrepform1-v2.docx





