A single synonymous nucleotide change impacts the male-killing phenotype of prophage WO gene wmk
Figures
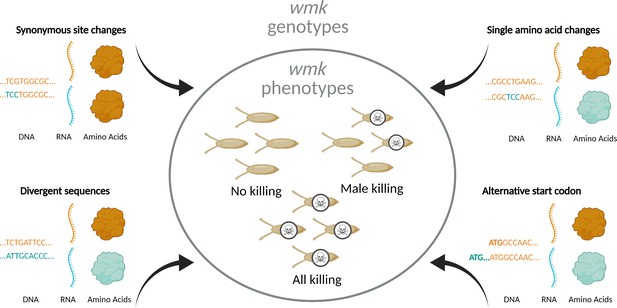
Overview of experimental design.
To investigate the genotype-phenotype landscape, we transgenically expressed wmk homologs with varying degrees of genetic changes. These sequences are codon-optimized based on different codon biases due to different tRNA abundances in the divergent bacterial source and eukaryotic destination species (Plotkin and Kudla, 2011; Gustafsson et al., 2004). Transgenic wmk in Drosophila melanogaster embryos results in three different phenotypes: no killing, male killing, and killing of males and females. Compared to wMel wmk, these transgenes were either divergent homologs from other Wolbachia strains, a homolog with a single amino acid change, homologs with an additional nine codons at the 5’ ends of the genes starting at an alternative upstream start codon, or variants with a single synonymous codon or nucleotide difference. These genotypes resulted in varying degrees of RNA sequence- and amino acid-level changes. Created with BioRender.com.
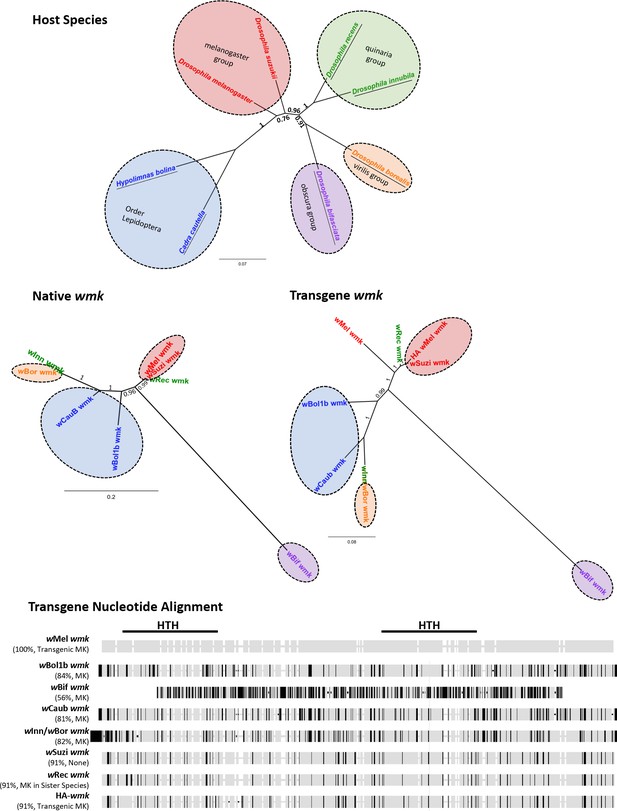
Homologs of wmk tested in this study include variation in native gene and transgene sequence identity as well as host species.
(Top) Bayesian nucleotide phylogeny of insect hosts based on 652 bp of the cytochrome oxidase subunit 1 (COI) gene from D. melanogaster, H. bolina, D. bifasciata, C. cautella, D. innubila, D. borealis, D. suzukii, and D. recens. Branch labels and scale bar indicate posterior probability. Species names are colored either by group within the Drosophila genus or by Order Lepidoptera. Underlines indicate hosts in which male-killing strains have been reported. Colored circles indicate host species group. (Middle) Bayesian nucleotide phylogenies of native (middle left, non-transgenic) or transgene (middle right) wmk sequences based on 690 or 686 bp, respectively. Label colors reflect groups from host phylogeny for comparison. Colored circles indicate host species group. Branch labels and scale bar indicate posterior probability. wBif wmk branches distantly due to a highly divergent sequence. wBor and wInn wmk share a branch because they share the same transgenic sequence. (Bottom) Nucleotide alignment of transgenes tested in this study as compared to the previously tested wMel wmk, with the regions encompassing the Helix-turn-helix (HTH) protein domains marked with black lines. Black ticks indicate sequence differences with the wMel wmk reference strain, light gray indicates sequence matches to the reference sequence, and white indicates an indel in at least one strain. In parentheses under strain names, percentages refer to nucleotide similarity compared to the wMel wmk transgene. MK indicates a male-killing strain in its native host, transgenic MK indicates the ability to induce a biased sex ratio transgenically only, and MK in sister species indicates the ability to kill males in a sister host species but not in the native host species. Alignment excludes the HA tag in the tagged strain, which is located between the two HTH domains.
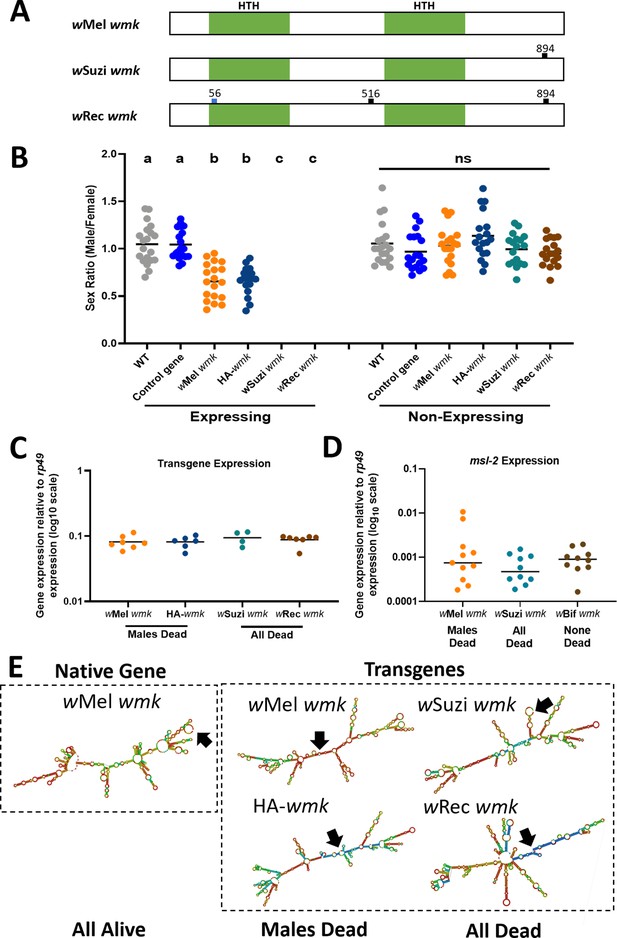
Transgenic expression of closelyrelated wmk homologs causes male-killing and all-killing phenotypes in D. melanogaster.
(A) Schematic of wMel, wSuzi, and wRec wmk native nucleotide sequences. The blue tick mark indicates a non-synonymous nucleotide difference. Black tick marks indicate synonymous nucleotide changes. Numbers indicate nucleotide position across the entire 912 nucleotide sequence. (B) Sex ratios of adult flies are shown for expressing (Act5c-Gal4) and non-expressing (CyO) embryonic offspring. Each sample point represents the adult offspring (N = 50–157, mean 86) produced by a replicate family of ten mothers and two fathers, with expressing and non-expressing flies of a given genotype being siblings. Bars represent the mean sex ratio. Statistics are based on a Kruskal-Wallis, one-way ANOVA followed by Dunn’s correction across either expressing or non-expressing flies. wRec and wSuzi wmk have no points in the expressing category due to death of most or all males and females. HA-wmk contains a 3 X HA tag in the linker region between the two helix-turn-helix domains. This experiment was performed twice. Data and statistical outputs are available in Figure 2—source data 1 and Figure 2—source data 2, respectively. (C) Gene expression in embryos 4–5 h AED of each indicated wmk transgene from (B), relative to Drosophila housekeeping gene, rp49. There is no significant difference in expression based on a Kruskal-Wallis one-way ANOVA followed by Dunn’s correction. Data and statistical outputs are available in Figure 2—source data 3 and Figure 2—source data 4, respectively. (D) Gene expression in embryos 4–5 h AED of the host msl-2 dosage compensation gene relative to rp49 under simultaneous expression of the indicated transgene. There is no significant difference in expression based on a Kruskal-Wallis one-way ANOVA followed by Dunn’s correction. Data and statistical outputs are available in Figure 2—source data 5 and Figure 2—source data 6, respectively. (E) Predicted RNA secondary structures of native wMel wmk and several transgene strains. Black arrows point to the location of the start codon within each structure.
-
Figure 2—source data 1
Data for sex ratios of closely-related homologs corresponding to Figure 2B.
- https://cdn.elifesciences.org/articles/67686/elife-67686-fig2-data1-v2.xlsx
-
Figure 2—source data 2
Statistical output of Kruskal-Wallis test corresponding to sex ratios of closelyrelated homologs in Figure 2B.
- https://cdn.elifesciences.org/articles/67686/elife-67686-fig2-data2-v2.xlsx
-
Figure 2—source data 3
Data for qPCR of closely-related transgenes corresponding to Figure 2C.
- https://cdn.elifesciences.org/articles/67686/elife-67686-fig2-data3-v2.xlsx
-
Figure 2—source data 4
Statistical output of Kruskal-Wallis test corresponding to qPCR for transgene expression in Figure 2C.
- https://cdn.elifesciences.org/articles/67686/elife-67686-fig2-data4-v2.xlsx
-
Figure 2—source data 5
Data for qPCR of msl-2 expression with transgene expression corresponding to Figure 2D.
- https://cdn.elifesciences.org/articles/67686/elife-67686-fig2-data5-v2.xlsx
-
Figure 2—source data 6
Statistical output of Kruskal-Wallis test corresponding to qPCR for msl-2 expression in Figure 2D.
- https://cdn.elifesciences.org/articles/67686/elife-67686-fig2-data6-v2.xlsx
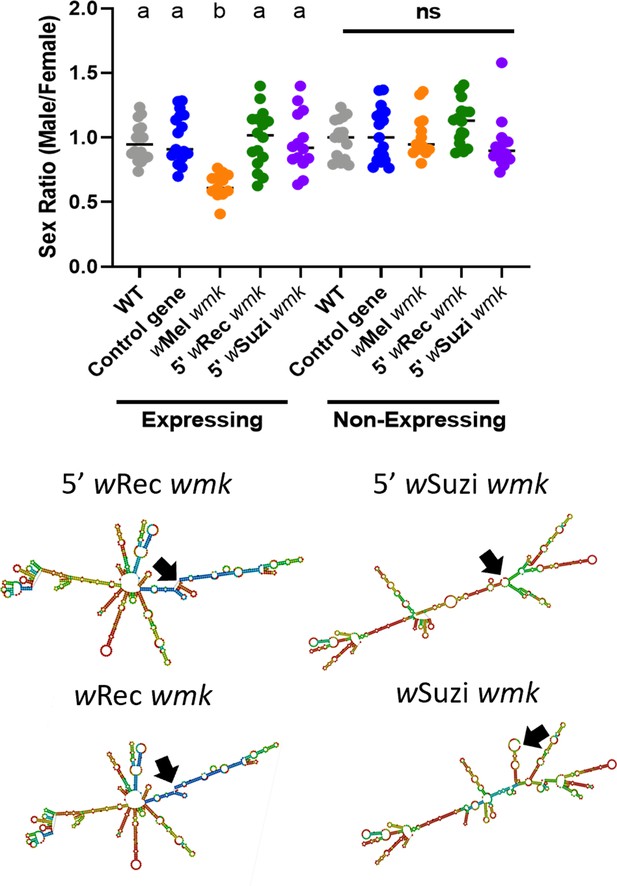
wRec and wSuzi transgenes expressed with an alternative start codon lose their transgenic phenotypes.
(Top) Sex ratios of adult flies are shown for expressing (Act5c-Gal4) and non-expressing (CyO) offspring. Each sample point represents the adult offspring (N = 50–120, mean 84) produced by a replicate family of 10 mothers and 2 fathers, with expressing and non-expressing flies of a given genotype being siblings. Bars represent the mean sex ratio. Statistics are based on a Kruskal-Wallis one-way ANOVA followed by Dunn’s correction across either expressing or non-expressing flies. This experiment was performed twice. Data and statistical outputs are available in and , respectively. (Bottom) Predicted RNA secondary structures of the wRec and wSuzi wmk transgenes with the additional 5’ sequence exhibit slight structural changes compared to their non-lengthened counterparts (included again from Figure 2 for ease of comparison). Black arrows point to the location of the start codon within each structure.
-
Figure 2—figure supplement 1—source data 1
Data for sex ratios of 5’ alternative start codon transgene expression corresponding to Figure 2—figure supplement 1.
- https://cdn.elifesciences.org/articles/67686/elife-67686-fig2-figsupp1-data1-v2.xlsx
-
Figure 2—figure supplement 1—source data 2
Statistical output of Kruskal-Wallis test corresponding to sex ratios of 5’ alternative start codon transgene expression in Figure 2—figure supplement 1.
- https://cdn.elifesciences.org/articles/67686/elife-67686-fig2-figsupp1-data2-v2.xlsx
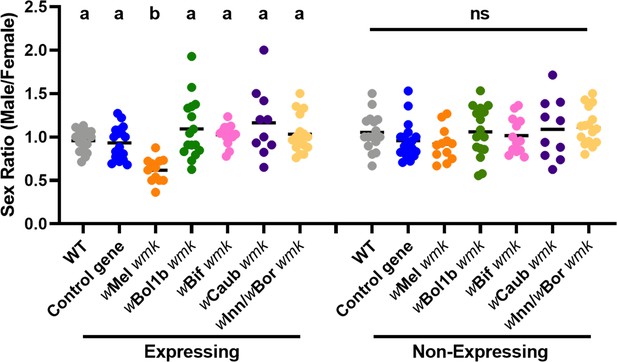
Divergent homologs of wmk from male-killing strains do not induce a biased sex ratio in D. melanogaster.
Sex ratios of adult flies are shown from either expressing (Act5c-Gal4) or non-expressing (CyO) offspring. WT refers to the background insertion line and Control gene refers to the WD0034 control transgene that induces no sex ratio bias. Each sample point represents the adult offspring (N = 50–132, mean 69) produced by a replicate family of ten mothers and two fathers, with expressing and non-expressing flies of a given genotype being siblings. Bars represent the mean sex ratio. Statistics are based on a Kruskal-Wallis one-way ANOVA followed by Dunn’s correction across either expressing or non-expressing flies. This experiment was performed twice. Data and statistical outputs are available in Figure 3—source data 1 and Figure 3—source data 2, respectively.
-
Figure 3—source data 1
Data for sex ratios of distantly-related homologs in Figure 3.
- https://cdn.elifesciences.org/articles/67686/elife-67686-fig3-data1-v2.xlsx
-
Figure 3—source data 2
Statistical output of Kruskal-Wallis test corresponding to sex ratios of divergent homologs in Figure 3.
- https://cdn.elifesciences.org/articles/67686/elife-67686-fig3-data2-v2.xlsx
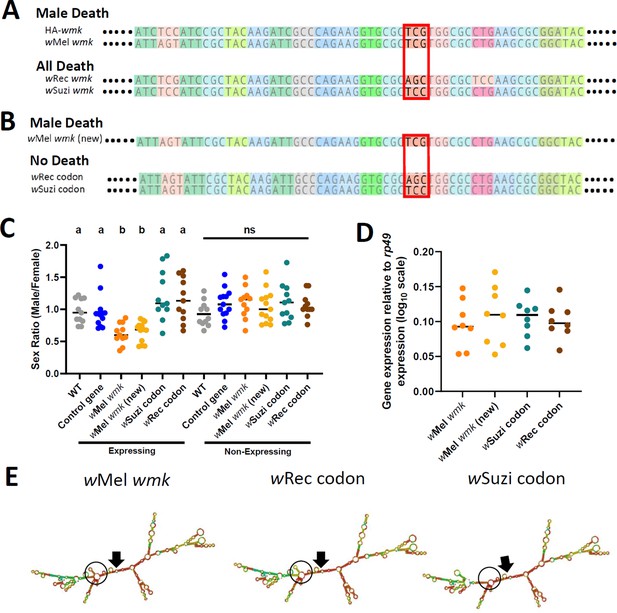
Synonymous nucleotide changes in the 16th codon position of wmk alters resulting phenotype.
(A) Sequence alignment of transgenic wmk homologs. The codon farthest on the left is the fourth codon in the sequence, and the highlighted codon is the 16th, with the farthest right representing the 23rd codon, and ellipses indicating codons continuing on either side. The red box outlines where the genotypes cluster by phenotype. The ‘HA-wmk’ and ‘wMel wmk’ genotypes share the same codon in this position, and both induce male-specific death. The ‘wRec wmk’ and ‘wSuzi wmk’ genotypes both exhibit different codons from the previous two and exhibit an all-killing phenotype. Colors correlate with amino acid identity. (B) Sequence alignment of transgenes with either the wMel wmk sequence made anew (wMel wmk new), or with the 16th codon (red box) replaced with the synonymous codons from the wRec and wSuzi wmk transgenes. The colors and symbols reflect those in (A). (C) Sex ratios of adult transgenic flies are shown for expressing (Act5c-Gal4) and non-expressing (CyO) offspring that include the original transgene wMel wmk strain used in previous figures, along with the newly created identical wMel wmk (new) transgene and the additional transgenes with the single codon swapped out for the indicated codons noted in (A). wSuzi codon and wRec codon refer to the strains that have the same sequence as the wMel wmk, but with one or three silent sites changed in the single codon at the 16th amino acid position. Each sample point represents the adult offspring (N = 50–161, mean 73) produced by a replicate family of 10 mothers and two fathers, with expressing and non-expressing flies of a given genotype being siblings. Bars represent the mean sex ratio. Statistics are based on a Kruskal-Wallis one-way ANOVA followed by Dunn’s correction across either expressing or non-expressing flies. This experiment was performed twice. Data and statistical outputs are available in Figure 4—source data 1 and Figure 4—source data 2, respectively. (D) Gene expression in embryos 4–5 h AED denotes expression of each transgene relative to that of rp49. There is no significant difference in expression based on a Kruskal-Wallis one-way ANOVA followed by Dunn’s correction. Data and statistical outputs are available in Figure 4—source data 3 and Figure 4—source data 4, respectively. (E) Predicted RNA secondary structures are shown for the wMel wmk transcript compared to both of the wRec or wSuzi codon transgenes exhibiting slight structural differences. The structure for transgene wMel wmk is included again from Figure 3 for ease of comparison. Black arrows point to the location of the start codon within each structure. Black circles highlight a key area of difference between the structures, with a stem absent in the wSuzi codon strain, and different base pair match probabilities calculated for each as indicated by color (blue to red, low to high probability). Within the black circle, the wSuzi codon transgene structure is missing a predicted stem that the others have. The stem in the wRec codon line, while present, has a weaker prediction as noted by the cooler colors, so there may be structural differences compared to the wMel wmk model.
-
Figure 4—source data 1
Data for sex ratios from expression of transgenes with single codon changes corresponding to Figure 4C.
Data for qPCR from expression of transgenes with single codon changes corresponding to Figure 4D.
- https://cdn.elifesciences.org/articles/67686/elife-67686-fig4-data1-v2.xlsx
-
Figure 4—source data 2
Data for qPCR from expression of transgenes with single codon changes corresponding to Figure 4D.
- https://cdn.elifesciences.org/articles/67686/elife-67686-fig4-data2-v2.xlsx
-
Figure 4—source data 3
Data for qPCR from expression of transgenes with single codon changes corresponding to Figure 4D.
- https://cdn.elifesciences.org/articles/67686/elife-67686-fig4-data3-v2.xlsx
-
Figure 4—source data 4
Statistical output of Kruskal-Wallis test corresponding to qPCR from expression of transgenes with single codon changes in Figure 4D.
- https://cdn.elifesciences.org/articles/67686/elife-67686-fig4-data4-v2.xlsx
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Wolbachia pipientis) | WD0626 | NCBI | NCBI:WD_RS02815 | Also known as wmk (WO-mediated killing) |
| Genetic reagent (D. melanogaster) | Act5c-Gal4/CyO | Bloomington Drosophila Stock Center | BDSC:3953; FlyBase FBti0012290 | P{AyGAL4}25 |
| Genetic reagent (D. melanogaster) | WT (y1w67c23; P[CaryP]P2) | Bloomington Drosophila Stock Center | BDSC:8622; FlyBase FBti0040535 | WT strain used in this study; P{CaryP}attP2 |
| Genetic reagent (D. melanogaster) | wMel wmk | This paper; Perlmutter et al., 2019; PMID:31504075 | Expresses codon- optimized transgene; UAS promoter | |
| Genetic reagent (D. melanogaster) | wBol1b wmk | This paper | Expresses codon- optimized transgene; UAS promoter | |
| Genetic reagent (D. melanogaster) | wBif wmk | This paper | Expresses codon- optimized transgene; UAS promoter | |
| Genetic reagent (D. melanogaster) | wCaub wmk | This paper | Expresses codon- optimized transgene; UAS promoter | |
| Genetic reagent (D. melanogaster) | wInn/wBor wmk | This paper | Expresses codon- optimized transgene; UAS promoter; wInn and wBor wmk have same exact sequence | |
| Genetic reagent (D. melanogaster) | wSuzi wmk | This paper | Expresses codon- optimized transgene; UAS promoter | |
| Genetic reagent (D. melanogaster) | wRec wmk | This paper | Expresses codon- optimized transgene; UAS promoter | |
| Genetic reagent (D. melanogaster) | HA-wmk | This paper | Expresses codon- optimized transgene; UAS promoter; 3 X HA tag epitope in linker between HTH domains of wMel wmk | |
| Genetic reagent (D. melanogaster) | 5’ wRec wmk | This paper | Expresses codon-optimized transgene; UAS promoter; Sequence has additional nine amino acids starting at upstream alternative start codon | |
| Genetic reagent (D. melanogaster) | 5’ wSuzi wmk | This paper | Expresses codon-optimized transgene; UAS promoter; Sequence has additional nine amino acids starting at upstream alternative start codon | |
| Genetic reagent (D. melanogaster) | wMel wmk (new) | This paper | Expresses codon-optimized transgene; UAS promoter; Same exact sequence as wMel wmk, newly transformed strain | |
| Genetic reagent (D. melanogaster) | wSuzi codon | This paper | Expresses codon-optimized transgene; UAS promoter; Same as wMel wmk, but with 16th amino acid position using TCC Serine codon from wSuzi wmk strain | |
| Genetic reagent (D. melanogaster) | wRec codon | This paper | Expresses codon-optimized transgene; UAS promoter; Same as wMel wmk, but with 16th amino acid position using AGC Serine codon from wRec wmk strain | |
| Recombinant DNA reagent | pTIGER (plasmid) | Ferguson et al., 2012; PMID:22328499 | Modified pUASp plasmid for enhanced germline expression under Gal4/UAS control | |
| Sequence-based reagent | Rp49_F | This paper | PCR primers | CGGTTACGGAT CGAACAAGC |
| Sequence-based reagent | Rp49_R | This paper | PCR primers | CTTGCGCTTCT TGGAGGAGA |
| Sequence-based reagent | wmk_homologs_opt_F | This paper | PCR primers | CTGTATGCCATTG CCGAGACCCT |
| Sequence-based reagent | wmk_homologs_opt_R | This paper | PCR primers | TCACCAGATCCTTG GCGATCTTCATC |
| Sequence-based reagent | Msl-2_F | This paper | PCR primers | GGATTAACGCGGT CTAAGCATGTGTAACTG |
| Sequence-based reagent | Msl-2_R | This paper | PCR primers | GTATGCCGTCTG GGCCATGATG |
| Commercial assay or kit | Direct-zol RNA MiniPrep Kit | Zymo | R2051 | |
| Commercial assay or kit | Superscript VILO cDNA Synthesis Kit | ThermoFisher | 11754050 | |
| Chemical compound, drug | DNase, RNase-free | Ambion, Life Technologies | AM2222 | |
| Chemical compound, drug | iTaq Universal SYBR Green Mix | Bio-Rad | 1725120 | |
| Software, algorithm | GraphPad Prism 8 | GraphPad Prism 8 | RRID:SCR_002798 | |
| Software, algorithm | Geneious Pro v.2019.2; Geneious Pro v.2020.2.4 | Geneious | RRID:SCR_010519 | |
| Software, algorithm | jModelTest | jModelTest | RRID:SCR_015244 | |
| Software, algorithm | RNAfold WebServer | University of Vienna, Gruber et al., 2008, Lorenz et al., 2011 | PMID:18424795; PMID:22115189 | http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi |
-
**Reagents source from this paper may be obtained from Bordenstein lab.
Additional files
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/67686/elife-67686-transrepform1-v2.docx
-
Source data 1
Source data for all graphical data sets and statistical tests performed for this study.
- https://cdn.elifesciences.org/articles/67686/elife-67686-supp1-v2.xlsx





