Injection with Toxoplasma gondii protein affects neuron health and survival
Figures
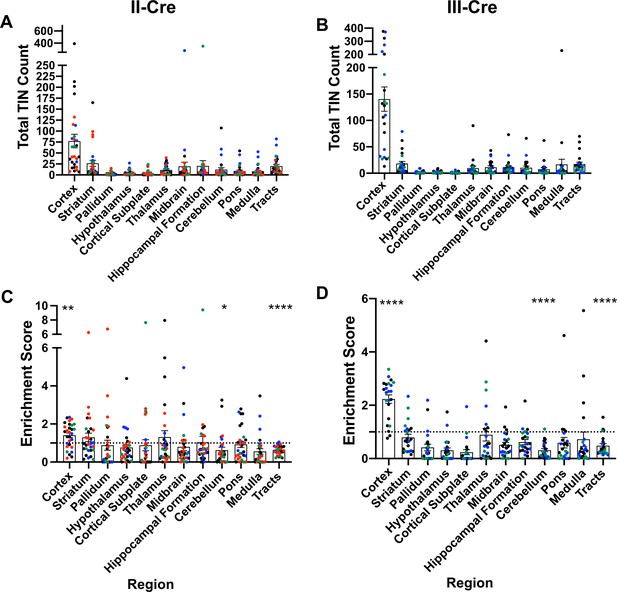
Toxoplasma-injected neurons (TINs) show a predilection for the cortex at 3 weeks post-infection.
Cre-reporter mice were infected with II-Cre or III-Cre Toxoplasma parasites as indicated. Brains were harvested, sectioned, labeled, and quantified as previously described (Mendez et al., 2018). (A, B) Graphs of the absolute numbers of TINs mapped to 12 regions of the brain. (C, D) Graphs of TINs/region normalized to the size of the region. The dashed line is 1, the value at which the distribution of TINs would be considered proportional to the region size. Bars, mean ± SEM. N = 16–19/sections/mouse. Individual colors denote animals from individual cohorts, N = 4–12 mice/cohort for II-Cre infected mice, 2–11 mice/cohort for III-Cre infected mice. (C, D) *p=0.0170, **p=0.0021, ****p≤0.0001, one-sample t-test. p-Values for all regions are in Supplementary file 2. Mice were excluded from analyses if GFP+ cells were not above background rate of GFP+ cells in saline-injected Cre reporter mice or if identified as an outlier by ROUT outlier test, which exclude both Cohort 1 (red) III-Cre infected mice. Figure 1—figure supplement 1 includes all mice and uses all Allen Institute sections for normalization/enrichment index.
-
Figure 1—source data 1
Raw data for TINs count for II-Cre and III-Cre cohort 1 (Red).
- https://cdn.elifesciences.org/articles/67681/elife-67681-fig1-data1-v3.xlsx
-
Figure 1—source data 2
Raw data for TINs count for II-Cre and III-Cre cohort 2 (Green).
- https://cdn.elifesciences.org/articles/67681/elife-67681-fig1-data2-v3.xlsx
-
Figure 1—source data 3
Raw data for TINs count for II-Cre and III-Cre cohort 3 (Blue).
- https://cdn.elifesciences.org/articles/67681/elife-67681-fig1-data3-v3.xlsx
-
Figure 1—source data 4
Raw data for 10 mice each for TINs count for II-Cre and III-Cre cohort 4 (Black).
- https://cdn.elifesciences.org/articles/67681/elife-67681-fig1-data4-v3.xlsx
-
Figure 1—source data 5
Raw data for remaining mice for TINs count for II-Cre and III-Cre cohort 4 (Black).
- https://cdn.elifesciences.org/articles/67681/elife-67681-fig1-data5-v3.xlsx
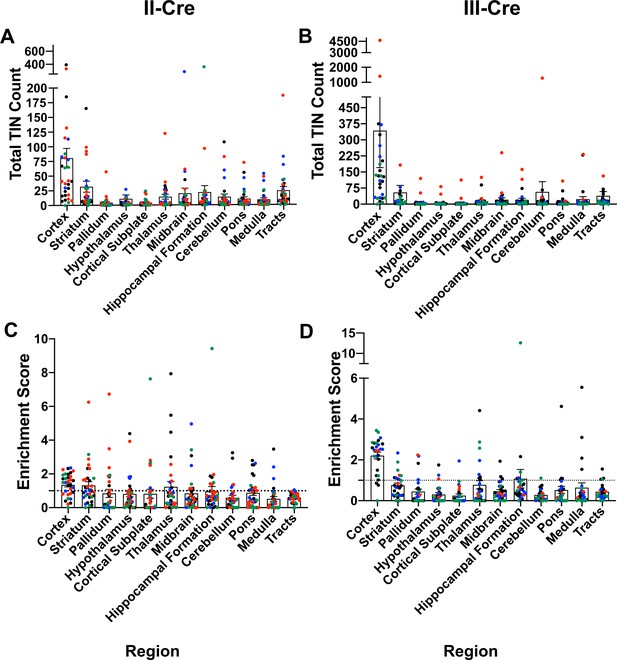
Original data with all mice and using full Allen Atlas area.
Cre-reporter mice were infected with II-Cre or III-Cre Toxoplasma parasites as indicated. Brains were harvested, sectioned, labeled, and analyzed as described in Figure 1. (A, B) Graphs of the absolute numbers of Toxoplasma-injected neurons (TINs) mapped to 12 regions of the brain. (C, D) Graphs of TIN numbers/region normalized to the size of the region. The dashed line indicates the value at which TINs distribution would be considered random and appropriate for the region size. Data presented is before the exclusion of any mice. Exclusion was determined by (i) the total number of TINs being less than or equal to the total GFP+ central nervous system (CNS) cells in saline-injected mice or (ii) identification as an outlier via the ROUT method. Bars, mean ± SEM.
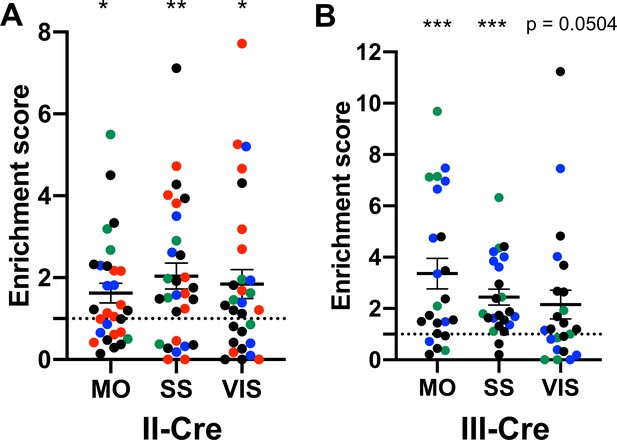
The visual, somatosensory, and motor cortices are highly enriched cortical regions containing Toxoplasma-injected neurons (TINs).
(A) Graph showing the normalized distribution of cortical TINs from II-Cre infected mice in motor (MO), somatosensory (SS), and visual (VIS) cortices. (B) As in (A) except for III-Cre infected mice. *p≤0.05, **p=0.01, ***p≤0.0005, by one-sample t-test. Individual colors denote mice from the same cohort. The dashed line indicates the value at which TINs distribution would be considered random and appropriate for the region size. Lines, mean ± SEM.
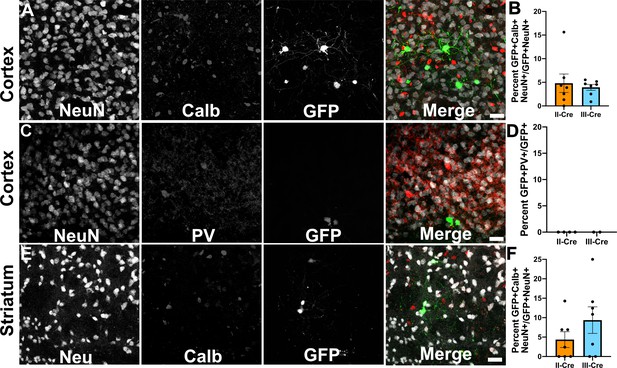
Toxoplasma-injected neurons (TINs) rarely co-localize with inhibitory interneurons.
Forty micron brain sections from II-Cre or III-Cre infected mice were stained with anti-NeuN antibodies and either anti-calbindin or anti-parvalbumin antibodies. Stained sections were then imaged by confocal microscopy to determine co-localization between TINs (GFP+) and calbindin (Calb) or parvalbumin (PV) staining. (A) Representative images of a cortical region from a section stained for Calb. (B) Quantification of the percentage of TINs that co-localized with Calb staining. (C) As in (A) except the images are of a cortical region stained for PV. (D) As in (B) except for PV quantification. (E) As in (A), but in the striatum. (F) As in (B) except for striatal Calb quantification. For (A, C, E) Merge images, gray = NeuN, red = Calb or PV, and green = GFP. N = 9 sections/mouse, seven mice/group for Calb, 2–4 mice/group for PV. (B, D, F) Bars, mean ± SEM. (B) For II-Cre infected mice, 24–127 TINs/mouse were analyzed; for III-Cre infected mice, 254–503 TINs/mouse were analyzed. (D) For II-Cre, 8–68 TINs/mouse, and for III-Cre, 281, 346 TINs/mouse were analyzed. (F) For II-Cre infected mice, 3–290 TINs/mouse were analyzed; for III-Cre infected mice, 21–214 TINs/mouse were analyzed. No significant differences were identified between groups, Student’s t-test.
-
Figure 2—source data 1
Raw numbers for quantification of Calb+ cells.
- https://cdn.elifesciences.org/articles/67681/elife-67681-fig2-data1-v3.xlsx
-
Figure 2—source data 2
Raw numbers for quantification of PV+ cells.
- https://cdn.elifesciences.org/articles/67681/elife-67681-fig2-data2-v3.xlsx
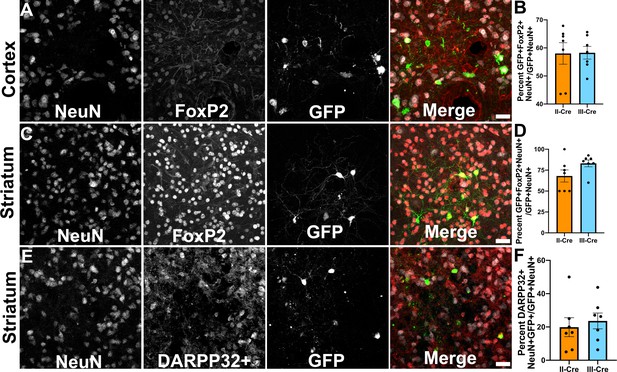
Toxoplasma-injected neurons (TINs) co-localize with FoxP2 and DARPP32 staining.
Brain sections from II-Cre or III-Cre infected mice were stained with anti-NeuN antibodies and anti-FoxP2 or anti-DARPP32 antibodies. DARPP32 staining was only done in the striatum. Stained sections were then analyzed by confocal microscopy to determine co-localization between TINs (GFP+) and FoxP2 or DARPP32 staining. (A) Representative image of a cortical region from a section stained as labeled. (B) Quantification of the percentage of TINs that co-localized with FoxP2 staining. (C) As in (A) except the images are of a striatal region. (D) As in (B) except for striatal TINs. (E) As in (C) except the tissue is stained with anti-DARPP32 antibodies (as labeled). (F) As in (D) except co-localization is between TINs and DARPP32 staining. (A, C, E) Merge images, gray = NeuN, red = FoxP2 or DARPP32, and green = GFP. Scale bar = 50 µm. (B, D, F) Bars, mean ± SEM. For (B, D, F) N = 9 sections/mouse, seven mice/group. (B) For II-Cre infected mice, a total of 24–127 TINs/mouse were analyzed; for III-Cre infected mice, 254–503 TINs/mouse were analyzed. (D) For II-Cre infected mice, a total of 28–68 TINs/mouse were analyzed; for III-Cre infected mice, 290–858 TINs/mouse were analyzed. (F) For II-Cre infected mice, 6–237 TINs/mouse were analyzed; for III-Cre infected mice, 16–198 TINs/mouse were analyzed. No significant differences were identified between groups, Student’s t-test.
-
Figure 3—source data 1
Raw numbers for quantification of DARPP32+ cells.
- https://cdn.elifesciences.org/articles/67681/elife-67681-fig3-data1-v3.xlsx
-
Figure 3—source data 2
Raw numbers for quantification of FoxP2+ cells.
- https://cdn.elifesciences.org/articles/67681/elife-67681-fig3-data2-v3.xlsx
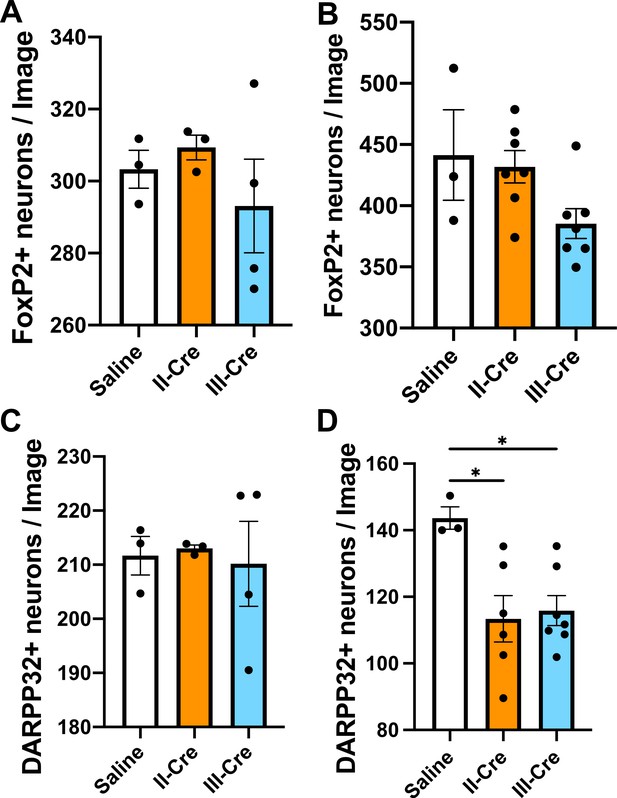
Quantification of FoxP2+ or DARPP32+neurons in the striatum.
Brain sections from saline or II-Cre or III-Cre inoculated mice were stained with anti-NeuN antibodies and anti-FoxP2 or anti-DARPP32 antibodies. Stained sections were then analyzed for the number of FoxP2+ or DARPP32+ neurons. (A) Average number of FoxP2+ neurons/image when randomly sampling the striatum. (B) Average number of FoxP2+ neurons when sampling near TINs. (C) As in (A), but for DARPP32+ neurons (random sampling). (D) As in (B) but for DARPP32+ neurons (near TINs). Bars, mean ± SEM. N = 9 sections/mouse, 31–44 images/mouse, 3–7 mice/group. *p<0.05, one-way ANOVA with Dunnett’s post-test.
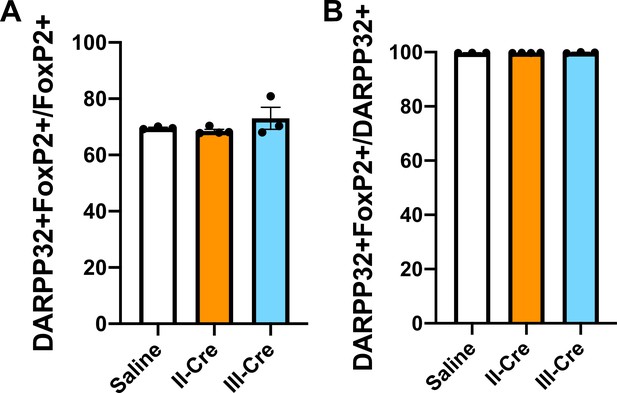
Baseline quantification for DARPP32+ and FoxP2+co-localization in the striatum.
Brain sections from saline, II-Cre, or III-Cre inoculated mice were stained for both FoxP2 and DARPP32. (A) Percentage of FoxP2+ neurons that co-localize with DARPP32 labeling. (B) Percentage of DARPP32+ neurons that co-localize with FoxP2 labeling. Bars, mean ± SEM. N = 9 sections/mouse, 31–44 images/mouse, 3–4 mice/group. No significant differences were identified between groups, one-way ANOVA with Dunnett’s post-test.
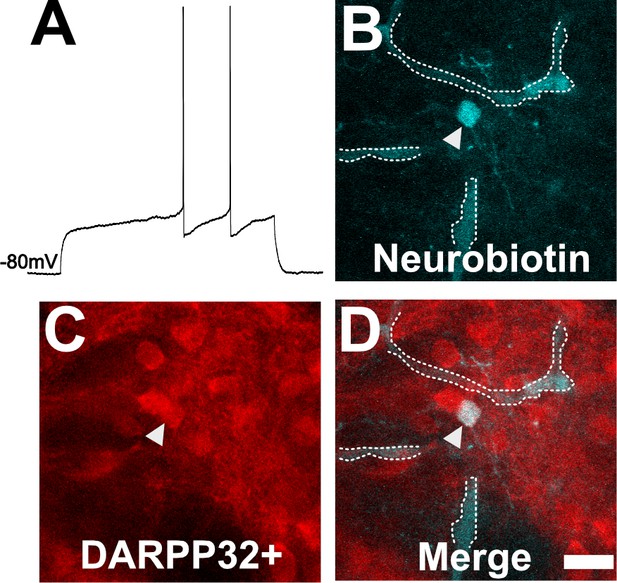
Neuron with electorphysiology of a medium spiny neuron (MSN) co-localizes with DARPP32 staining.
(A) Sample tracing from the shown neuron. Note the hyperpolarized resting membrane potential of −80 mV and the delayed time to the first action potential. (B) Image of patched neuron filled with neurobiotin. Arrowhead points to filled neuron. Dashed white line denotes biotin-filled blood vessels. (C) Image of DARPP32 staining. After recording and filling, the section was fixed and counter-labeled with anti-DARPP32 antibodies. (D) Merge of images B and C. (B–D) Section imaged on a Zeiss 880 confocal microscope. The shown images are a maximum projection of 12, 1 µm step images, from a 100 µm z-stack. Scale bar = 50 µm.
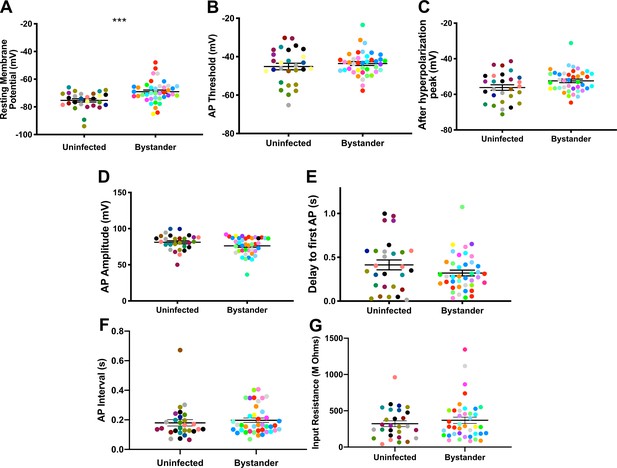
Bystander medium spiny neurons (MSNs) show similar electrophysiology as MSNs from uninfected mice.
(A) Graph of the resting membrane potential of whole cell patch clamped MSNs in uninfected mice (−74.9 ± 2.1 mV) and bystander MSNs in infected mice (−68.7 ± 4.0 mV). (B) Action potential (AP) threshold, (C) after hyperpolarization peak, (D) AP amplitude, (E) delay to first AP, (F) AP interval, and (G) input resistance in MSNs from uninfected mice and bystander MSNs in infected mice. Dots represent individual MSNs. Matching color dots denote cells from the same mouse. Uninfected MSNs, N = 28 cells recorded from 14 mice, 1–5 cells recorded/mouse. Bystander MSNs, N = 40 cells recorded from 18 mice, 1–5 cells recorded/mouse. Line, mean ± SEM. ***p=0.0007, Mann-Whitney U-test.
-
Figure 5—source data 1
Raw electrophysiology recording data for MSNs from uninfected mice and bystander MSNs and TINs from infected mice (source data for Figures 5 and 6A).
- https://cdn.elifesciences.org/articles/67681/elife-67681-fig5-data1-v3.xlsx
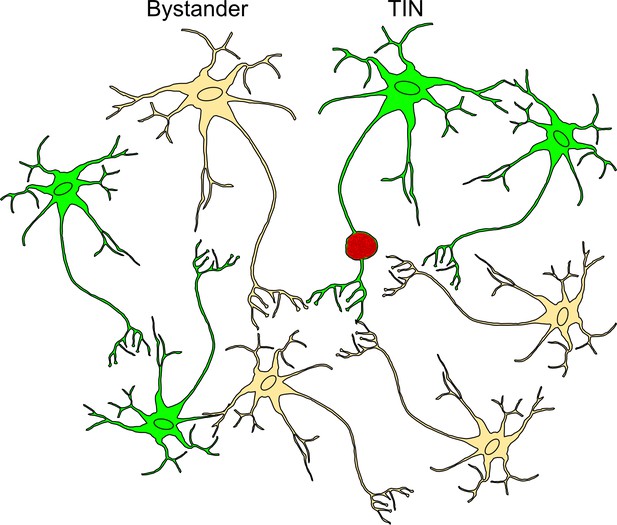
Schematic of bystander and Toxoplasma-injected neurons (TINs) during recording procedure.
Bystander neurons are neurons that are in the same inflammatory microenvironment as TINs but have not been injected with Toxoplasma protein. TINs (GFP+) include both infected and uninfected neurons, with over 90% of TINs not harboring parasites (Koshy et al., 2012). During recordings, no TINs with a cyst were observed. Bystanders = beige neuron, TINs = green neuron, red vacuole = Toxoplasma cyst.
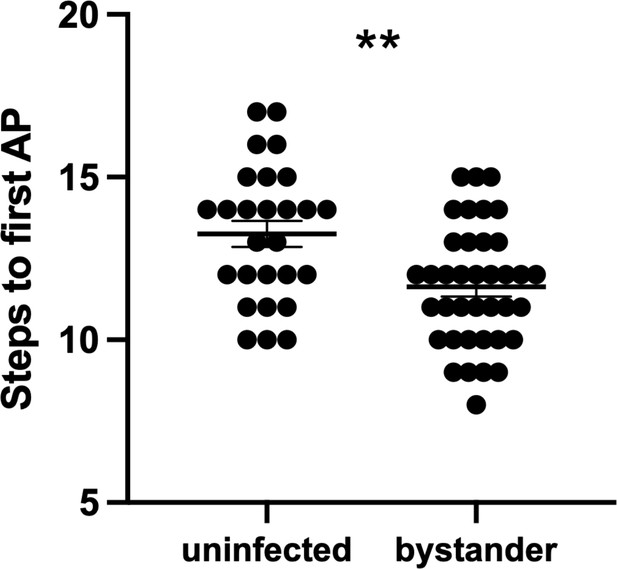
Bystander medium spiny neurons (MSNs) fire the first action potential (AP) sooner than MSNs from uninfected controls.
To initiate AP firing in patch-clamped neurons, 20 steps of both hyperpolarizing and depolarizing current are injected into a patched neuron (Wang et al., 2019). The graph shows the step number at which each neuron fired its first AP. Dots represent individual MSNs. Lines, mean ± SEM. **p=0.0014 by unpaired two-tailed Student’s t-test.
-
Figure 5—figure supplement 2—source data 1
Raw data for the number of steps quantified to reach first action potential.
- https://cdn.elifesciences.org/articles/67681/elife-67681-fig5-figsupp2-data1-v3.xlsx
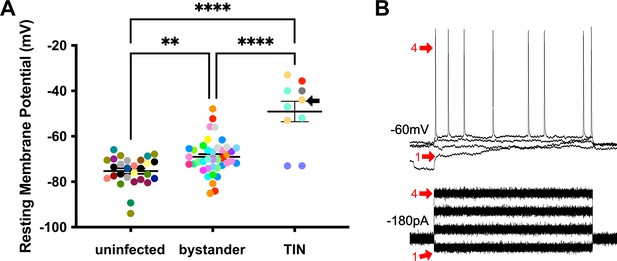
Unlike bystander neurons, Toxoplasma-injected neurons (TINs) have highly abnormal electrophysiology.
(A) Graph of the resting membrane potential of whole cell patch clamped medium spiny neurons (MSNs) in uninfected mice (−74.9 ± 2.1 mV) and bystander MSNs (−68.7 ± 4.0 mV) and TINs (−49.1 ± 4.5 mV) in infected mice. **p=0.0086, ****p=0.001, one-way ANOVA with Tukey’s post-test. For TINs, N = 10 cells recorded from six mice; uninfected and bystander described in Figure 5. Black arrow identifies TIN shown in (B). Lines, mean ± SEM. (B) Top: Example traces from TIN after hyperpolarization. Bottom: Visualization of a portion of the current injection protocol. The identified TIN was hyperpolarized to a resting membrane potential of −60 mV by the injection of −180 pA of current. The red arrow denotes a single hyperpolarized step. The numbers (1, 4) match the voltage measurement (tracings) with the steps of increasing current. Note that an action potential is finally generated at the fourth step of increasing current.
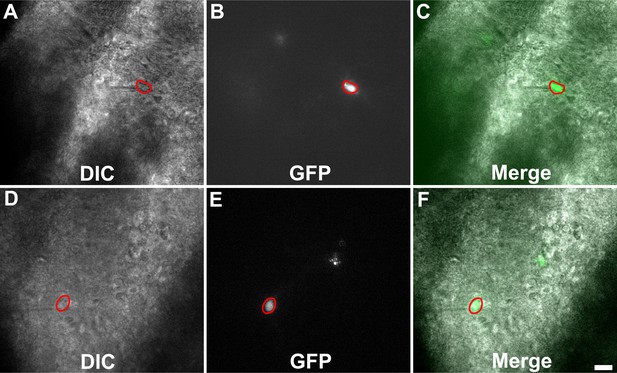
Representative images of Toxoplasma-injected neurons (TINs) used for recording.
Images are from 200-µm-thick brain sections from III-Cre infected mice. Images were obtained on an Olympus BX51. Red circles outline TINs that were patched. Scale bar = 20 µm.
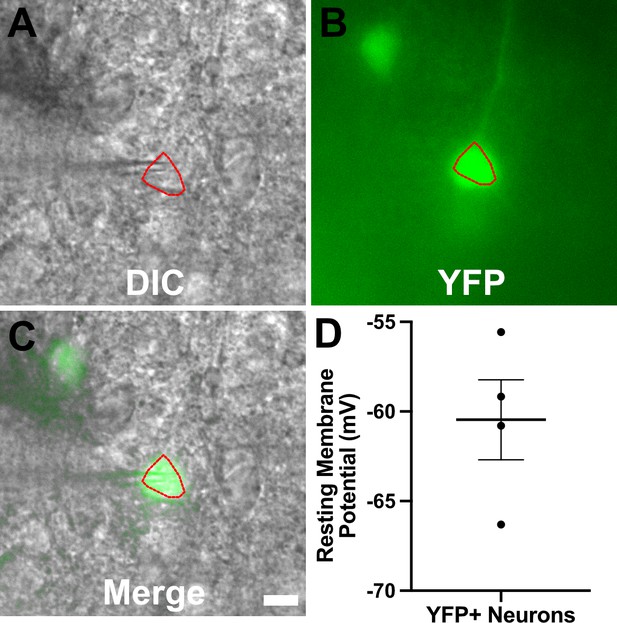
YFP+ cortical neurons from infected mice show typical resting membrane potentials.
Recording procedure carried out as for Figures 4–6 except that brain slices came from infected thy1-YFP mice (Feng et al., 2000). (A–C) Representative images for a YFP+ neuron used for recording. Images are from a 200-µm-thick brain section from a III-Cre infected thy1-YFP mouse. Images were captured as in Figure 6—figure supplement 1. Scale bar = 10 µm. (D) Graph of the resting membrane potential of whole cell patch clamped YFP+ cortical neurons, N = 4, from two mice. Lines, mean ± SEM.
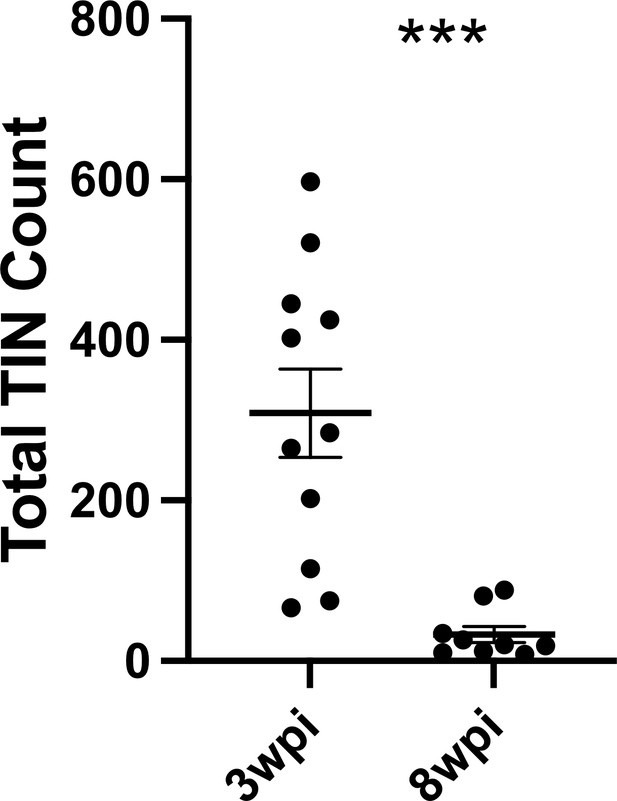
Number of Toxoplasma-injected neurons (TINs) decrease by 8 weeks post-infection (wpi).
Mice were infected and, at 3 or 8 wpi, analyzed as in Figure 1 except that total TINs numbers were quantified. Lines, mean ± SEM. ***p≤0.0001, Mann-Whitney U-test; 16–19 sections/mouse, N = 9–11 mice/time point.
-
Figure 7—source data 1
Raw data for quantification of TINs count at 3 and 8 wpi.
- https://cdn.elifesciences.org/articles/67681/elife-67681-fig7-data1-v3.xlsx
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Toxoplasma gondii) | Type II (Prugniaud) | Koshy et al., 2012 | II-Cre | Expresses mCherry and Cre recombinase |
| Strain, strain background (Toxoplasma gondii) | Type III (CEP) | Christian et al., 2014 | III-Cre | Expresses mCherry and Cre recombinase |
| Strain, strain background (Mus musculus) | Ai6 mouse, ZsGreen1 | Jackson Laboratories | Stock # 007906 RRID:IMSR_JAX:007906 | |
| Strain, strain background (Mus musculus) | thy1-YFP-H mouse | Jackson Laboratories | Stock # 003782 RRID:IMSR_JAX:003782 | |
| Antibody | Anti-ZsGreen, (rabbit polyclonal) | Clontech | Cat# 632474 RRID:AB_2491179 | IHC (1:10,000) |
| Antibody | Anti-rabbit (goat polyclonal biotinylated conjugated) | Vector Labs | Cat# BA-1000 RRID:AB_2313606 | IHC (1:500) |
| Antibody | Anti-NeuN B-clone A60 (mouse biotin conjugated) | Millipore | Cat# MAB377 RRID:AB_177621 | IF (1:500) |
| Antibody | Anti-calbindin, (rabbit polyclonal) | Sigma-Aldrich | Cat# C2724 RRID:AB_258818 | IF (1:500) |
| Antibody | Anti-parvalbumin, (rabbit polyclonal) | Abcam | Cat# ab11427 RRID:AB_298032 | IF (1:1000) |
| Antibody | Anti-FoxP2, (rabbit polyclonal) | Abcam | Cat# ab16046 RID:AB_2107107 | IF (1:1000) |
| Antibody | Anti-DARPP32, (rabbit polyclonal) | Abcam | Cat# ab40801 RRID:AB_731843 | IF (1:500) |
| Antibody | Anti-rabbit IgG Alexa Fluor 568 (goat polyclonal) | Invitrogen | Cat# A11011 RRID:AB_143157 | IF (1:500) |
| Antibody | Cy5 streptavidin | Invitrogen | Cat# SA1011 | IF (1:500) |
| Commercial Assay or kit | Avidin-biotin complex kit | Thermo Fisher Scientific | Cat# 32020 | |
| Commercial Assay or Kit | 3,3’-Diaminobenzidine (DAB) Vectastain | Vector Labs | SK-4100 RRID:AB_2336382 | |
| Other | Neurobiotin | Vector Labs | SP1120 RRID:AB_2336606 | 0.6% |
| Other | DAPI stain | Invitrogen | D1306 RRID:AB_2629482 | (1 µg/ml) |
| Software | pCLAMP | Molecular Devices | v10.7 RRID:SCR_011323 | |
| Software | Prism | GraphPad | v9.1.0 RRID:SCR_002798 | |
| Software | MATLAB | MathWorks | v2015a |
Additional files
-
Supplementary file 1
Area size and percentages of whole brain for quantified regions.
- https://cdn.elifesciences.org/articles/67681/elife-67681-supp1-v3.xlsx
-
Supplementary file 2
p-Values for enrichment scores for the 12 quantified regions.
- https://cdn.elifesciences.org/articles/67681/elife-67681-supp2-v3.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/67681/elife-67681-transrepform-v3.docx





