A deep learning algorithm to translate and classify cardiac electrophysiology
Figures
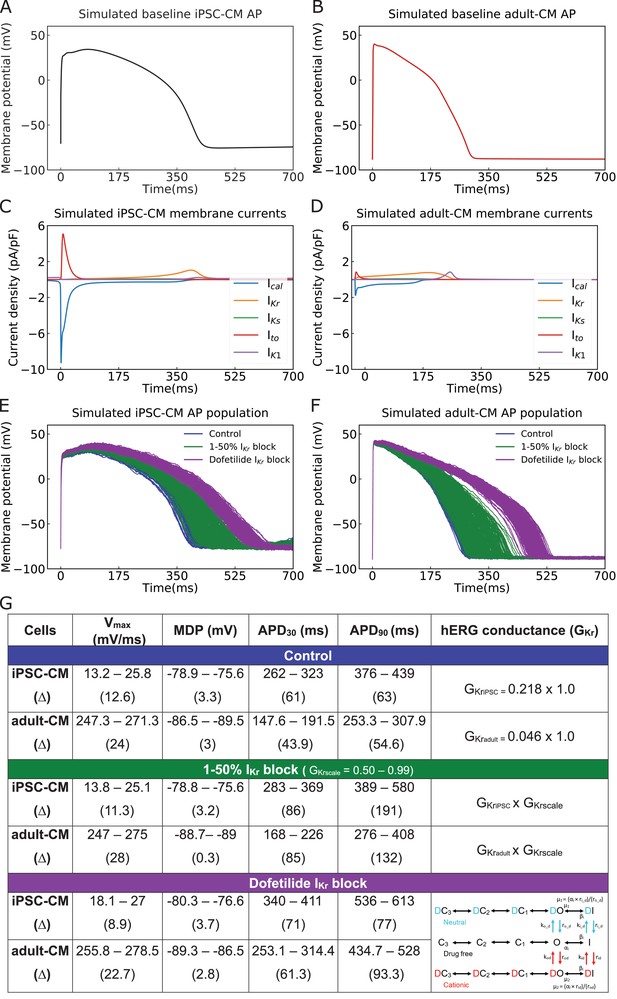
Cellular action potential (AP) and ionic currents for induced pluripotent stem cell-derived cardiomyocytes (iPSC-CMs) and adult cardiomyocytes (adult-CMs) (O’Hara–Rudy human ventricular APs).
Comparison of cellular APs in the baseline model of (A) iPSC-CMs and (B) adult-CMs at a matched cycle length of 982 ms. (C, D) Simulated ionic current (ICaL, IKr, IKs, Ito, IK1) profiles during (C) iPSC-CM and (D) adult-CM APs. (E) APs of spontaneously beating iPSC-CM cells (n = 208) and (F) adult-CM APs at matched cycle lengths were simulated after incorporating physiological noise currents as drug-free (blue) and drugged IKr modeled as simple GKr reduction by 1–50% IKr block (green) and a complex model of conformation-state dependent IKr block in the presence of 2.72 ng/mL dofetilide (purple). (G) Comparison between iPSC-CM and adult-CM drug-free and drugged models with simple and complex IKrblock model schemes (as indicated in the right column), including upstroke velocity (Vmax), maximum diastolic potential (MDP), and action potential duration (APD).
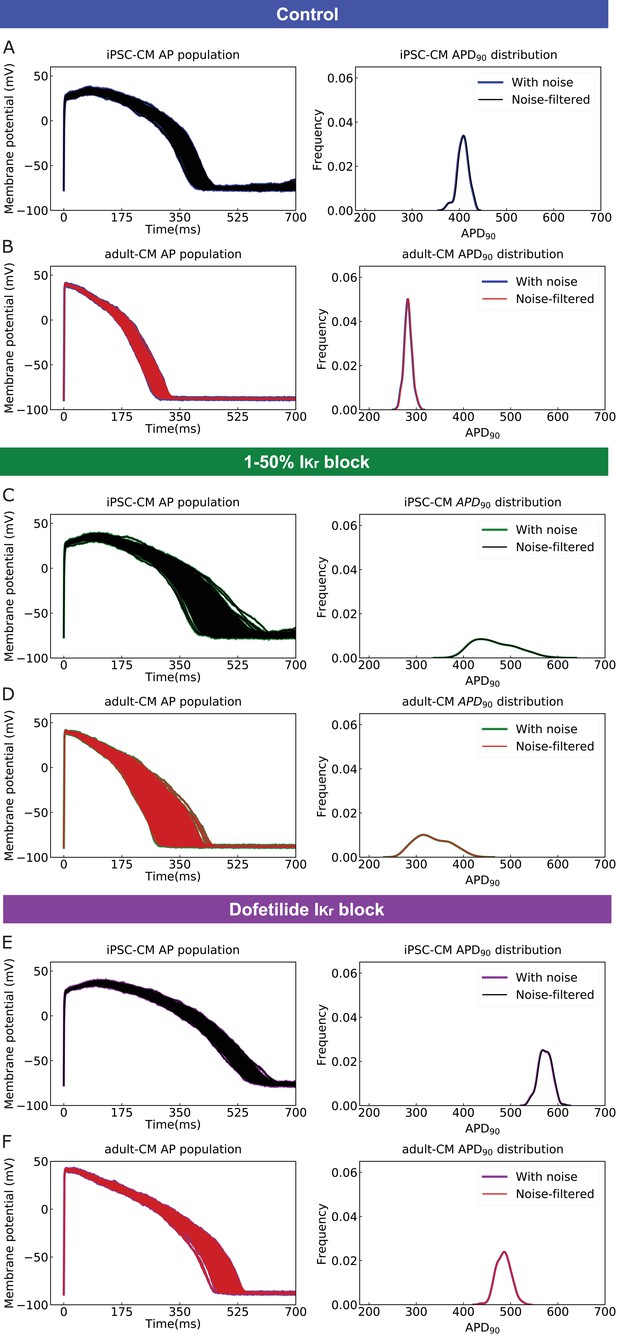
Application of a digital forward and backward data filtering technique to simulated induced pluripotent stem cell-derived cardiomyocyte (iPSC-CM) and adult cardiomyocyte (adult-CM) action potentials (APs) population (left panels) indicates zero phase distortion for APD90 value distributions (right panels).
(A) drug-free iPSC-CM APs with physiological noise in blue and after applying the noise filtering technique in black; (B) drug-free adult-CM APs – blue and red traces; (C) drugged iPSC-CM APs with 1–50% IKr block – green and black traces; (D) drugged adult-CM APs with 1–50% IKr block – green and red traces; (E) drugged iPSC-CM APs with 2.72 ng/mL dofetilide – purple and black traces; and (F) drugged adult-CM APs with 2.72 ng/mL dofetilide –purple and red traces.
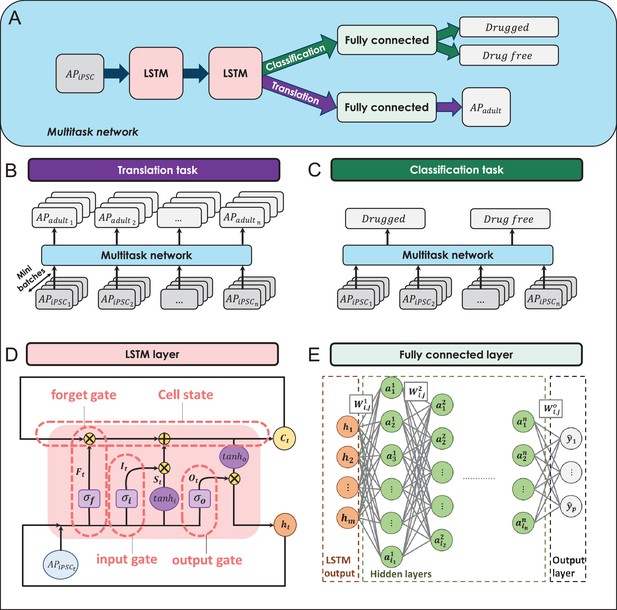
The building blocks of the multitask network.
(A) The general overview of the multitask network presented in this study. (B) The translation task to reconstruct adult cardiomyocyte (adult-CM) action potentials (APs) from the corresponding induced pluripotent stem cell-derived cardiomyocyte (iPSC-CM) APs. (C) The classification task to classify iPSC-CM APs into drug-free and drugged categories. (D) The logic flow process in the long-short-term-memory (LSTM) layers. (E) The architecture of the implemented fully connected layers in the multitask network.
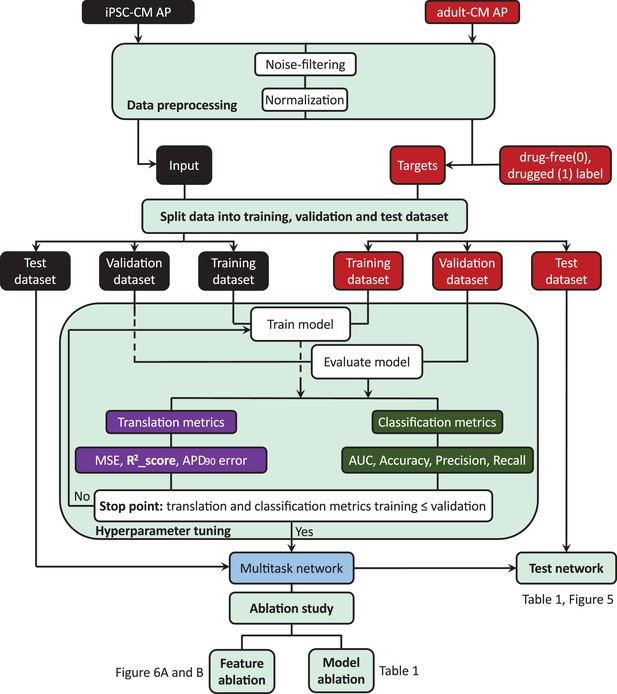
Machine learning workflow in this study.
(1) Data preprocessing includes noise filtering and normalization of the drug-free and drugged induced pluripotent stem cell-derived cardiomyocyte (iPSC-CM) and adult cardiomyocyte (adult-CM) action potentials (APs). (2) Incorporating the preprocessed iPSC-CM APs as input and adult-CM APs and corresponding labels (drug-free [0] and drugged [1]) of iPSC-CM APs as targets into the multitask network. (3) Splitting the input and target data into training, validation and test set, and using training and validation set for training and tuning the network hyperparameters. (4) Comparing the network performance for training set and validation set to decide when to stop training and tuning the network hyperparameters. (5) Testing the overall multitask network performance using holdout test dataset and removing the long-short-term-memory (LSTM) layers, classification task (model ablation), and iPSC-CM AP values at different time frames (feature ablation) to study the performance of the network in the absence of its building blocks.
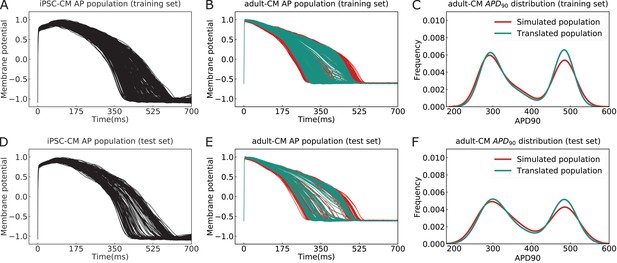
The performance of the multitask network for translating induced pluripotent stem cell-derived cardiomyocyte (iPSC-CM) action potentials (APs) into adult cardiomyocyte (adult-CM) APs.
(A) The iPSC-CM APs used for training the multitask network contained a variety of drug-free and drugged action potential morphologies (training set). (B) Comparison between simulated (red) and translated adult-CM APs (cyan) in the training set. (C) Comparison between the histogram distribution of APD90 values for simulated and translated adult-CM APs in the training set. (D) Dedicated iPSC-CM APs for testing the performance of the multitask network (test set). (E) Comparison between simulated (red) and translated adult-CM APs (cyan) in the test set. (F) Comparison between histogram distribution of APD90 values for simulated and translated adult-CM APs in the test set.
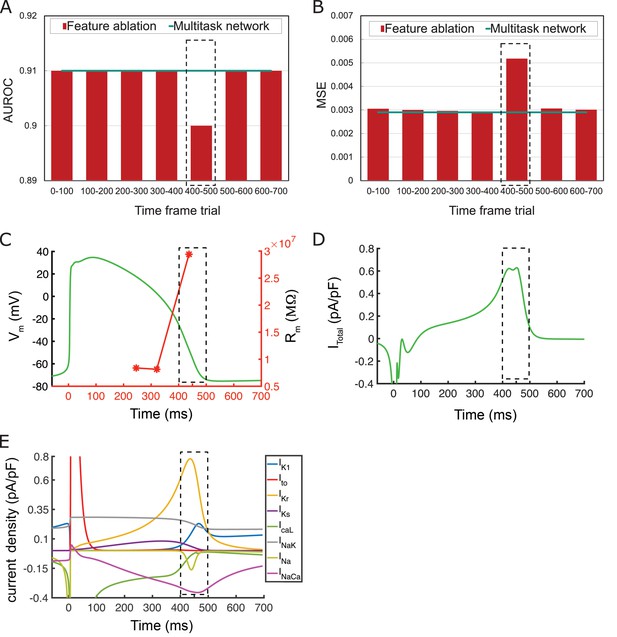
The feature ablation study on the proposed multitask network is performed by removing induced pluripotent stem cell-derived cardiomyocyte (iPSC-CM) action potential (AP) values during different time frames and evaluating their importance on drug-free and drugged iPSC-CM AP classification and adult cardiomyocyte (adult-CM) AP translation.
The largest effect (most important information) is observed at 400–500 ms interval (dotted black line). (A) Comparison between intact multitask network area under the receiver operating characteristic curve (AUROC) and obtained AUROC values for drug-free and drugged iPSC-CM AP classification during removal of indicated time frames within iPSC-CM APs. (B) Comparison between intact multitask network mean-squared error (MSE) (cyan line) and obtained MSE values for adult-CM AP translation during removal of indicated time frames within iPSC-CM APs (red bars). (C) AP trace (green) and membrane resistance (red) as a function of simulation time indicating very high values (as dI → 0, dV/dI → ∞) for the latter at 400–500 ms. (D) Total current density, Itotal, demonstrates a plateau followed by a rapid decline at 400–500 ms. (E) Individual current densities indicate a period of inward and outward current balance followed by rapid changes in IKrand other repolarizing components at 400–500 ms time interval.
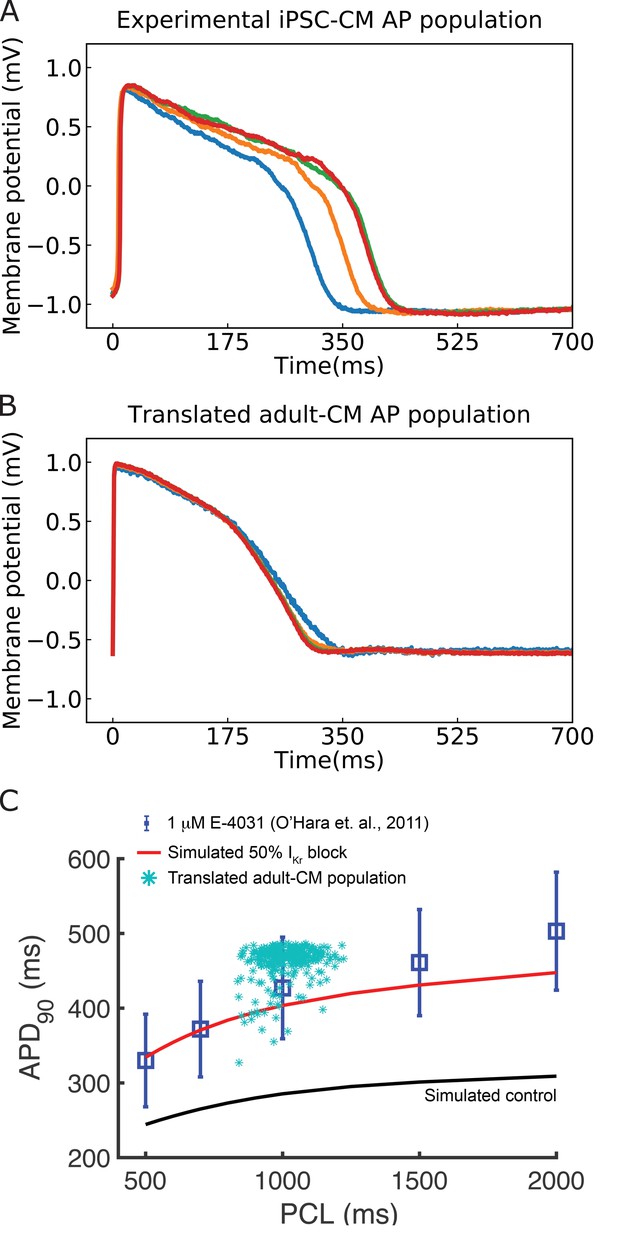
Translation of experimentally recorded induced pluripotent stem cell-derived cardiomyocyte (iPSC-CM) action potentials (APs) into adult cardiomyocyte (adult-CM) APs to demonstrate the deep learning network applied to experimental data and validate the multitask network performance.
(A) Experimentally recorded iPSC-CM APs from the Kurokawa lab. (B) Translated adult-CM APs from experimentally recorded iPSC-CM APs via the multitask network. (C) Comparing translated adult-CM APD90 values with 50% IKrblock from spontaneously beating simulated iPSC-CMs around 1000 ms beating frequency (turquoise asterisks) with previously published simulated (red curve for drugged and black for drug-free control) and experimental (blue squares) values from the O’Hara–Rudy study (Shaheen et al., 2018) indicates that predictions fall within the range of experimentally reported values at 1 Hz.
Tables
Statistical measures for evaluating the performance of the multitask network for both iPSC-CM AP trace classification into drug-free and drugged categories and their translation into adult-CM APs for training and test datasets as well as the effect of removing LSTM layers and classification task on the network performance.
| Translation | |||||||
|---|---|---|---|---|---|---|---|
| Performance metrics | MSE | R2 score | Error in APD90 prediction | ||||
| Training dataset | 0.0027 | 0.992 | 3.41% | ||||
| Test dataset | 0.0029 | 0.991 | 3.60% | ||||
| Remove LSTM layers test dataset | 0.0031 | 0.991 | 3.78% | ||||
| Remove classification task test dataset | 0.0034 | 0.990 | 4.33% | ||||
| Classification | |||||||
| Performance metrics | AUROC | Accuracy | Recall | Precision | |||
| Training dataset | 0.93 | 92% | 0.92 | 0.93 | |||
| Test dataset | 0.91 | 92% | 0.92 | 0.92 | |||
| Remove LSTM layers test dataset | 0.90 | 92% | 0.90 | 0.91 | |||
| iPSC-CM: induced pluripotent stem cell-derived cardiomyocyte; AP: action potential; adult-CM: adult cardiomyocyte; AP: action potential; AUROC: area under the receiver operating characteristic curve; LSTM: long-short-term-memory. | |||||||





