The human amniotic epithelium confers a bias to differentiate toward the neuroectoderm lineage in human embryonic stem cells
Figures
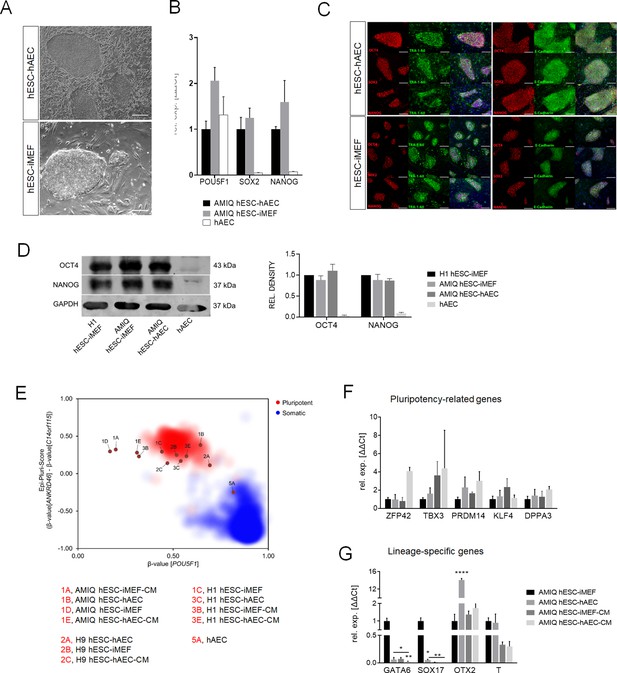
Characterization of pluripotency in alternative human embryonic stem cell (hESC)-human amniotic epithelial cell (hAEC) and conventional hESC-inactivated mouse embryonic fibroblast (iMEF).
(A) Representative phase-contrast micrographs of Amicqui-1 hESC on hAEC feeder layer (AMIQ hESC-hAEC) and Amicqui-1 hESC on iMEF feeder layer (AMIQ hESC-iMEF). Scale bar, 200 µm. (B) Expression of the ‘core pluripotency’ genes in AMIQ hESC-hAEC, AMIQ hESC-iMEF, and hAEC alone by quantitative Polymerase Chain Reaction (qPCR). Data are mean ± SE, n=three biological samples per group, and three repetitions per sample. (C) Representative epifluorescence micrographs of AMIQ hESC-iMEF and AMIQ hESC-hAEC colonies with double immunostaining for each OCT4, SOX2, and NANOG transcription factor with TRA-1–60 surface antigen or E-cadherin cell-cell adhesion molecule. Scale bar, 50 µm. (D) Detection and semiquantitative analysis of OCT4 and NANOG proteins in H1 hESC-iMEF, AMIQ hESC-iMEF, AMIQ hESC-hAEC, and HAEC alone by western blot. n=three biological samples per group. (E) Epi-Pluri-Score reveals that hESC lines are pluripotent regardless of different culture conditions. DNA methylation (DNAm) was analyzed at three specific CpG sites. One of these CpGs was localized within the pluripotency-associated gene POU5F1 (also known as OCT4). Furthermore, the difference in DNAm levels (β-values) of CpGs in ANKRD46 and C14orf115 was determined and combined as Epi-Pluri-Score (Lenz et al., 2015). The red and blue clouds refer to DNAm profiles (all Illumina HumanMethylation27 BeadChip platform) of 264 pluripotent and 1951 non-pluripotent cell preparations, respectively. (F) Analysis of pluripotency-related genes (F) and lineage-specific genes (G) expression in Amicqui-1 maintained on iMEF feeder layer (AMIQ hESC-iMEF) by reverse transcription qPCR (RT-qPCR); Amicqui-1 on hAEC feeder layer (AMIQ hESC-hAEC); feeder-free Amicqui-1 with conditioned media of iMEF (AMIQ hESC-iMEF-CM); feeder-free Amicqui-1 with conditioned media of hAEC (AMIQ hESC-hAEC-CM). Data are mean ± SE, n=three biological samples per group, and three repetitions per sample. * p<0.05, ** p<0.01, and **** p<0.0001 with AMIQ hESC-iMEF as control.
-
Figure 1—source data 1
Raw unedited blots (replicate one) for detection of NANOG and OCT4 in human embryonic stem cells (hESCs).
Left: replica one of the western blot for NANOG (green, ∼37 kDa) and internal control GAPDH (red, ∼37 kDa). Right: replica one of the western blot for OCT4 (green, ∼42 kDa) and internal control GAPDH (red, ∼37 kDa). The first left lane of each blot corresponds to the molecular weight ladder; second lane, H1 hESC-inactivated mouse embryonic fibroblast (iMEF); third lane, AMIQ hESC-iMEF; fourth lane, AMIQ hESC-human amniotic epithelial cell (hAEC); fifth lane, hAEC.
- https://cdn.elifesciences.org/articles/68035/elife-68035-fig1-data1-v2.zip
-
Figure 1—source data 2
Raw unedited blots (replicate two) for detection of OCT4 and NANOG in human embryonic stem cells (hESCs).
Left: replica two of the western blot for OCT4 (red, ∼42 kDa) and internal control GAPDH (green, ∼37 kDa). Right: replica two of the western blot for NANOG (red, ∼37 kDa) and internal control GAPDH (green, ∼37 kDa). The first left lane of each blot corresponds to the molecular weight ladder; second lane, H1 hESC-inactivated mouse embryonic fibroblast (iMEF); third lane, AMIQ hESC-iMEF; fourth lane, AMIQ hESC-human amniotic epithelial cell (hAEC); fifth lane, hAEC.
- https://cdn.elifesciences.org/articles/68035/elife-68035-fig1-data2-v2.zip
-
Figure 1—source data 3
Raw unedited blots (replicate three) for detection of NANOG and OCT4 in human embryonic stem cells (hESCs).
Left: replica one of the western blot for NANOG (green, ∼37 kDa) and internal control GAPDH (red, ∼37 kDa). Right: replica one of the western blot for OCT4 (green, ∼42 kDa) and internal control GAPDH (red, ∼37 kDa). The first left lane of each blot corresponds to the molecular weight ladder; second lane, H1 hESC-inactivated mouse embryonic fibroblast (iMEF); third lane, AMIQ hESC-iMEF; fourth lane, AMIQ hESC-human amniotic epithelial cell (hAEC); fifth lane, hAEC.
- https://cdn.elifesciences.org/articles/68035/elife-68035-fig1-data3-v2.zip
-
Figure 1—source data 4
Uncropped blots with the bands labeled of representative western blot Figure 1D.
Left: representative bands labeled for NANOG (green, ∼37 kDa). Right: representative bands labeled for OCT4 (green, ∼42 kDa) and internal control GAPDH (red, ∼37 kDa). First left lane for each blot, molecular weight ladders (M.W.); second lane, H1 human embryonic stem cell (hESC)-inactivated mouse embryonic fibroblast (iMEF); third lane, AMIQ hESC-iMEF; fourth lane, AMIQ hESC-human amniotic epithelial cell (hAEC); fifth lane, hAEC.
- https://cdn.elifesciences.org/articles/68035/elife-68035-fig1-data4-v2.zip
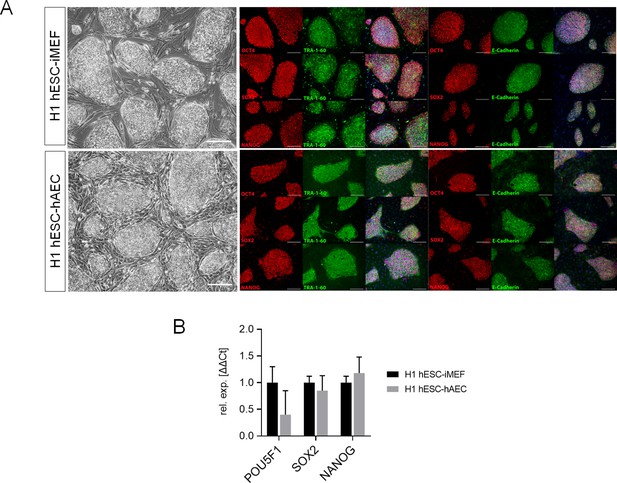
Detection of pluripotency markers in H1 human embryonic stem cell (hESC) on inactivated mouse embryonic fibroblast (iMEF) or human amniotic epithelial cell (hAEC).
(A) Representative micrographs for H1 maintained on iMEF or hAEC feeder layer (H1 hESC-iMEF or H1 hESC-hAEC) colonies of phase-contrast (scale bar, 200 µm) and epifluorescence (scale bar, 50 µm) with double immunostaining for each OCT4, SOX2, and NANOG transcription factor with TRA-1–60 surface antigen or E-cadherin cell-cell adhesion molecule. (B) Expression of the ‘core pluripotency’ genes in H1 hESC-iMEF and H1 hESC-hAEC by quantitative PCR (qPCR). Data are mean ± SE, n=three biological samples per group and three repetitions per sample.
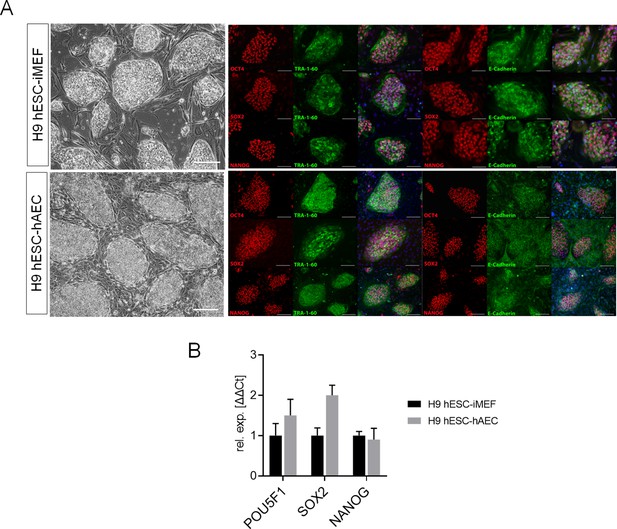
Detection of pluripotency markers in H9 human embryonic stem cell (hESC) on inactivated mouse embryonic fibroblast (iMEF) or human amniotic epithelial cell (hAEC).
(A) Representative micrographs for H9 hESC maintained on iMEF or hAEC feeder layer (H9 hESC-iMEF or H9 hESC-hAEC) colonies of phase-contrast (scale bar, 200 µm) and epifluorescence (scale bar, 50 µm) with double immunostaining for each OCT4, SOX2, and NANOG transcription factor with TRA-1–60 surface antigen or E-cadherin cell-cell adhesion molecule. (B) Expression of the ‘core pluripotency’ genes in H9 hESC-iMEF and H9 hESC-hAEC by quantitative PCR (qPCR). Data are mean ± SE, n=three biological samples per group and three repetitions per sample.
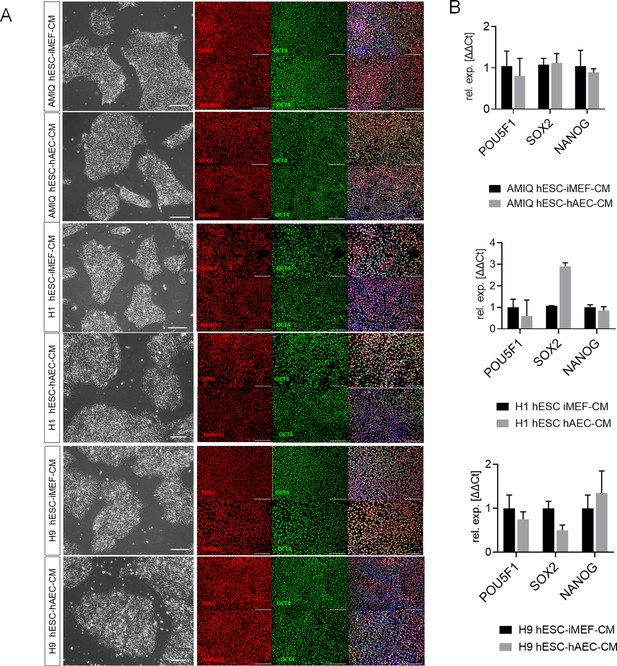
Characterization of human embryonic stem cell (hESC) lines maintained on feeder-free conditions.
(A) Representative micrographs for Amicqui-1, H1, and H9 on Matrigel with conditioned media of inactivated mouse embryonic fibroblast (iMEF) (hESC-iMEF-CM) or human amniotic epithelial cell (hAEC) (hESC-hAEC-CM) of phase-contrast (scale bar, 100 µm) and epifluorescence (scale bar, 50 µm) with double immunostaining for SOX2/OCT4 or NANOG/OCT4. (B) Reverse transcription quantitative PCR (RT-qPCR) analysis of the expression of the ‘core pluripotency’ genes in AMIQ, H1, and H9 on feeder-free conditions (hESC-iMEF-CM and hESC-hAEC-CM). Data are mean ± SE, n=three biological samples per group and two repetitions per sample.
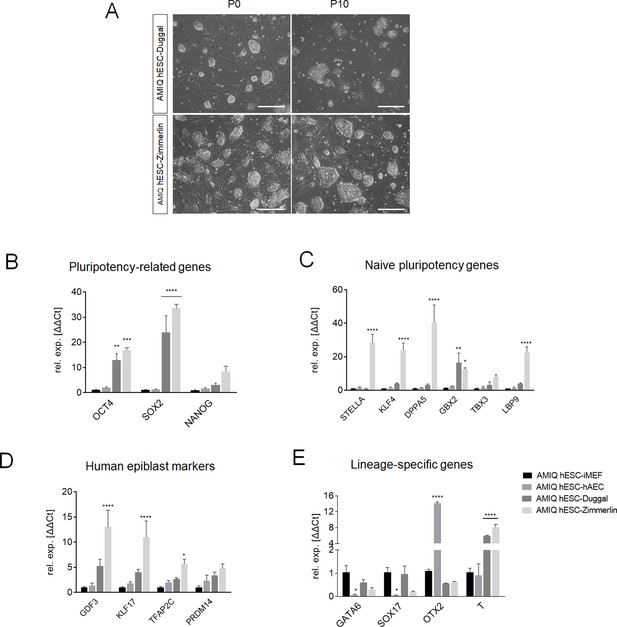
Comparison of AMIQ human embryonic stem cell (hESC)-human amniotic epithelial cell (hAEC) between primed and naïve conditions in the expression of pluripotency-related genes and lineage-specific genes.
(A) Representative phase-contrast micrographs showing the morphology of Amicqui-1 colonies in naïve conditions reported by Duggal et al., (AMIQ hESC-Duggal) and Zimmerlin (AMIQ hESC-Zimmerlin) at passage 0 and passage 10. Reverse transcription quantitative PCR (RT-qPCR) analysis to compare the expression of the ‘core pluripotency’ genes (B), naïve pluripotency markers (C), human epiblast markers (D), and lineage-specific genes (E) in Amicqui-1 maintained on conventional conditions (AMIQ hESC-inactivated mouse embryonic fibroblast [iMEF]), alternative hAEC feeder layer (AMIQ hESC-hAEC), and naïve conditions (AMIQ hESC-Duggal and AMIQ hESC-Zimmerlin). * p<0.05, ** p<0.01, ***p<0.001, and **** p<0.0001 with AMIQ hESC-iMEF as control. For each gene in A, B, and D–G, data are mean ± SE, n=three biological samples per group and three repetitions per sample.
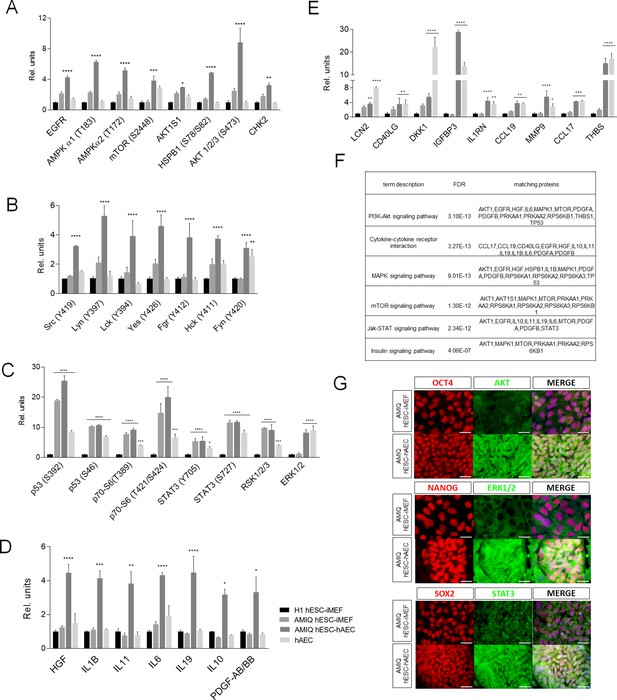
Different signaling pathways related to pluripotency and development converge in AMIQ human embryonic stem cell (hESC)-human amniotic epithelial cell (hAEC).
(A) Phospho-kinases levels increased in Amicqui-1 hESC maintained on hAEC feeder layer (AMIQ hESC-hAEC). (B) Phospho-kinases levels of SRC family in AMIQ hESC-hAEC. (C) Phospho-kinases levels increased in both AMIQ hESC-hAEC and hAEC. (D) Cytokines levels increased in AMIQ hESC-hAEC. (E) Cytokines levels increased in both AMIQ hESC-hAEC and hAEC. * p<0.05, ** p<0.01, ***p<0.001, and **** p<0.0001 using H1 hESC maintained on inactivated mouse embryonic fibroblast (iMEF) as control (H1 hESC-iMEF). For each phospho-kinase and cytokine detected, data are mean ± SE, n=three biological samples per group, and two repetitions per sample. (F) List of selected Kyoto Encyclopedia of Genes and Genomes (KEGG) and Reactome pathways obtained from Gene ontology (GO) analysis based on the proteins identified in proteome arrays. (G) Presence of AKT, ERK1/2, and STAT3 in AMIQ hESC-hAEC. Representative epifluorescence micrographs of double immunostaining for OCT4 with AKT, NANOG with ERK1/2, and SOX2 with STAT3 in AMIQ hESC-iMEF and AMIQ hESC-hAEC. All the images were obtained with an epifluorescence microscope with the same gain and exposure parameters. Scale bar, 50 µm.
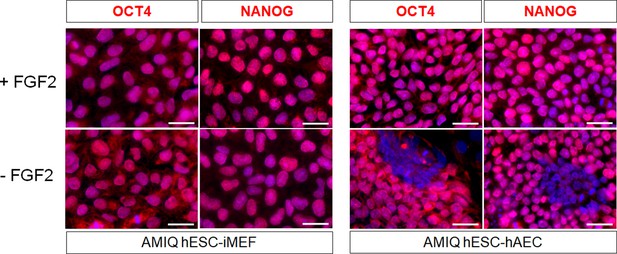
AMIQ human embryonic stem cell (hESC)-human amniotic epithelial cell (hAEC) depends on FGF2 exogenous to maintain pluripotency-related transcription factors.
Representative epifluorescence micrographs of immunostaining for OCT4 and NANOG in Amicqui-1 hESC maintained on inactivated mouse embryonic fibroblast (iMEF) feeder layer (AMIQ hESC-iMEF) or hAEC feeder layer (AMIQ hESC-hAEC) with exogenous FGF2 or 48 hr without FGF2 (scale bar, 50 µm).
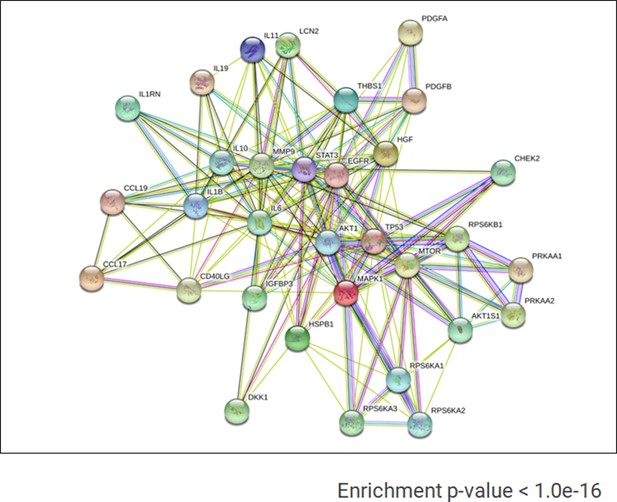
Known and predictive interactions of cytokines and kinases present in AMIQ human embryonic stem cell (hESC)-human amniotic epithelial cell (hAEC).
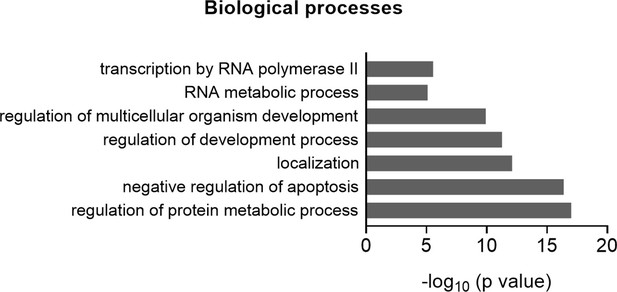
Gene ontology (GO) term analyses for biological processes based on the proteins identified in the proteome arrays.
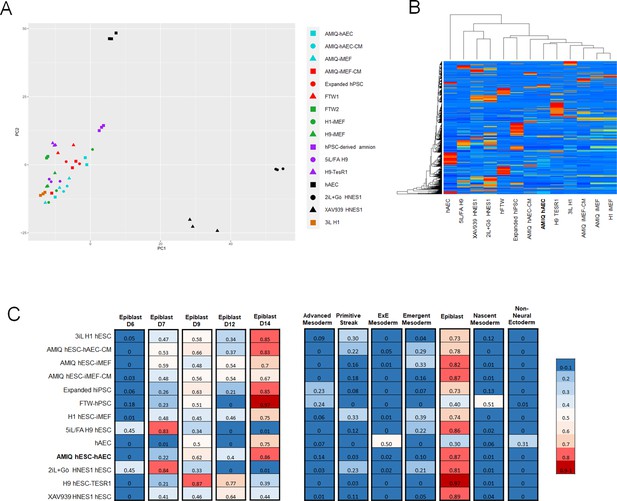
Comparison of the genome-wide expression of AMIQ human embryonic stem cell (hESC)-human amniotic epithelial cell (hAEC) with other human pluripotent stem cell (hPSC) lines and human embryonic lineages.
(A) Principal component analysis (PCA) of bulk RNA-seq data of hPSC lines. AMIQ hAEC, Amiqui hESC on alternative hAEC feeder layer; AMIQ hAEC-CM, feeder-free Amicqui-1 hESC with hAEC-conditioned media; AMIQ hESC-inactivated mouse embryonic fibroblast (iMEF), Amicqui-1 hESC on conventional iMEF feeder layer; AMIQ hESC-iMEF-CM, feeder-free Amicqui-1 hESC with iMEF-conditioned media; expanded hPSC, expanded human induced PSC (hiPSC); FTW1 and FTW2, formative hiPSCs; H1 iMEF, H1 hESC on conventional iMEF feeder layer; H9 hESC-iMEF, H9 hESC on conventional iMEF; hPSC-derived amnion, amnion derived from H9 hESC; 5iL/FA H9, naïve 5iL/FA H9 hESC; H9 hESC-TesR1, feeder-free H9 hESC with TesR1 medium; hAEC, hAEC without co-culture with hESC; 2iL +Go HNES1, naïve 2iL +Go HNES1 hESC; XAV939 HNES1, formative XAV939 HNES1 hESC; 3iL H1, 3iL H1 hESC. (B) Heatmap of signature matrix for differential expression genes of hPSC lines. (C) Relative similarity of the bulk RNA-seq of the hPSC lines with the single cell RNA sequencing (scRNA-seq) of human embryos.
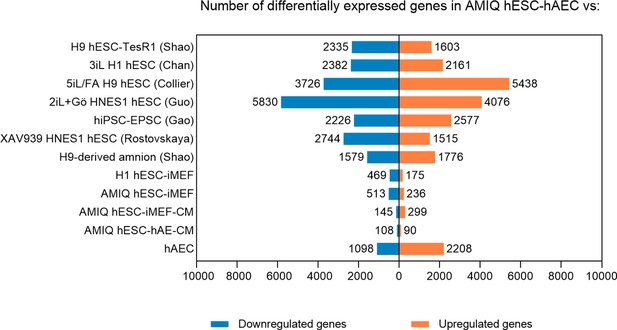
The number of genes differentially expressed in AMIQ human embryonic stem cell (hESC)-human amniotic epithelial cell (hAEC) versus human pluripotent stem cell (hPSC) lines.
Fold Change <1.5 and p-value<0.05.
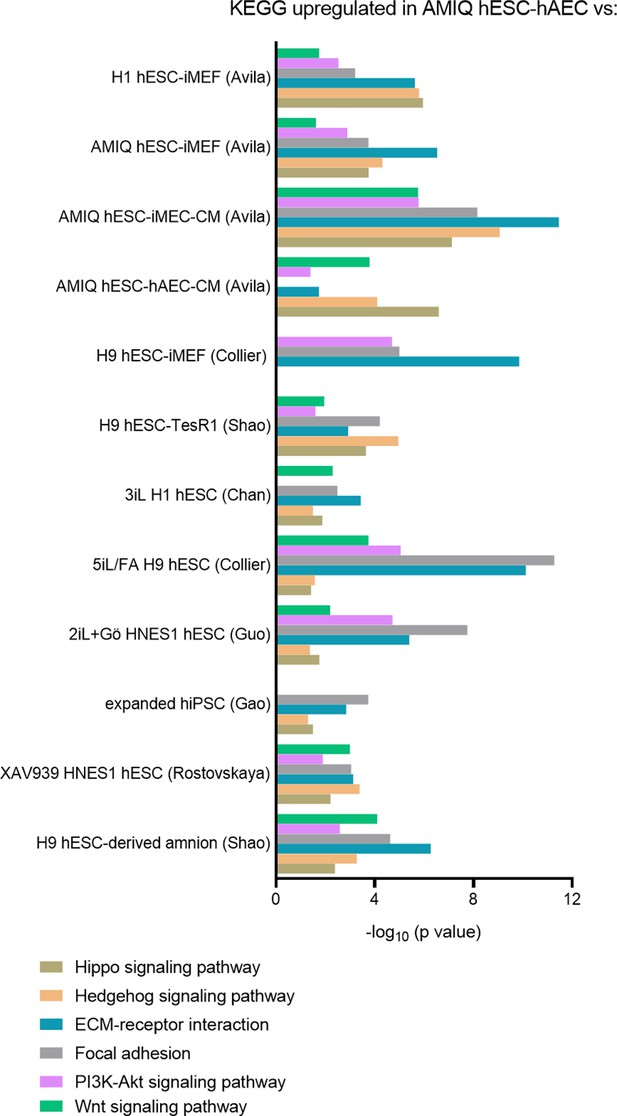
KEGG pathways upregulated in AMIQ human embryonic stem cell (hESC)-human amniotic epithelial cell (hAEC) versus human pluripotent stem cell (hPSC) lines.
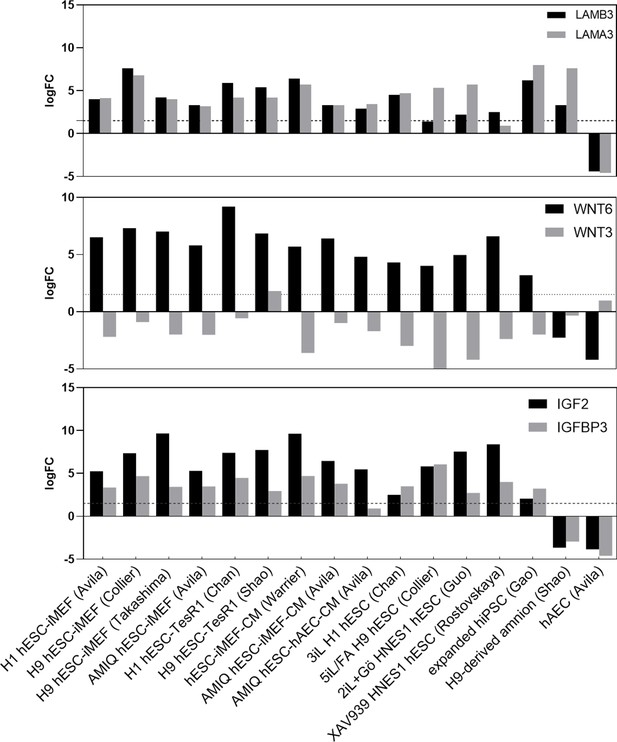
LogFC from differential gene expression (DEG) analysis (FC<1.5 and p-value<0.05) for genes in AMIQ human embryonic stem cell (hESC)-human amniotic epithelial cell (hAEC) compared with conventional primed, feeder-free, naïve, expanded, and XAV939 conditions as well as versus H9-derived amnion and hAEC isolated from membranes at term.
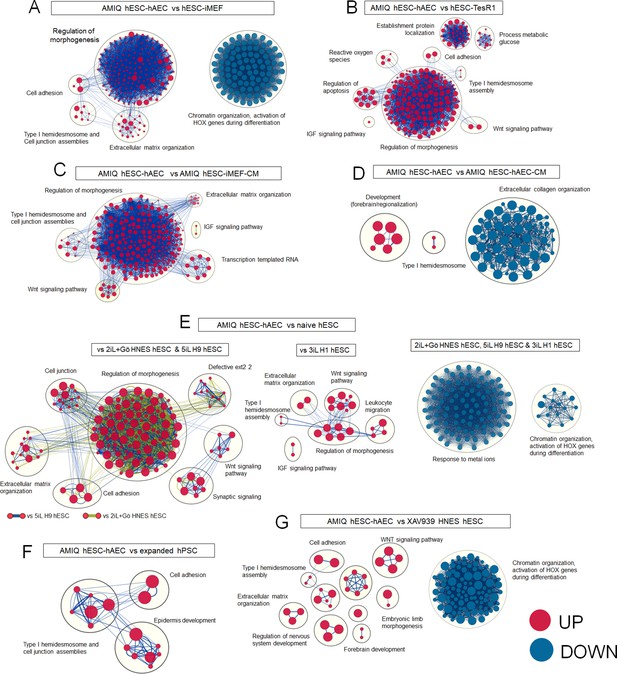
Enrichment maps of biological processes that differ in AMIQ human embryonic stem cell (hESC)-human amniotic epithelial cell (hAEC) versus several pluripotency conditions.
Clusters of nodes were created using AutoAnnotate Cytoscape application; the red and blue nodes represent upregulated and downregulated pathways in Amicqui-1 hESC maintained on the hAEC feeder layer (AMIQ hESC-hAEC), respectively; (A) versus Amicqui-1 hESC on inactivated mouse embryonic fibroblast (iMEF) feeder layer (AMIQ hESC-iMEF); (B) versus feeder-free H9 hESC growing with defined medium TesR1; (C) versus feeder-free Amicqui-1 hESC with conditioned media of iMEF (blue edges) and conditioned media of hAEC (green edges); (D) versus naïve hESC (2iL +Gö HNES hESC, 5iL H9 hESC, and 3iL H1 hESC); (E) versus expanded human pluripotent stem cell (hPSC) (H1 hESC and hiPSC); (F) versus XAV939 HNES hESC. Parameters: False discovery rate (FDR) q-value<0.01 and combined coefficient >0.375.
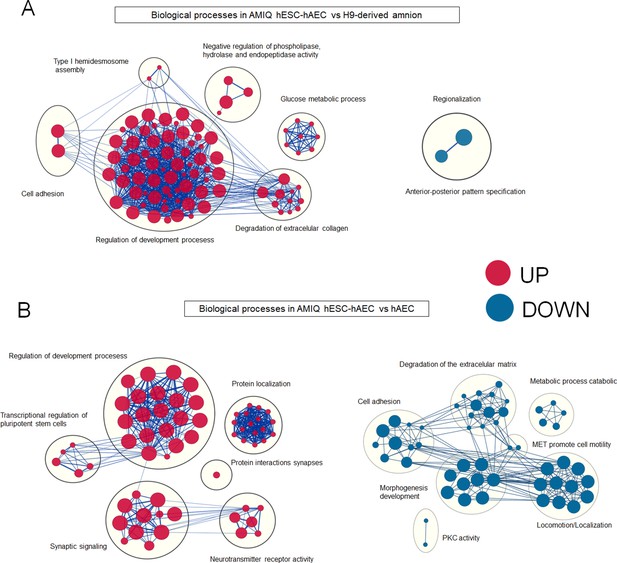
Enrichment maps of biological processes differ in AMIQ human embryonic stem cell (hESC)-human amniotic epithelial cell (hAEC) versus amnion-like cells derived from H9 (A) and hAEC isolated from amnion at term (B).
Parameters: FDR q-value<0.01 and combined coefficient >0.375. Clusters of nodes were created using AutoAnnotate Cytoscape application; the red and blue nodes represent upregulated and downregulated pathways in hESC-hAEC, respectively.
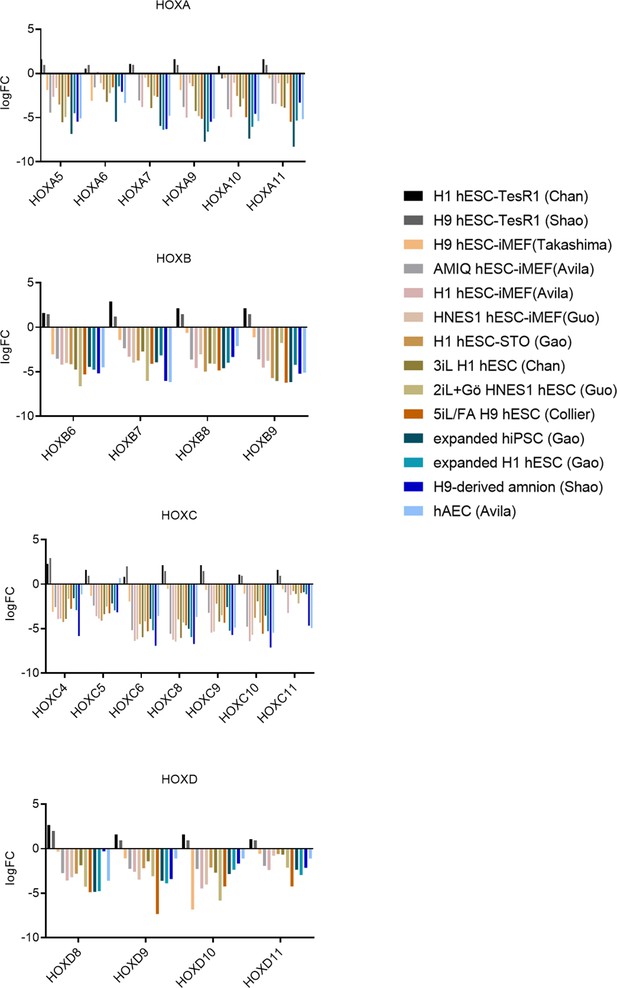
Differential expression of HOX genes in AMIQ human embryonic stem cell (hESC)-human amniotic epithelial cell (hAEC).
LogFC of differential expression of HOX genes in Amicqui-1 hESC on hAEC feeder layer (AMIQ hESC-hAEC) versus other conditions. The conditions compared with hESC-hAEC were: hESC on iMEF (H1, H9, and Amicqui-1) or STO feeder layer (H1); feeder-free hESC with TesR1 (H1 and H9); hESC on naïve conditions (3iL H1, 2iL +Go HNES, and 5iL/FA H9); expanded hPSC (H1 hESC and hiPSC); amnion-like cells derived from H9 hESC; and hAEC isolated from amnion at term (parameters FC<−1.0 with p<0.05).
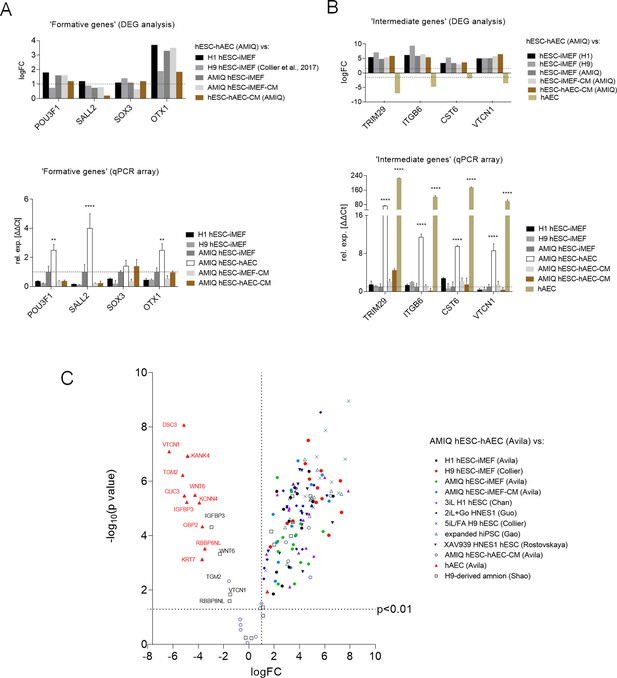
Expression of formative pluripotency-associated genes and ‘intermediate genes’ in AMIQ human embryonic stem cell (hESC)-human amniotic epithelial cell (hAEC).
(A) Differential gene expression (DEG) analysis and validation by a quantitative PCR (qPCR) array of ‘formative genes’ upregulated in Amicqui-1 hESC maintained on hAEC feeder layer (AMIQ hESC-hAEC). Data are mean ± SE, n=three biological samples per group, and two repetitions per sample. ****p<0.0001 using Amicqui-1 hESC on inactivated mouse embryonic fibroblast (iMEF) feeder layer (AMIQ hESC-iMEF) as a control for qPCR. FC<1.0 and p<0.05 for DEG. (B) DEG analysis and validation by a qPCR array of ‘intermediate genes’ upregulated in AMIQ hESC-hAEC. Data are mean ± SE, n=three biological samples per group, and two repetitions per sample. ****p<0.0001 using AMIQ hESC-iMEF control for qPCR. FC<1.0 and p<0.05 for DEG. (C) AMIQ hESC-hAEC upregulation of genes associated with an ‘intermediate population’. List of genes: C1orf21, CERCAM, DSC3, CLIC3, GBP2, KCNN4, RBBP8NL, WNT6, IGFBP3, KANK4, KRT7, ODAM, TGM2, and VTCN1. FC<1.5 and p<0.05. The conditions compared to AMIQ hESC-hAEC were: H1, H9, and Amicqui-1 hESC on iMEF feeder layer; feeder-free H9 hESC with TesR1; hESC on naïve conditions (3iL H1, 2iL +Go HNES1, and 5iL/FA H9); expanded hPSC; and hESC in formative transition (HNES1 line with XAV939 inhibitor).
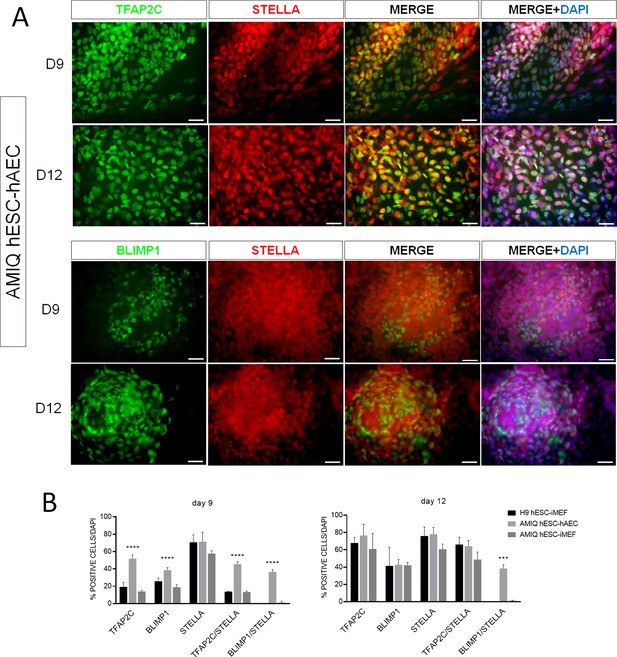
Human embryonic stem cell (hESC)-human amniotic epithelial cell (hAEC) is competent to differentiate into germline-like cells.
(A) Detection of germline markers (TFAP2C+/STELLA+ and BLIMP1+/STELLA) in day 9 and day 12 human primordial germ cells (hPGC)-like cells derived from Amicqui-1 hESC on hAEC. (B) Analysis of immunofluorescence TFAP2C+, BLIMP1+, STELLA+, TFAP2C+/STELLA + and BLIMP1+/STELLA+ in hPGC-like cells derived from hESC on hAEC or on inactivated mouse embryonic fibroblast (iMEF) on day 9 and day 12. Data are mean ± SE. n=two biological samples per group and two repetitions per sample. *** p=0.0001 and **** p<0.0001 with H9 hESC-iMEF as control.
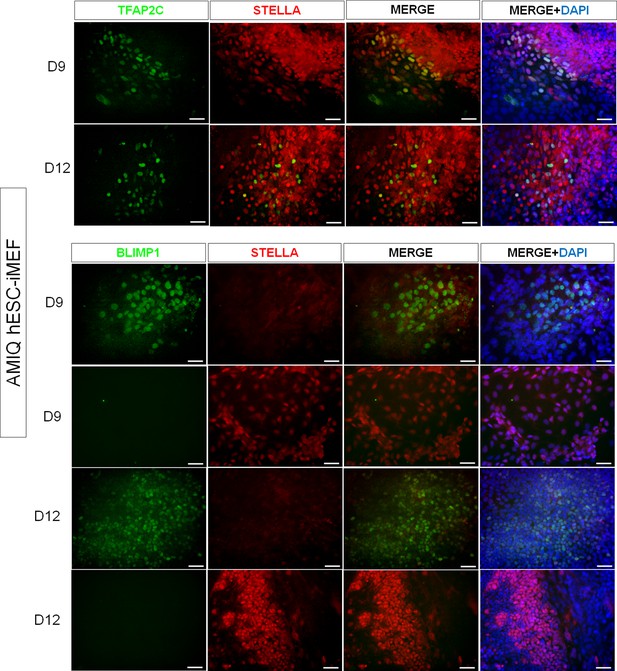
Representative immunodetection micrographs of Amicqui-1 human embryonic stem cell (hESC) on inactivated mouse embryonic fibroblast (iMEF)-derived TFAP2C+, BLIMP1+, and STELLA+ cells on day 9 and day 12.
No double labeling for BLIMP1+/STELLA+ was detected.
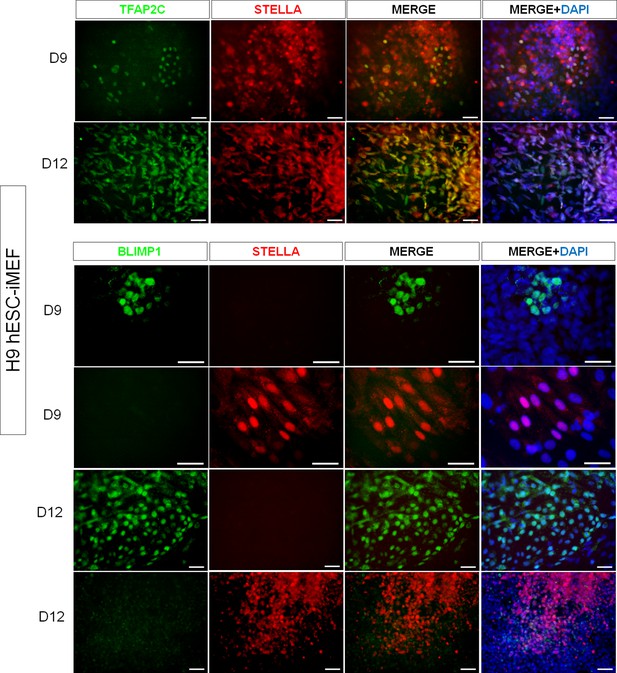
Representative immunodetection micrographs of H9 human embryonic stem cell (hESC) on inactivated mouse embryonic fibroblast (iMEF)-derived TFAP2C+, BLIMP1+, and STELLA+ cells on day 9 and day 12.
No double labeling for BLIMP1+/STELLA+ was detected.
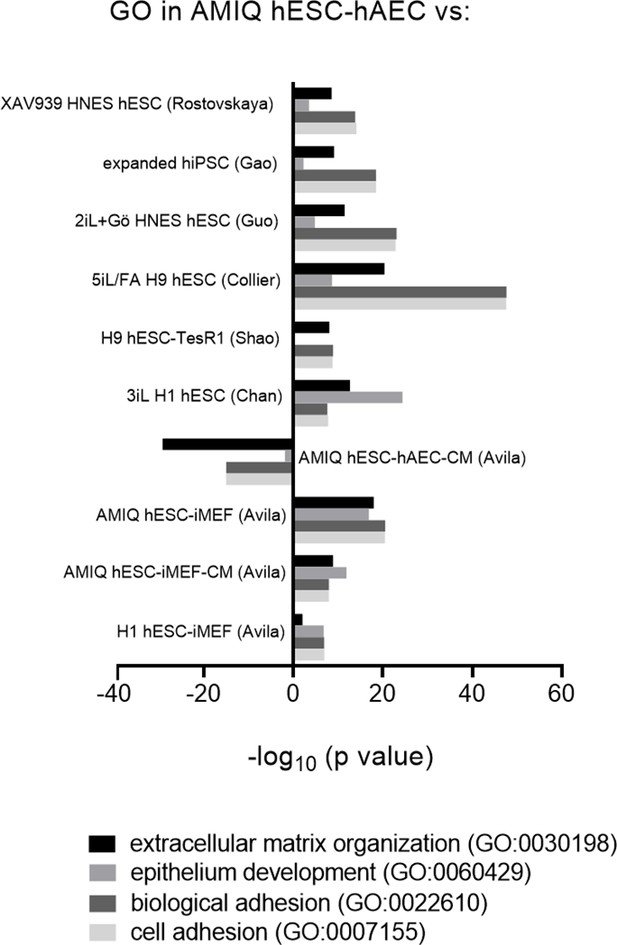
Biological processes involved in epithelialization and extracellular matrix organization upregulated in AMIQ human embryonic stem cell (hESC)-human amniotic epithelial cell (hAEC).
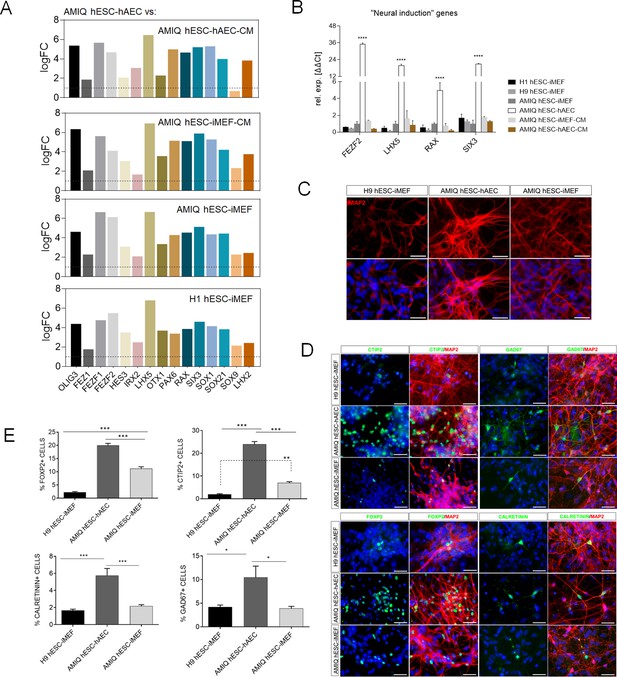
Upregulation of neural genes associated with forebrain development in human embryonic stem cell (hESC)-human amniotic epithelial cell (hAEC).
(A) Expression of genes involved in forebrain development as signature molecular in Amicqui-1 hESC maintained on hAEC feeder layer (AMIQ hESC-hAEC). Log2 fold change determined using Limma-method from neural genes upregulated in AMIQ hESC-hAEC. FC>1.0 and p<0.05. (B) Validation by quantitative PCR (qPCR) array of selected transcripts upregulated in AMIQ hESC-hAEC. Data are mean ± SE and n=three biological samples per group. ****p<0.0001 using H1 hESC-inactivated mouse embryonic fibroblast (iMEF) as control. Lines and conditions used for (A–B) analysis: AMIQ hESC-hAEC, Amiqui hESC on alternative hAEC feeder layer; H1 hESC-iMEF, H1 hESC on conventional iMEF feeder layer; H9M hESC-iMEF, H9 hESC on conventional iMEF feeder layer; AMIQ hESC-iMEF, Amiqui hESC on conventional iMEF feeder layer; AMIQ hESC-iMEF-CM, feeder-free Amicqui-1 hESC with iMEF-conditioned media; AMIQ hESC-hAEC-CM, feeder-free Amicqui-1 hESC with hAEC-conditioned media. (C) Micrographs of the immunodetection of the mature neuron marker MAP2 in cells derived from H9 hESC-iMEF, AMIQ hESC-hAEC, and AMIQ hESC-iMEF on day 25 (D25). Scale bar, 50 µm. (D) Micrographs of the immunodetection of FOXP2+, CTIP2+, CALRETININ+, and GAD67+ neurons derived from H9 hESC-iMEF, AMIQ hESC-hAEC, and AMIQ hESC-iMEF. Scale bar, 50 µm. (E) Quantitative analysis of FOXP2+ or CTIP+ cells on day 45 (D45) and CALRETININ+ or GAD67+ on day 55 (D55). Data are mean ± SE. ** p<0.01 and *** p<0.001 with H9 hESC-iMEF as control. n=three biological samples per group and two repetitions per sample.
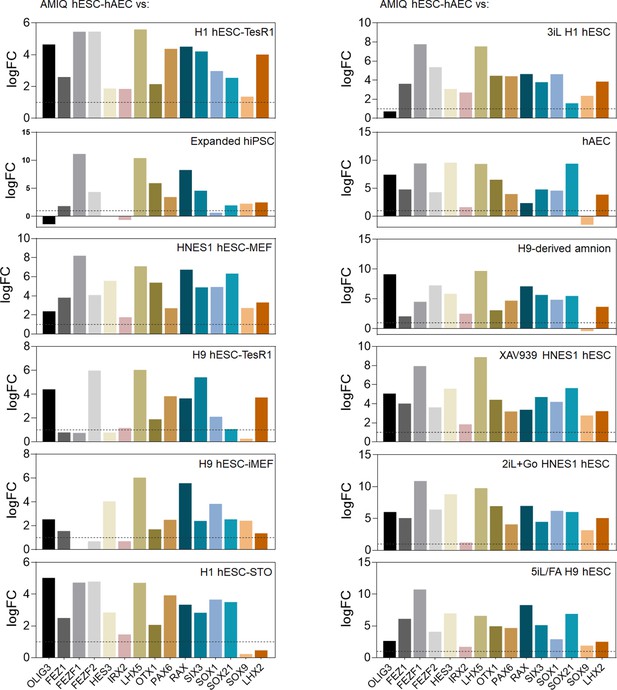
Differential gene expression (DEG) analysis of genes involved in forebrain development as signature molecular AMIQ in human embryonic stem cell (hESC)-human amniotic epithelial cell (hAEC) compared with conventional, naïve, expanded, and XAV939 conditions.
The Log2 fold change was determined using Limma-method. FC>1.0 and p<0.05.
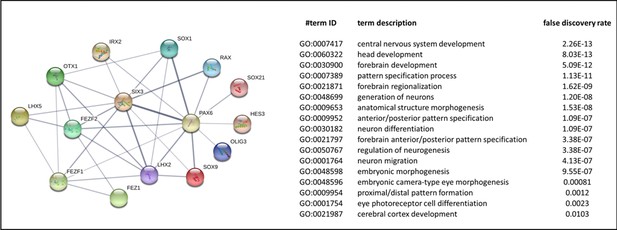
Interaction network and biological process (Gene ontology [GO]) of coding proteins of neural genes upregulated in AMIQ human embryonic stem cell (hESC)-human amniotic epithelial cell (hAEC).
p<0.05.
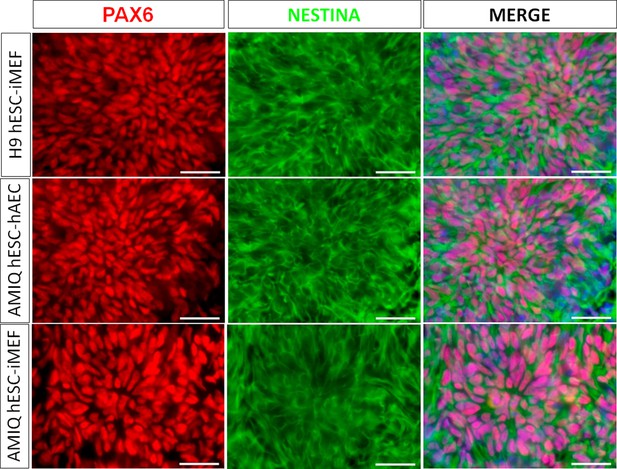
Micrographs of immunodetection of neural rosettes PAX6+/NESTIN+ derived from H9 human embryonic stem cell (hESC)-inactivated mouse embryonic fibroblast (iMEF), AMIQ hESC-human amniotic epithelial cell (hAEC), and AMIQ hESC-iMEF on day 16 (D16) of start the neural induction protocol.
Scale bar, 50 µm.
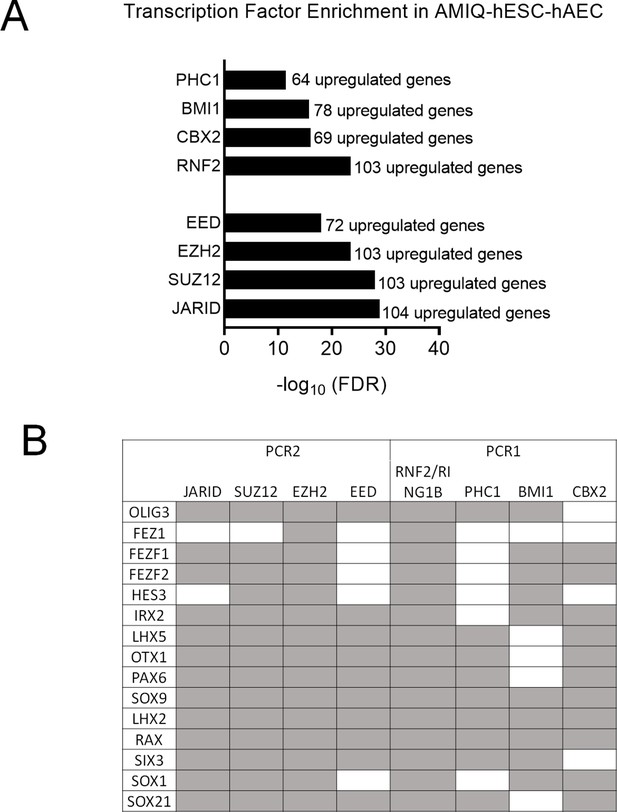
(A) Enrichment of transcription factors whose target genes (experimentally validated targets in the ChEA database) are upregulated in AMIQ hESC-hAEC compared to AMIQ hESC-iMEF.
(B) Neurodevelopmental regulatory genes upregulated in AMIQ human embryonic stem cell (hESC)-human amniotic epithelial cell (hAEC) and targeted (gray box) by the subunits of the PCR1 and polycomb repressive complex (PCR2) repressor complexes.
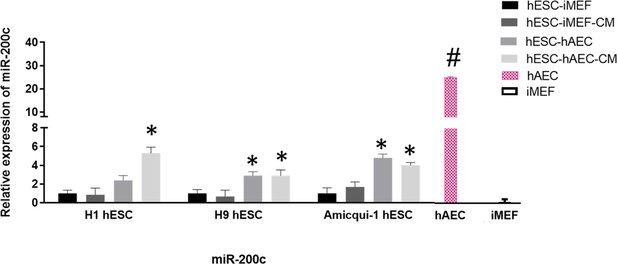
miR-200c relative expression in human embryonic stem cell (hESC) lines (Amicqui-1, H1, and H9) cultured on different feeder layers (inactivated mouse embryonic fibroblast [iMEF] or human amniotic epithelial cell [hAEC]) or feeder-free conditions (iMEF-CM or hAEC-CM).
Data are mean ± SE, n=five biological samples per group and three repetitions per sample. * p<0.001 with hESC-iMEF (H1, H9 or Amicqui-1) as control. # p<0.0001 with Amicqui-1 hESC-iMEF as control.
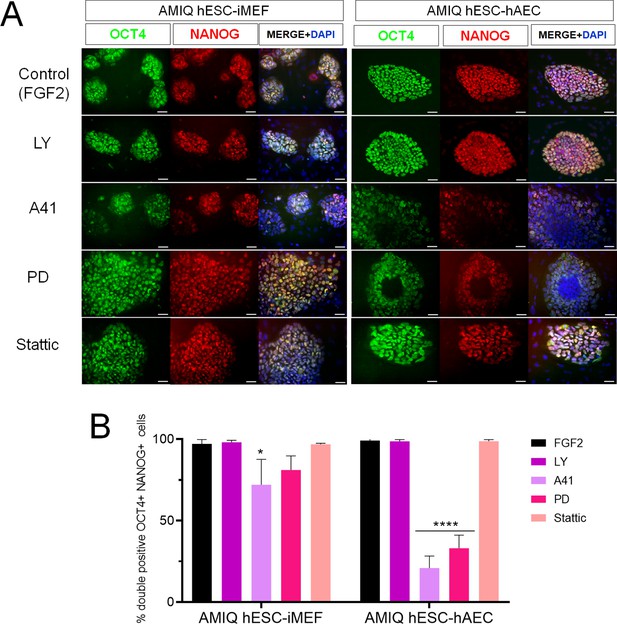
Inhibition of SRC and ERK2 disrupted expression pluripotency-related factors in human embryonic stem cell (hESC)-human amniotic epithelial cell (hAEC).
(A) Immunofluorescence for the detection of NANOG+/OCT4+ colonies in hESC maintained on hAEC (AMIQ hESC-hAEC) and on inactivated mouse embryonic fibroblast (iMEF) (AMIQ hESC-iMEF) after treatment for 48 hr with inhibitors for specific signaling pathways. Control, hESC medium supplemented with dimethyl sulfoxide (DMSO) (1 µ/mL) and FGF2 (10 ng/mL); LY, hESC medium supplemented with FGF2 (10 ng/mL) and PI3K inhibitor LY294002 (5 µM); A41, hESC medium supplemented with FGF2 (10 ng/mL) and SRC inhibitor A419259 (1 µM); PD, hESC medium supplemented with FGF2 (10 ng/mL) and ERK2 inhibitor PD0325901 (1 µM); Stattic, hESC medium supplemented with FGF2 (10 ng/mL) and STAT3 inhibitor Stattic (2.5 µM). (B) Quantification of double positive OCT4+NANOG+ cells for each treatment in hESC on iMEF or hAEC. Data are mean ± SD. n=two biological samples per group and two repetitions per sample (20 colonies for each condition were analyzed). * p=0.0125 using AMIQ hESC-iMEF without any inhibitor as control and **** p<0.0001 using AMIQ hESC-hAEC without any inhibitor as control.
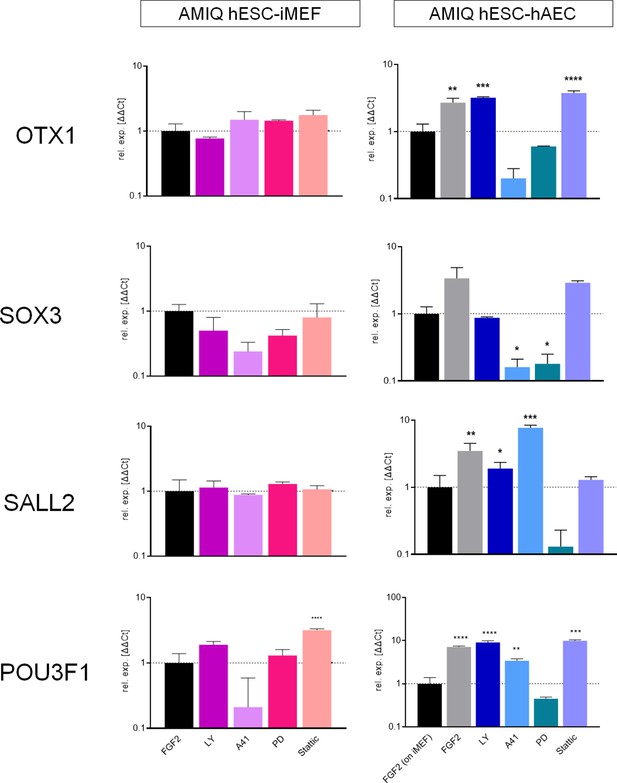
Analysis of formative pluripotency-related genes by quantitative PCR (qPCR) array in Amicqui-1 maintained on inactivated mouse embryonic fibroblast (iMEF) feeder layer (AMIQ human embryonic stem cell [hESC]-iMEF) or human amniotic epithelial cell (hAEC) feeder layer (AMIQ hESC-hAEC), under conditions of inhibition of specific signaling pathways for 48 hr.
Treatments: FGF2, hESC medium supplemented with DMSO (1 µ/mL) and FGF2 (10 ng/mL); LY, hESC medium supplemented with FGF2 (10 ng/mL) and PI3K inhibitor LY294002 (5 µM); A41, hESC medium supplemented with FGF2 (10 ng/mL) and SRC inhibitor A419259 (1 µM); PD, hESC medium supplemented with FGF2 (10 ng/mL) and ERK2 inhibitor PD0325901 (1 µM); Stattic, hESC medium supplemented with FGF2 (10 ng/mL) and STAT3 inhibitor Stattic (2.5 µM). Data are mean ± SD from technical duplicates. * p<0.05, ** p<0.01, *** p<0.001, and **** p<0.0001 with AMIQ hESC-iMEF without any treatment as control.
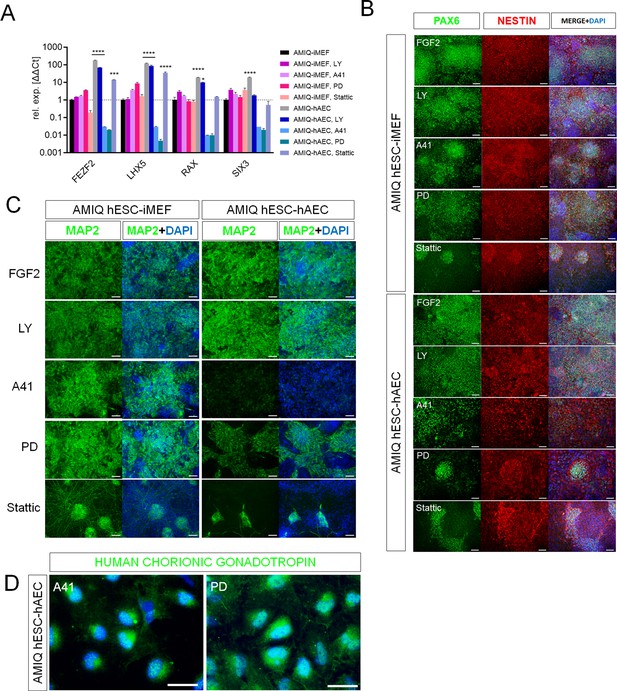
SRC and ERK2 inhibition alter biased neural potential of human embryonic stem cell (hESC)-human amniotic epithelial cell (hAEC).
(A) Expression of selected neurodevelopmental regulatory gene (NRG) by quantitative PCR (qPCR) array in AMIQ hESC on inactivated mouse embryonic fibroblast (iMEF) (AMIQ iMEF) or hAEC (AMIQ hAEC) after 48 hr treatment with inhibitors of specific signaling pathways. Data are mean ± SD from technical duplicates. * p<0.05, ***p<0.001, and **** p<0.0001 with AMIQ hESC-iMEF as control. (B) Micrographs of immunodetection of neural rosettes PAX6+/NESTIN+ derived from AMIQ hESC-iMEF and AMIQ hESC-hAEC on day 20 of start the neural induction protocol and with a 48 hr pre-treatment with signaling pathway inhibitors. Scale bar, 25 µm. (C) Micrographs of immunodetection of mature neurons MAP2+ derived from AMIQ hESC-iMEF and AMIQ hESC-hAEC on day 30 of start the neural induction protocol and with a 48 hr pre-treatment with signaling pathway inhibitors. Scale bar, 25 µm. (D) Micrographs of immunodetection of human chorionic gonadotropin+ cells derived from AMIQ hESC-hAEC on day 30 of start the neural induction protocol and with a 48 hr pre-treatment with SRC and ERK2 signaling pathway inhibitors. Scale bar, 25 µm.
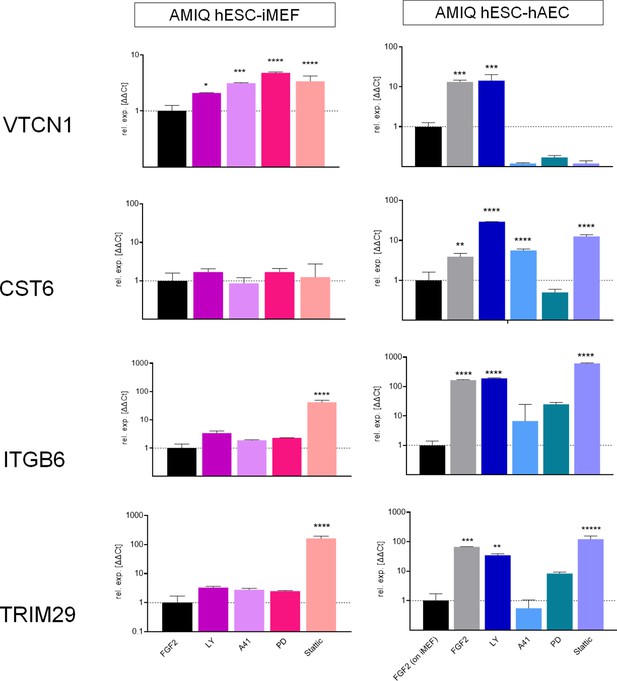
Analysis of ‘intermediate genes’ by quantitative PCR (qPCR) array in Amicqui-1 maintained on inactivated mouse embryonic fibroblast (iMEF) feeder layer (AMIQ human embryonic stem cell [hESC]-iMEF) or human amniotic epithelial cell (hAEC) feeder layer (AMIQ hESC-hAEC), under conditions of inhibition of specific signaling pathways for 48 hr.
Treatments: FGF2, hESC medium supplemented with dimethyl sulfoxide (DMSO) (1 µ/mL) and FGF2 (10 ng/mL); LY, hESC medium supplemented with FGF2 (10 ng/mL) and PI3K inhibitor LY294002 (5 µM); A41, hESC medium supplemented with FGF2 (10 ng/mL) and SRC inhibitor A419259 (1 µM); PD, hESC medium supplemented with FGF2 (10 ng/mL) and ERK2 inhibitor PD0325901 (1 µM); Stattic, hESC medium supplemented with FGF2 (10 ng/mL) and STAT3 inhibitor Stattic (2.5 µM). Data are mean ± SD from technical duplicates. * p<0.05, ** p<0.01, *** p<0.001, and **** p<0.0001 with AMIQ hESC-iMEF without any inhibitor as control.
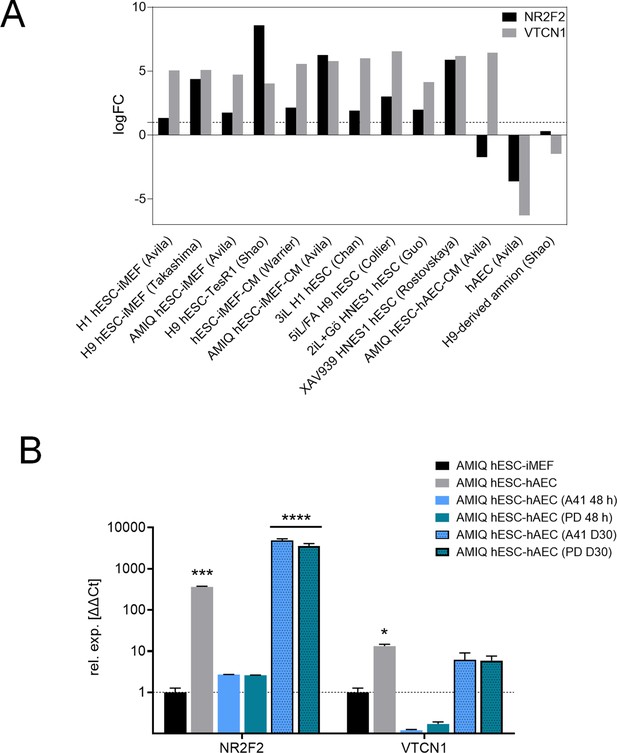
(A) LogFC from differential gene expression (DEG) analysis (FC<1.0, p-value<0.05) for NR2F2 and VTCN1 genes in AMIQ human embryonic stem cell (hESC)-human amniotic epithelial cell (hAEC) compared with conventional primed, feeder-free, naïve, expanded, and XAV939 conditions as well as versus H9-derived amnion and hAEC isolated from membranes at term.
(B) Analysis for NR2F2 and VTCN1 genes by quantitative (qPCR) array in AMIQ hESC-inactivated mouse embryonic fibroblast (iMEF) and AMIQ hESC-hAEC under conventional conditions (FGF2 10 ng/mL + dimethyl sulfoxide [DMSO] 1 µL 48 hr), treatments with inhibitors at 48 hr and 30 days (D30) of neural differentiation after treatment with inhibitors. PD, FGF2 10 ng/mL + ERK2 inhibitor PD0325901 1 µM; A41, FGF2 10 ng/mL + SRC inhibitor A419259 1 µM. Data are mean ± SD from technical duplicates. * p<0.05, *** p<0.001, and **** p<0.0001 with AMIQ hESC-iMEF without any inhibitor as control.
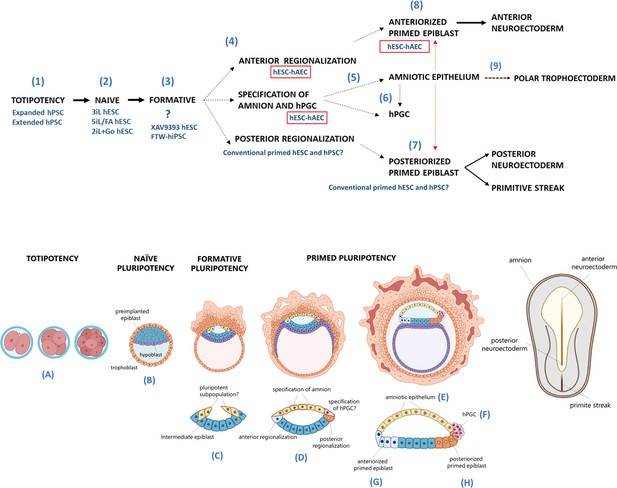
Proposed model for human embryogenesis with amnion as an organizer.
Simplified scheme of embryonic stages in humans and its in vitro counterparts of human pluripotent stem cell lines (hPSC) represent or possess some characteristics of each state. Totipotency can be represented by the expanded and extended hPSC (1). Different conditions to maintaining naïve hPSC have been characterized, which resemble the pre-implantation epiblast (2). The formative pluripotency has recently been proposed as an intermediate period between the naïve and primed states. hPSC treated with the WNT inhibitor XAV939 and FTW-hiPSC could exhibit some characteristics of this phase, although there is still no established consensus of the in vitro conditions that represent this period (3). At peri-implantation, pluripotency could be spatially disaggregated for epiblast anterior-posterior regionalization and specification of amnion and human primordial germ cells (hPGC) (4). The amnion could arise from a subpopulation of pluripotent cells of the formative or primed epiblast (5). Primordial germ cells (PGCs) could be derived from another subpopulation (called ‘germinal pluripotency’ by Chen et al., 2019), although it has been suggested that they could also be specified from amnion (6). The anteriorized post-implanted epiblast will generate the anterior structures such as the neuroectoderm (7). The posteriorized post-implanted epiblast will give rise to the primitive streak and the most posterior region of the neuroectoderm (8). Recent reports suggest that the polar trophectoderm derives from pluripotent cells that transit through amnion-like cells (9). The condition of human embryonic stem cells growing on inactivated mouse embryonic fibroblasts (hESC-iMEF) could share more characteristics of the posterior epiblast given the interaction with a fibroblast cell type to favor the epithelium-mesenchyme transition. hESC-human amniotic epithelial cells (hAEC) have a more significant bias to differentiate into anterior neuroectoderm, but they also have the capacity to form germline cells and probably polar trophectoderm. Therefore, hESC-hAEC could be composed of subpopulations that form a continuum from the formative phase, as well as an intermediate population for specification of the amnion and germ line, to finally segregate to the anterior part of the epiblast. The amnion secretes signals or interacts directly with the primed epiblast until the onset of gastrulation promotes the neural induction or primitive streak formation (dotted arrow, probably route of differentiation; dotted red arrow, probably interactions between lineages).
Additional files
-
Supplementary file 1
List of genes with differential expression in AMIQ human embryonic stem cells (hESC)-human amniotic epithelial cells (hAEC) compared to other culture conditions.
- https://cdn.elifesciences.org/articles/68035/elife-68035-supp1-v2.xlsx
-
Supplementary file 2
KEGG pathways upregulated in AMIQ human embryonic stem cells (hESC)-human amniotic epithelial cells (hAEC).
- https://cdn.elifesciences.org/articles/68035/elife-68035-supp2-v2.xlsx
-
Supplementary file 3
‘Intermediate genes’ upregulated in AMIQ human embryonic stem cells (hESC)-human amniotic epithelial cells (hAEC).
- https://cdn.elifesciences.org/articles/68035/elife-68035-supp3-v2.xlsx
-
Supplementary file 4
Transcription factors whose targets are overrepresented in the upregulated and downregulated genes identified in AMIQ human embryonic stem cells (hESC)-human amniotic epithelial cells (hAEC).
- https://cdn.elifesciences.org/articles/68035/elife-68035-supp4-v2.xlsx
-
Supplementary file 5
List of antibodies used in this study.
- https://cdn.elifesciences.org/articles/68035/elife-68035-supp5-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/68035/elife-68035-transrepform1-v2.docx





