Spatial modulation of individual behaviors enables an ordered structure of diverse phenotypes during bacterial group migration
Figures
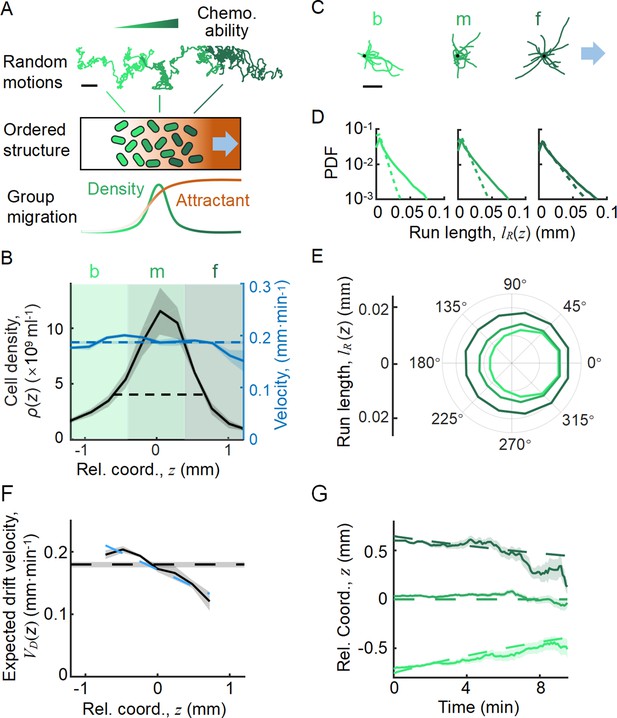
Statistics of single-cell behavior during collective group migration.
(A) Bacterial population of diverse phenotypes sorted according to their chemotactic abilities (increasing from light green to dark green) during collective migration following the self-generated attractant gradient (brown color). Meanwhile, individual cells perform run-and-tumble random motions biased toward the migration direction. Scale bar reflects 0.1 mm. (B) Bacterial density profile is stable (black solid line) in the moving coordinate . Where the width, represented by the black dashed-dotted line, is defined by two times the standard deviation of the bacterial relative position ( black dashed line). The instantaneous velocity () (blue solid line) is uniform and is equal to the average group velocity (blue dashed line). This observation was independent of the sampling time interval (Appendix 1—figure 2). (C) Sample runs of bacteria aligned by their initial positions (black dots) from the three regions (defined by colored regions in B) showed that cells at the back of the group tended to run forward, compared with cells in other regions. b, m, and f stand for the back, middle, and front of the migration group, respectively. Scale bar reflects 5 µm. (D) The exponential distributions of forward runs (solid lines) and backward runs (dashed lines) suggested the increasing efficiency of runs from the back to the front. (E) The mean run length in different directions confirmed the increasing efficiency of runs. (F) The expected drift velocity (black solid line) decreased from the back to the front, and is linearly fitted by , with and . (z) crossing with the average group velocity (black dashed line), implied that bacteria perform mean reversion motions. The profile was cut to present the majority of cells (~90%) (). (G) The time evolution of the average position (, solid lines) of cells starting from the back, middle, and front of the migration group (color code was defined in B) confirmed the reversion behavior. Shaded area represents the s.e.m. of more than 450 cells (see Materials and methods). Analytically, the OU-type model predicted that (dashed lines), where can be fitted by the starting position (see supplementary text). In panels B, F, and G, the shaded area represents the s.e.m. of three biological replicates. The spatial bin size was 240 µm .
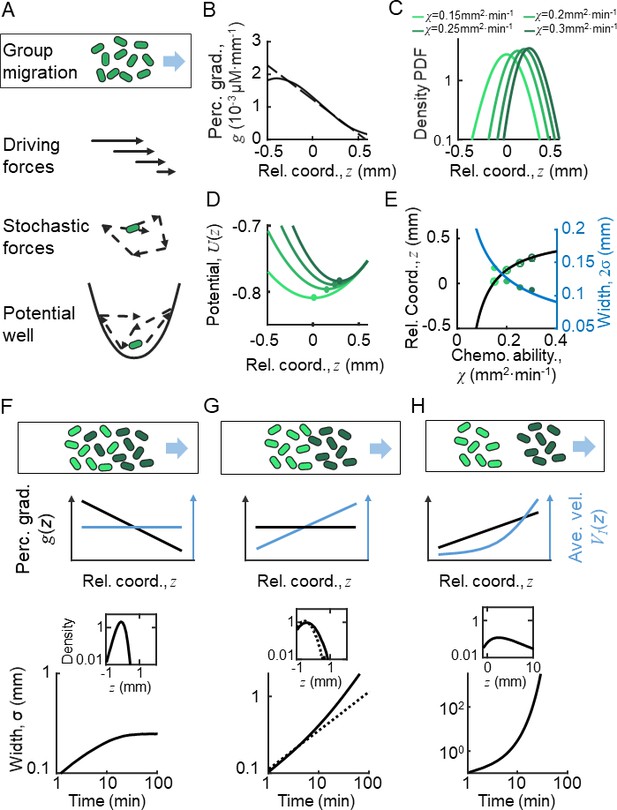
Ordered location of phenotypes of active particles in a moving gradient as a result of a pushed wave.
(A) Illustration of the Langevin-type model where the migrating bacteria are modeled by active particles under a deterministic decreasing driving force and a stochastic force. These forces allow the particles to form collective migration. On the moving coordinate of the migration group, active particles are bounded in a potential well generated by the driving force and the motion relative to the moving group (). (B) The perceived gradient (blue line) is deduced from , which is further calculated from the experimentally measured density profile (Appendix 1—figure 4D). This gradient profile was then transformed to a moving gradient , and applied in the simulations of different active particles following Langevin dynamics. A linear fit of around is plotted (dashed blue line) and is applied to the Ornstein-Uhlenbeck (OU)-type model. (C) Particles migrate collectively with a moving gradient, while they are located in ordered locations according to their phenotype (green lines). Colors from light to dark represent increasing . (D) The effective potential wells generated by the effective force () are spatially ordered. Circles present the minimum of the potential wells. (E) The peak positions (cross) and the mean positions (circles) increase with . These were predicted by the OU-type model (Equation S15) (black line). The width of the density profile is defined by two times the standard deviation of all cells (), which decreases with , which was consistent with the prediction of Equation S18 (blue line). (F–H) Group migration of mixed phenotypes under decreasing, invariant, and increasing gradient. Simulations were performed in one dimension (1D) with the chemotaxis ability of the group following a Gaussian distribution with a mean of 0.11 mm2 min−1 and a variation of 0.02 mm2 min−1 (Appendix 1—figure 5C). The force field used in the simulation followed the linearized force field, as in Equation 1, while in F, in G, and in H. All force fields were set as moving with velocity . The dashed line in G represents a group with a single phenotype of . Insert plots represent the wave front in the moving coordinate after 10 min of simulation. The width of the moving group, defined by the standard deviation of all particles, converges under decreasing gradient (F) while it diverges in cases of G and H.
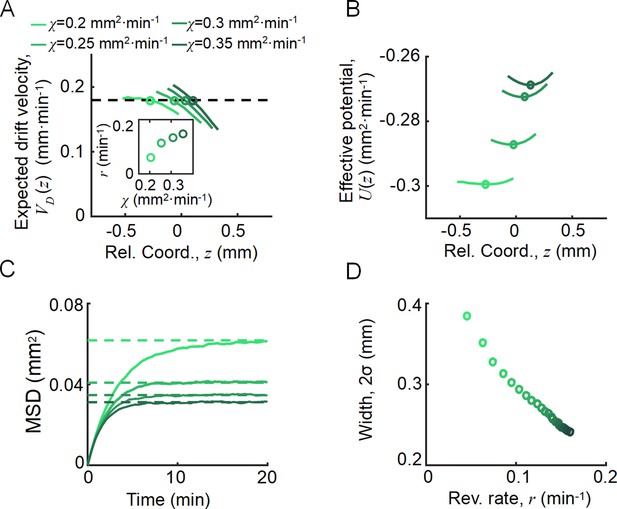
Agent-based simulations recapture the ordered behavior of individuals.
(A) The expected drift velocity of simulated bacteria decreased from the back to the front of the migration group, where the chemotactic ability ranged from 0.2 to 0.35 mm2min-1 , which was consistent with the experimental results shown in Figure 1F. The intersections between the curves with the preset group velocity (black dashed line) shifted toward the back of the migration group as decreased (circles). The different colors of the lines and circles correspond to different chemotactic abilities , as shown in the legend. The same color coding also applies to (B–D). (B) The reversion rate increased with chemotactic ability. (C) The effective potential well calculated by . Positions of the potential minimum are marked as circles. As illustrated, for a lower chemotaxis ability , the potential well is shallower and shifts toward the back of the migration group. (D) The width of the density profile (measured by , see Figure 1B) decreases with the reversion rate as well as the chemotaxis ability . The mean square displacement (MSD) of bacteria (insert, solid lines) is bound to (insert, dashed lines) (see supplementary text). In panel (A, C), curves were cut to present the majority of cells (90%) (). More details on the results of this simulation are presented in Appendix 1—figure 7.
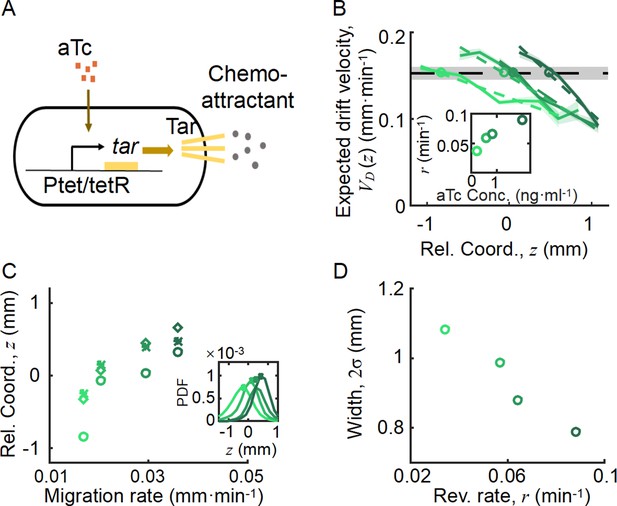
Spatial ordered structure emerged from the behavioral modulation of cells with different chemoreceptors.
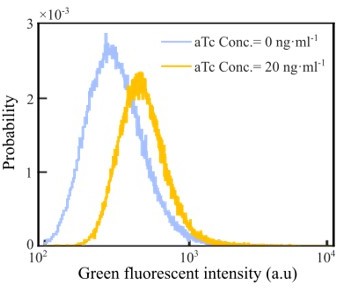
(A) Genetic circuit of the Tar-titratable strain. In the experiments, the expression level of Tar (a chemoattractant receptor protein) was titrated by the concentration of external inducer (aTc). The chemotactic ability of bacteria was then determined by the expression level of Tar (Adler, 1969). (B) The expected drift velocity of the Tar-titratable strain JCY20 (colored solid line) was spatially modulated and decreased from the back to the front of the migration group and intersected with a group migration velocity (black dashed line). The linear fit of (colored dashed lines) intersected with at positions (circles) determined by the corresponding Tar expression level. Colors from dark to light green corresponded to inducer (aTc) concentrations of . The black shaded area of represents the s.d. of four experiments, while the colored shaded area of the curves presents the s.e.m. of the counted runs. (C) In the experiment, the positions of the intersections (circles, illustrated in B), together with the peaks (stars, illustrated in the insert figure), and the average positions (diamond) of the bacterial density profiles, all shifted toward the front of the migration group for strains with higher Tar expression levels, which had higher chemotactic abilities and migrated faster on agar plates (x-axis, see Materials and methods; Appendix 1—figure 9). The related density profiles (PDF) were shown in the insert plot and the color coding of lines/symbols in both panels C and D was the same as that in B. (D) The width of density profiles () of Tar-titrated bacteria decreased with the reversion rate .
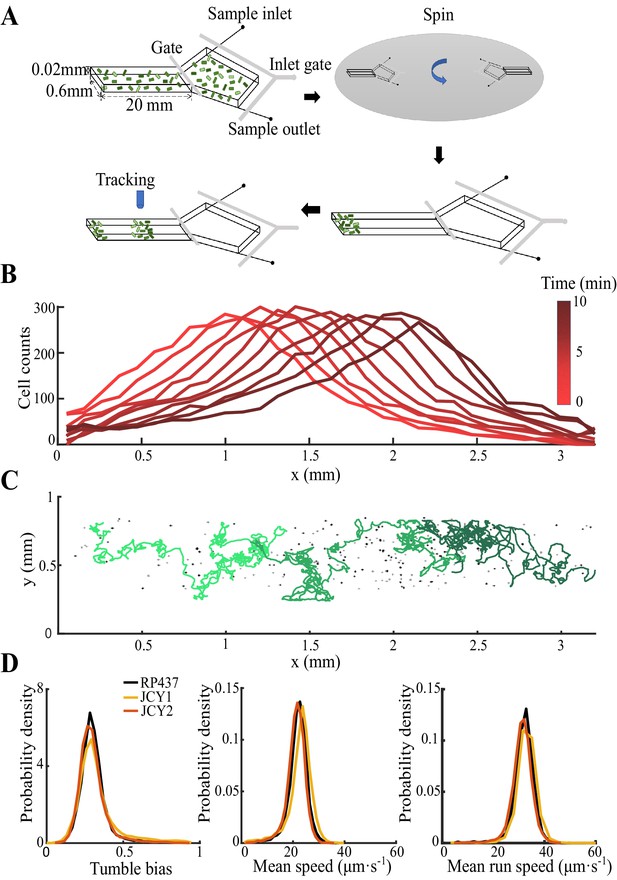
Experiment protocol, sample tracks, and fluorescent labeled cells.
(A) The microfluidic chip used in this study is consisted of a long channel (20 mm×0.6 mm×0.02 mm) and a large chamber (3 mm× 3 mm). Prepared bacteria are first loaded in the chip through inlets with M9 motility buffer (see Materials and methods). Next, the chip is spun at 3000 rpm for 15 min so that the bacteria are concentrated to one end of the channel. Bacteria consume aspartate (Asp) as the single chemoattractant to form a moving gradient that attracts the group migration. Finally, the bacterial motions are tracked under microscope of 4× objective by frame rate of 9 fps. (B) Cells expressing fluorescent protein (strain JCY2) are premixed into strain RP437 by 1:400 and counted each frame. Cell counts in nine successive frames () were plotted within bin size of . Colors from light to dark red represent time from 1 to 10 min. (C) Sample tracks of bacteria (colored lines) and bacterial signal (black dots) on the last captured frame. (D) Fluorescent labeled cells (strain JCY1 and JCY2) and the wild-type strain (RP437) were tracked separately on growth medium. All three strains show the same distribution of tumble bias (left), mean speed (middle), and mean run speed (right) of each track. All these quantities are average of more than 9000 tracks, weighted by their trajectory time. Each curve show data of one experiment.
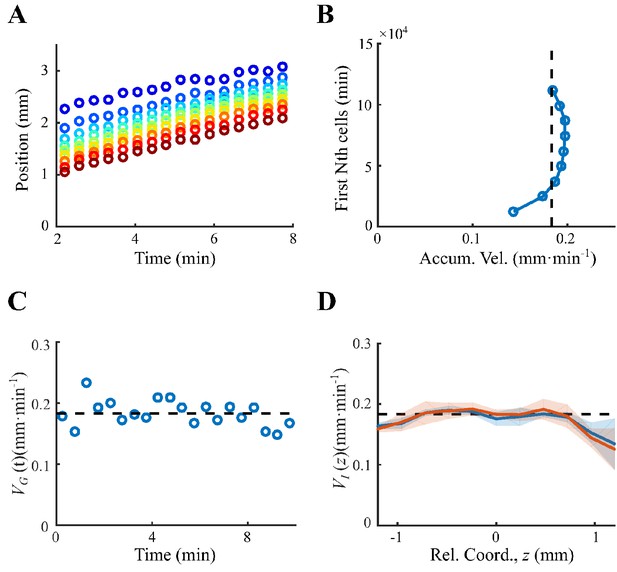
Determination of coherent migration group.
(A) Positions of first n-th cell (counted from the front, from blue to red ) increase linearly with time, of which the slope gives the traveling speeds. (B) The traveling speeds of the first n-th cells are close to the group velocity (black dashed line), representing a coherent migration group. (C). The instantaneous group velocity was constant over time. was averaged for all tracks over 30 s (blue dots). Black dashed line represents the average group velocity for all tracks on all times . (D). The constant instantaneous velocity for sampling time (blue solid line) and (red solid line), where shaded area was s.e.m. of three replicates. Both curves equal to the group velocity , indicating the migration group is coherent. Data in (A–C) was extracted from one experiment.
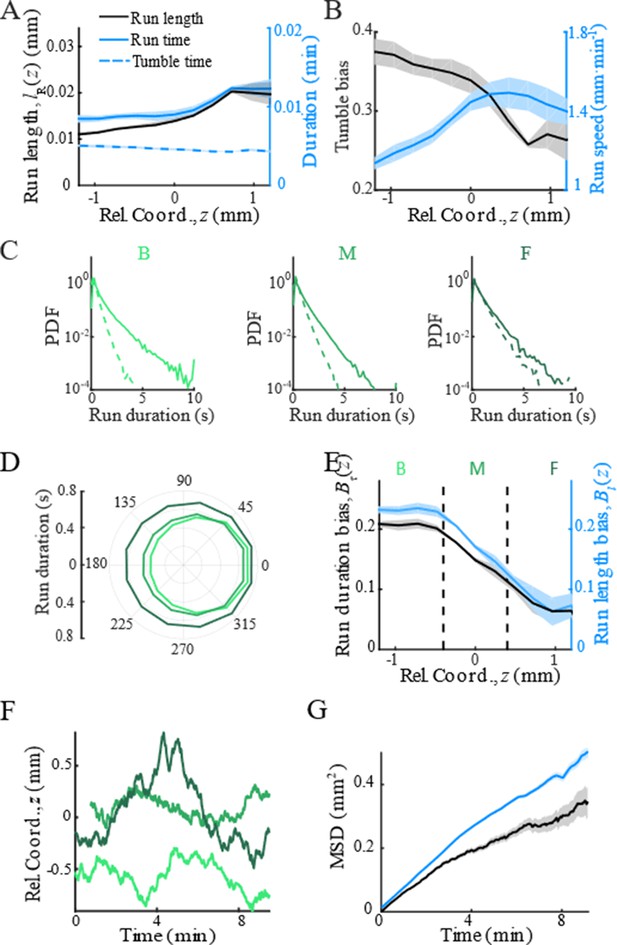
Modulation of bacterial behaviors.
(A) Distribution of forward run duration (solid line) and backward run duration (dash line) in three regions defined in Figure 1D and Appendix 1—figure 3E. (B) Mean run duration in different directions of bacteria in three regions. s.e.m. of four replicates is smaller than thickness of lines. Angular bin size is 15°. (C) Spatial structure of adapted tumbled bias (blue) and run speed (red). (D) The mean run length in different directions, with the angular bin size of 15°, also shows that cells in the back were better skewed to run forward. (E) The run length bias (black solid line) and the run time bias (blue solid line) both decreased from the back to the front of the migration group. In panels C–E, shaded area represents s.e.m. of three biological replicates. The spatial bin size is 240 µm. (F) Representative examples of single-cell trajectories (three colors represent three different tracks) showed the reversion behavior of bacteria around their mean positions. (G) The mean reversion behavior results in limited diffusion of bacteria in the moving frame. So that the mean square displacement (MSD) in the direction of migration (black curve) was lower than the pure diffusion of bacteria measured on free liquid with M9 motility buffer (blue curve). The shaded area on black curve represents the s.e.m. of more than 2900 tracks, while the shaded area on blue curve represents the s.e.m. of more than 1900 tracks.
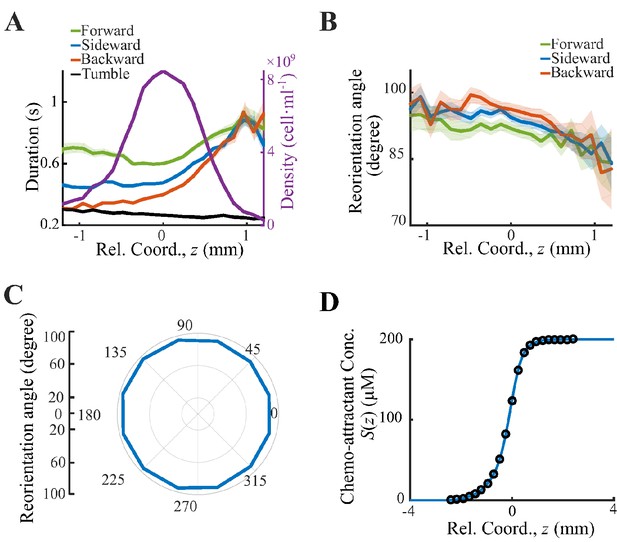
Behavior analysis and deduction of the chemoattractant profile.
(A) All cell tracks are divided into three sectors: forward () (green line), sideward () (blue line), backward () (red line). The run durations in all three sectors increase form back to front. (B) Spatial distribution of reorientation angle. (C) Reorientation angles defined in Saragosti et al., 2011 are plotted over the direction of previous runs. Such angles are biased to the back, indicating that bacteria running down the gradient reorient more in the next tumble. Shaded area shows s.e.m. of 14,892 tracks. The s.e.m. in panel C is smaller than the thickness of the curve. (D) The chemoattractant profile used for the simulation is interpolated by (blue line) from the experimental (black circles). While the experimental chemoattractant is deduced from the averaged density profile (Figure 1B).
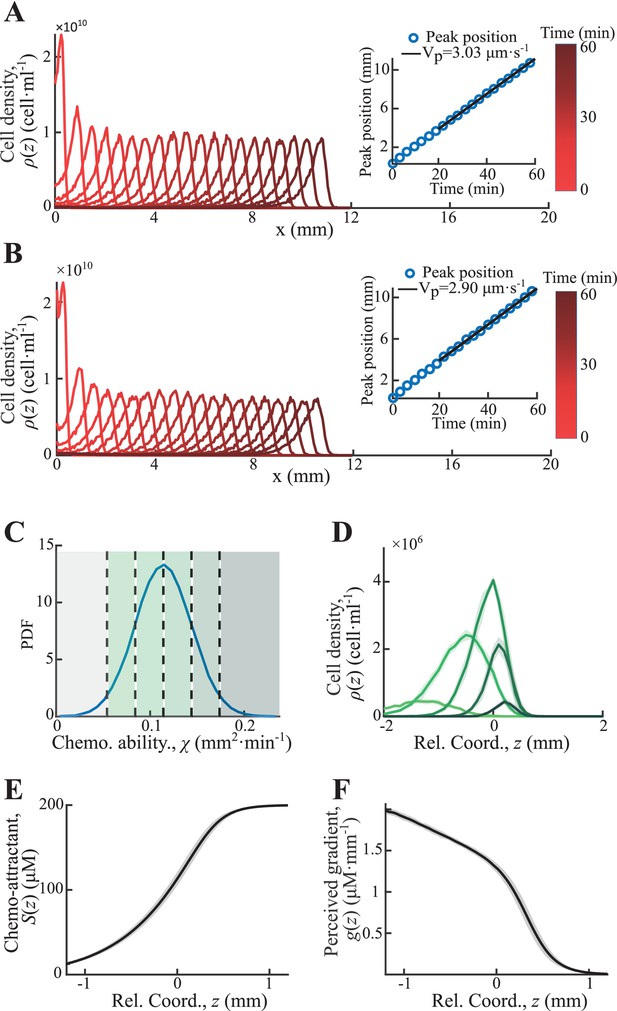
Particle-based simulation with chemoattractants consumption.
(A) Simulated density profiles of active particles of single phenotype of . over time. The peak positions increase linearly with time (insert plot). (B) Simulated density profiles of active particles of heterogenous phenotypes distributed in (C). The peak positions follow a stable speed of (insert plot). (D) Time-shifted density profiles of different phenotypes exhibit spatially ordered in moving coordinate. Cells in the gray area drop the group during migration. Time-shifted chemoattractant profile increases from back to front (E) while the perceived gradient decreases with a quasi-linear region around 0. (F) Shaded area in D, E, F represents s.d. of 40 profiles recorded at stable region (from 21 to 60 min) with time step of 1 min.
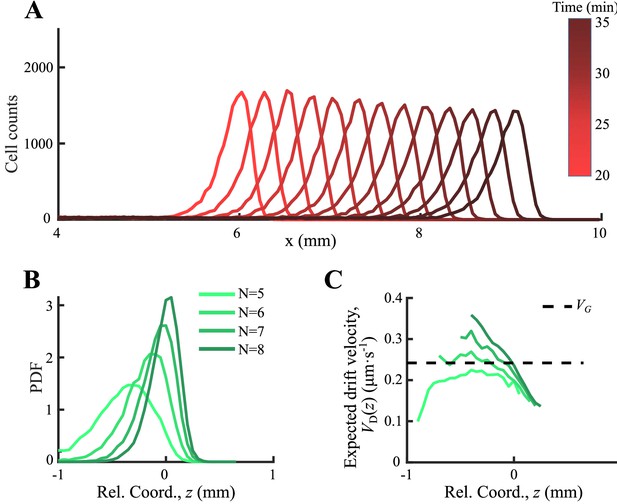
Agent-based simulations with consumption of chemoattractant.
(A) Simulated density profiles of bacteria integrated with chemotactic pathway show stable migration after 20 min simulation. Tracks were obtained in from 30 to 32 min, and were calibrated into a moving coordinate through group migration speed. (B) Four subgroups of different receptor gain N were orderly distributed. (C) The expected drift velocity is decreasing and is spatially ordered.
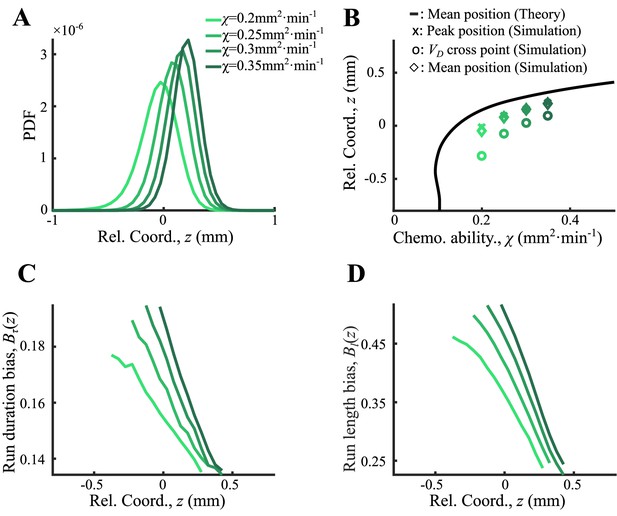
Agent-based simulations of cells with different receptor gains.
(A) Bacteria of different chemotatic ability () form ordered structure in a moving chemoattractant field as in Appendix 1—figure 4D with fixed velocity . (B) The average positions of each phenotype (diamonds), the peak positions (crosses), and the cross positions between their curves and the group velocity (circles) increase with . These positions follow the same trend as predicted by Ornstein-Uhlenbeck (OU) model (black line). (C, D) Run duration bias and run length bias of different subgroups decrease from back to front. Both curves were also spatially ordered by (color from light to dark green).
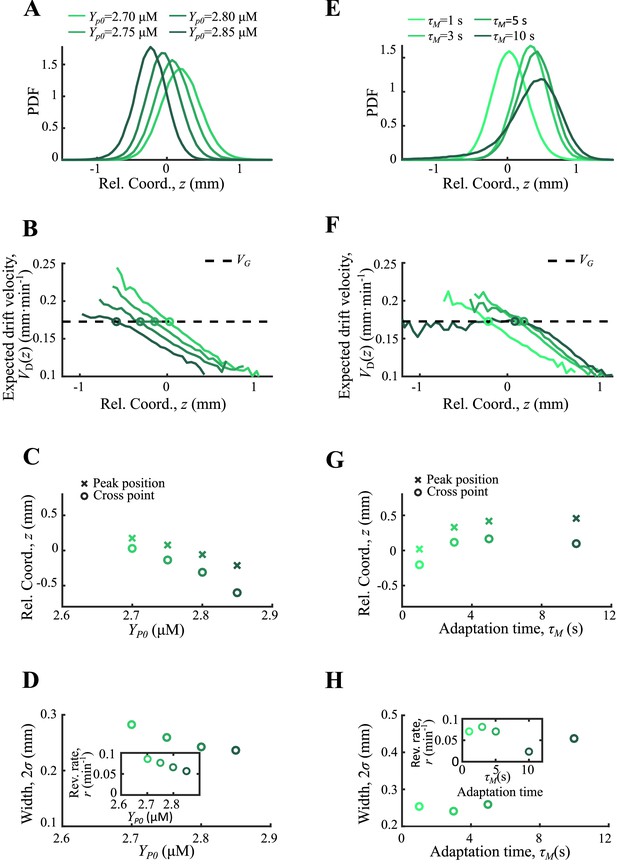
Agent-based simulations of cells with different CheY-P levels and adaptation times.
Simulations results for CheY-P concentration ranging from 2.70 to 2.85 µM (A–D) and for adaptation time ranging from 1 to 10 s (E,H) are presented. In both cases the cell density (A, E) and the expected drift velocity (B, F) are spatially ordered according to the varying parameter. The mean position (crossed) together with the cross position between and (circles) increase with (C) while they show non-monotonic trend to (G). The width of the density profile (defined by s.d. of all cells) and the reversion rate decrease with (D). And they showed a non-monotonic dependence on . The receptor gain is set to 24 in both cases, while is set to 2.77 µM for the varying case and is 1 s for the varying case. Simulations are performed under a steeper moving chemoattractant gradient of .
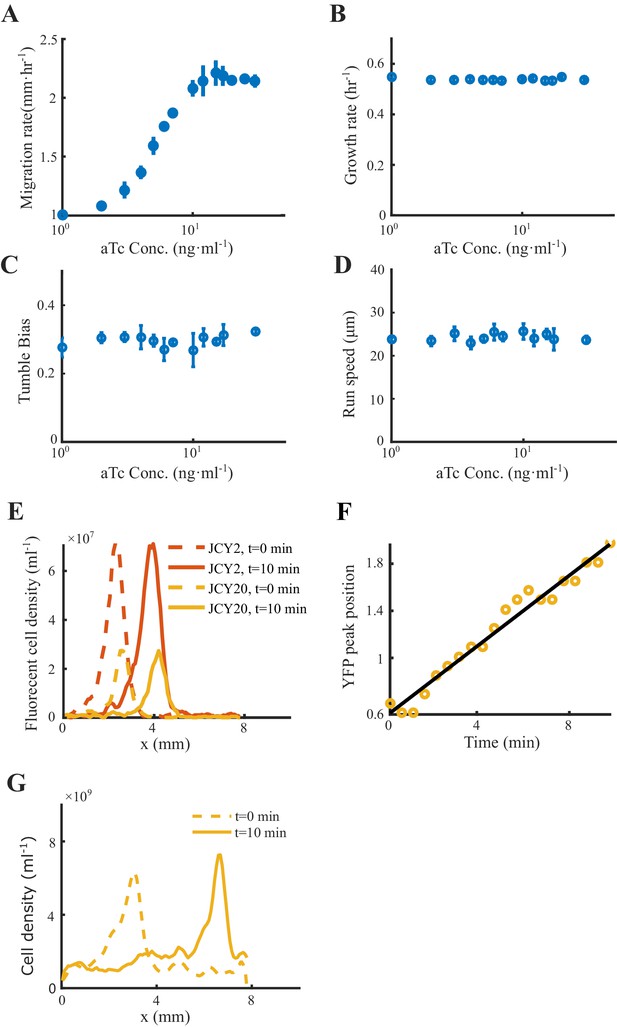
Migration rates, growth rates, motilities and coherent migration of the Tar-titratable strain.
(A) Tar-titratable strain (JCY20) allows us to change its chemotaxis ability (measured by its migration rate on agar plate) by adding inducer ([aTc]) of different concentrations. (B) This strain has constant growth rates measured in liquid batch culture. (C–D) Motility statistics were extracted from bacterial tracks in free liquid. The growth rate, tumble bias, and mean run speed were found constant with all tested aTc concentrations. Error bar represents s.d. of three biological replicates. (E) The density profile of the wild-type (WT) strain with red fluorescent protein (RFP) (strain JCY2) (red lines) and Tar-titratable strain with yellow fluorescent protein (YFP) (strain JCY20) (yellow lines) are plotted over space at time before (dashed lines) and after tracking (solid lines). Because the density profiles of both strains before and after tracking are considered to be identical, the migration group is stable during the tracking period (10 min ). The fluorescent strains (JCY2 and JCY20) are mixed with WT non-fluorescent strain (RP437) by ratio of 8:1:400. (F) During the 10 min tracking period, only the Tar-titratable strain (JCY20) is recorded. For each half a minute, we plotted the density profiles and find their peak positions (yellow circles). Such peak positions increase linearly with time (fitted by the black line). Data were extracted from one experiment. (G) The density profile of the Tar-only strain with YFP (strain UU1624 from Prof. Parkinson) are plotted over space at time before (dashed lines) and after 10 min (solid lines).
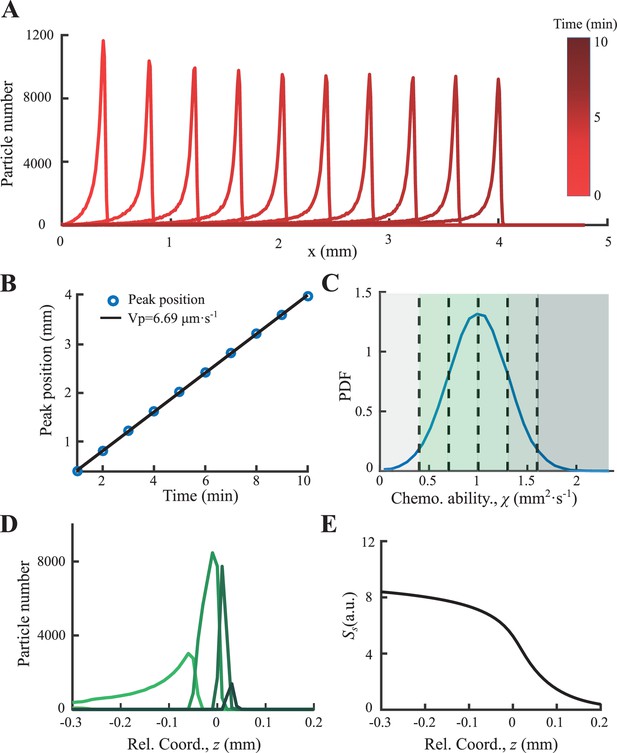
Particle based simulation of the signal secretion model.
(A) Simulated density profiles of heterogenous phenotypes distributed in C show (B) stable group migration with its peak positions following a constant speed of . (C) The phenotypic diversity was set by a Gaussian distribution on , with mean value of 1 and s.d. of 0.3. (D) Time-shifted density profiles of different phenotypes exhibit spatially ordered in moving coordinate. Color code follows C, while cells in the gray area drop from the group. (E) Time-shifted signal profile , which plays the role of ‘global driving force’, decreases from back to front.
Videos
Bacterial tracking in the migration group.
Each dot represents a bacterium captured under microscope. Lines represent typical tracks. The colors of tracks from light green to dark green represent the mean positions of the tracks from the back to the front of the migrating group.
Tables
Summary of quantities.
| Quantities | Definition | Formulations |
|---|---|---|
| Group velocity | ||
| Moving coordinate | ||
| Instantaneous velocity | ||
| Expected drift velocity | ||
| Cell density | ||
| Chemoattractant concentration | ||
| Perceived gradient |
Strains and plasmids used in this study.
| Strains | Description | Source |
|---|---|---|
| RP437 | Wild-type strain for chemotaxis | Fu et al., 2018 |
| JCY1 | RP437+ a Cmr plasmid with YFP | This study |
| JCY2 | RP437+ a Kanr plasmid with mRFP1 | This study |
| JCY20 | Construction based on RP437, tar<> loxp, bla:Ptet-tetR-tar at attB site,+ a Cmr plasmid with YFP | This study |
| UU1624 | Δaer-1 Δtsr-7028 Δtap-3654 Δtrg-100 | Gosink et al., 2006 |
Summary of parameters in agent-based simulation.
| Symbol | Definition | Value | Source |
|---|---|---|---|
| Chemotaxis ability | Defined in context | This study | |
| Consumption rate of chemoattractant | Fitted parameter for migration speed | This study | |
| Upper limit of chemoattractant sensing | 1 mM | Fu et al., 2018 | |
| Lower limit of chemoattractant sensing | 1 µM | Si et al., 2012 | |
| Diffusion coefficient of chemoattractant | 1000 µm2/s | Fu et al., 2018 | |
| Run speed of bacteria | 30 µm/s | Keller and Segel, 1971b | |
| Adaptation time | 1 s | Waite et al., 2016 | |
| Receptor gain | Deduced from chemotactic ability | SI notes in Section 7 | |
| Motor number | 5 | Sneddon et al., 2012 | |
| Basic CheY-P level | 2.77 µM | Correspond to |