Structurally distributed surface sites tune allosteric regulation
Figures
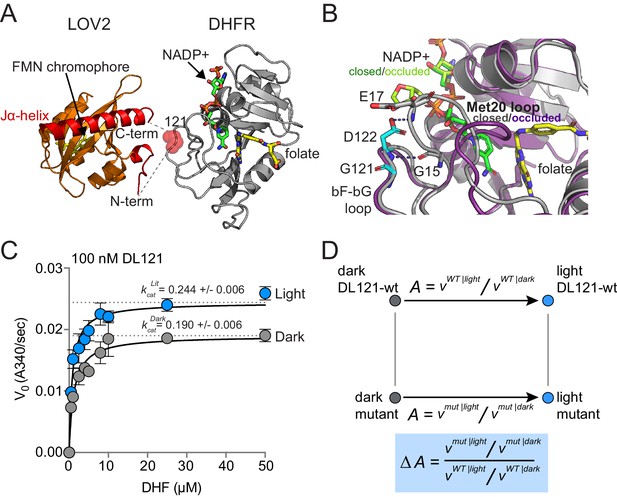
The DL121 DHFR/LOV2 fusion.
(A) Composite structures of the individual DHFR and LOV2 domains (PDB ID: 1R × 2 and 2V0U), indicating the LOV2 insertion site between positions 120 and 121 of DHFR (Sawaya and Kraut, 1997; Halavaty and Moffat, 2007). DHFR is in gray cartoon, NADP co-factor in green sticks, and folate substrate in yellow sticks. In LOV2 signaling, blue light triggers the formation of a covalent adduct between a cysteine residue (C450) and a flavin mononucleotide (FMN, yellow sticks) (Salomon et al., 2001; Crosson and Moffat, 2002; Swartz et al., 2002) and associated unfolding of the C-terminal Jα-helix (red cartoon); this order-to-disorder transition is used for regulation in several synthetic and natural systems (Pudasaini et al., 2015; Glantz et al., 2016). (B) DHFR loop conformational changes near the LOV2 insertion site. While the mechanism of DHFR regulation by LOV2 is currently unknown, inspecting the native DHFR structure provides some insight. The substrate-bound Michaelis complex of native DHFR is in the ‘closed’ conformation (gray cartoon), while the product ternary complex is in the ‘occluded’ state (purple cartoon). The βF-βG loop, where LOV2 is inserted, is highlighted in cyan. In native DHFR, hydrogen bonds between this loop (Asp122) and the Met20 loop (Gly15, Glu17) are thought to stabilize the closed conformation (Sawaya and Kraut, 1997; Schnell et al., 2004). Mutations to positions 121 and 122 reduce activity and cause the enzyme to prefer the occluded conformation (Cameron and Benkovic, 1997; Mhashal et al., 2018; Miller and Benkovic, 1998). (C) Steady state Michaelis Menten kinetics for the DL121 fusion under lit (blue) and dark (gray) conditions. The kcat of DHFR increases 28% in response to light; the difference in Km is statistically insignificant (Supplementary file 1a). Error bars represent standard deviation for three replicates. (D) Quantifying the allosteric effect of mutation. Allostery for the DL121 fusion is reported as the ratio between lit and dark velocity. The effect of a mutation on allostery is then computed as the ratio of mutant allostery to wt-DL121 allostery (bottom blue box).
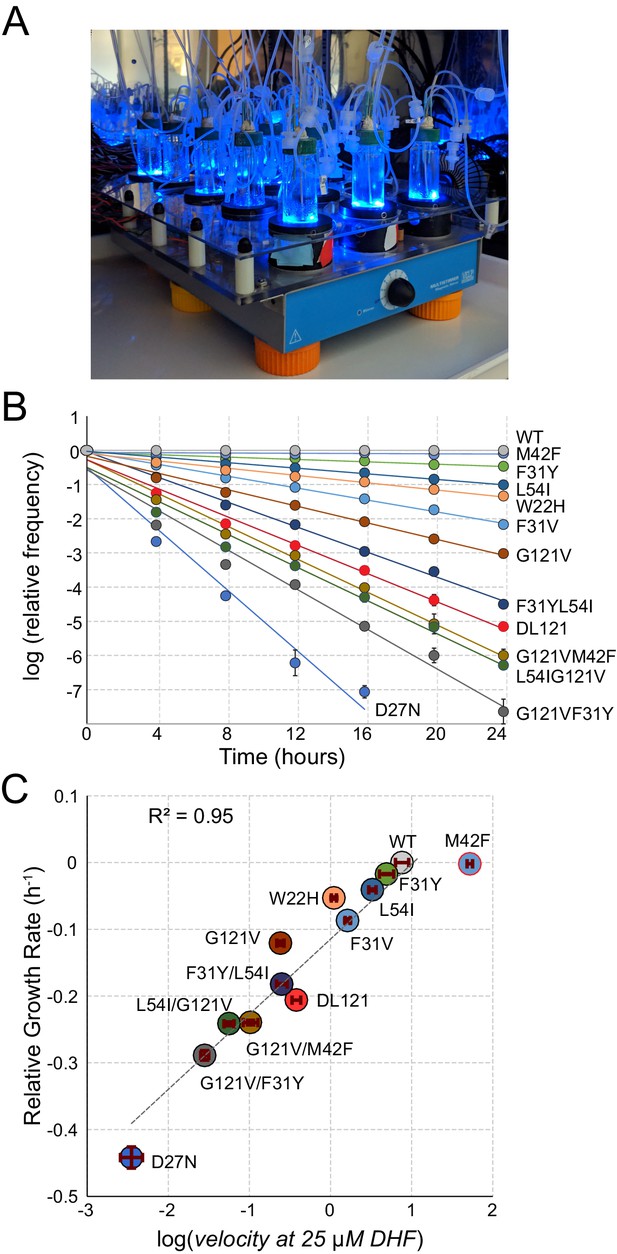
A high-throughput, high-resolution assay for DHFR activity.
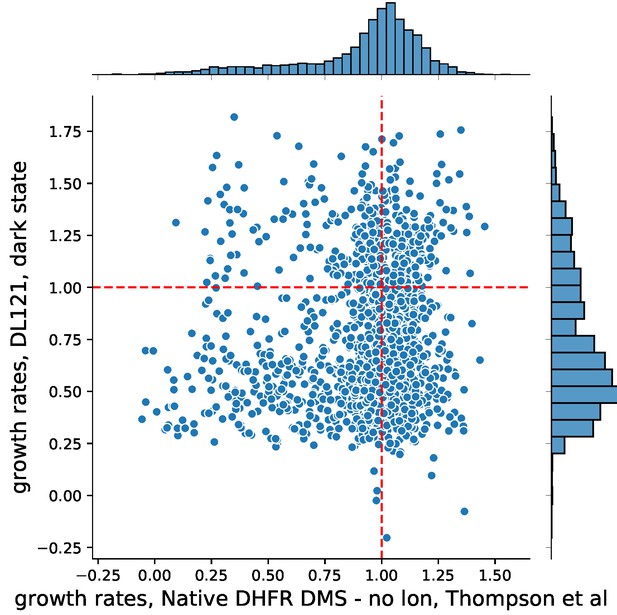
(A) The turbidostat. The instrument has 15 individual growth chambers (vials), positioned on a stir plate inside an incubator. Illumination was provided by blue LEDs in each vial holder. (B) Log-normalized relative allele frequency over time for 11 DHFR point mutations of known catalytic activity and the DL121 fusion. Allele frequency (colored circles) was determined by next-generation sequencing of mixed-population culture samples at each time point. All frequencies were normalized to t = 0 and WT DHFR (no LOV2 insertion). Error bars reflect standard error across four measurements, they are sometimes obscured by the marker. The slope for each line of best fit provides the growth rate of each mutant allele relative to WT DHFR. (C) Relative growth rate vs. log10(velocity) for the 11 DHFR mutants and DL121 as characterized in panel B. Color coding of mutations is matched to panel B. Error bars reflect standard error of the mean over four replicates. The dashed line was fit by linear regression to all mutants in the linear regime (M42F excluded).
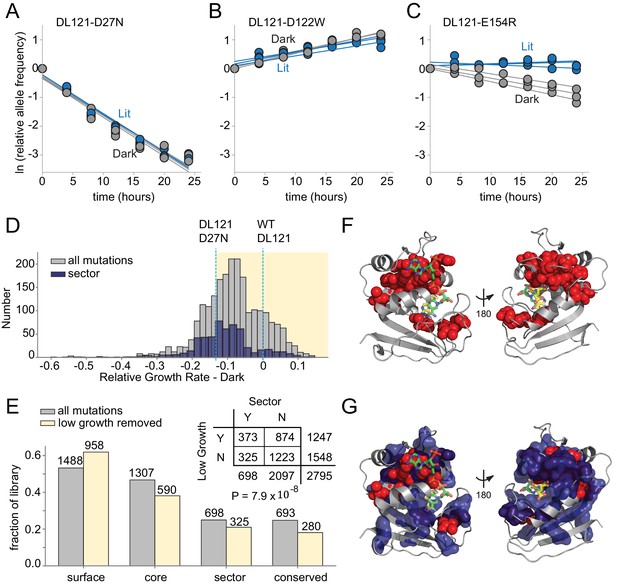
The effect of DL121 DHFR mutations on growth rate.
(A-C) Representative relative growth rate trajectories for three mutations. (A) DL121 D27N was deleterious in both lit and dark conditions. (B) DL121 D122W was advantageous under both lit and dark conditions. (C) DL121 E154R was deleterious in the dark, and near neutral in the light. Solid lines were obtained by linear regression; the slope of these provides the difference in growth rate relative to the unmutated DL121 construct. Relative growth rates were measured in triplicate for each mutant under lit (blue) and dark (gray) conditions. (D) Distribution of relative growth rates under dark conditions. The distribution for all mutations with measurable growth rate effects is in gray (‘null data’ and ‘no data’ excluded); the distribution for sector mutations is in navy. The relative growth rate of DL121 D27N, a mutation that severely disrupts catalytic activity, is indicated with a cyan dashed line. (E) The fraction of DL121 mutations with measurable growth rates that can be categorized as: DHFR surface, core, sector, and evolutionarily conserved (see Materials and methods for definitions). The fraction is shown for both the complete library (gray bars), and the library after removing mutations with low growth (growth rate <= DL121 D27N). The absolute number of mutations is shown above each bar. A contingency table summarizes the overlap between mutations in the sector (at a p-value cutoff of 0.010), and the mutations that yield low growth (growth rate <= DL121 D27N). (F) Structural distribution of positions enriched for mutations with growth rates as low as or lower than DL121 D27N (red spheres). The DHFR backbone is in gray cartoon, the folate substrate in yellow sticks, and the NADP co-factor in green sticks. (G) Relationship of the sector (navy blue surface) to positions enriched for growth-rate disrupting mutations (red spheres, same as in F).
-
Figure 3—source data 1
Relative growth rates under lit and dark conditions for DL121 point mutations as determined by next-generation sequencing.
Column 1 is the mutation name, columns 2–4 are relative growth rates in the light (three replicates), column 5 is the average lit relative growth rate, and column 6 is the standard deviation across lit replicates. Columns 7–9 are relative growth rates in the dark (three replicates), column 10 is the average dark relative growth rate, and column 11 is the standard deviation across dark replicates. Relative growth rate values of −999 indicate mutations with insufficient counts to fit a reliable growth rate (‘null data’), values of −1000 indicate mutations missing from the library at t = 0 (‘no data’), respectively.
- https://cdn.elifesciences.org/articles/68346/elife-68346-fig3-data1-v2.csv
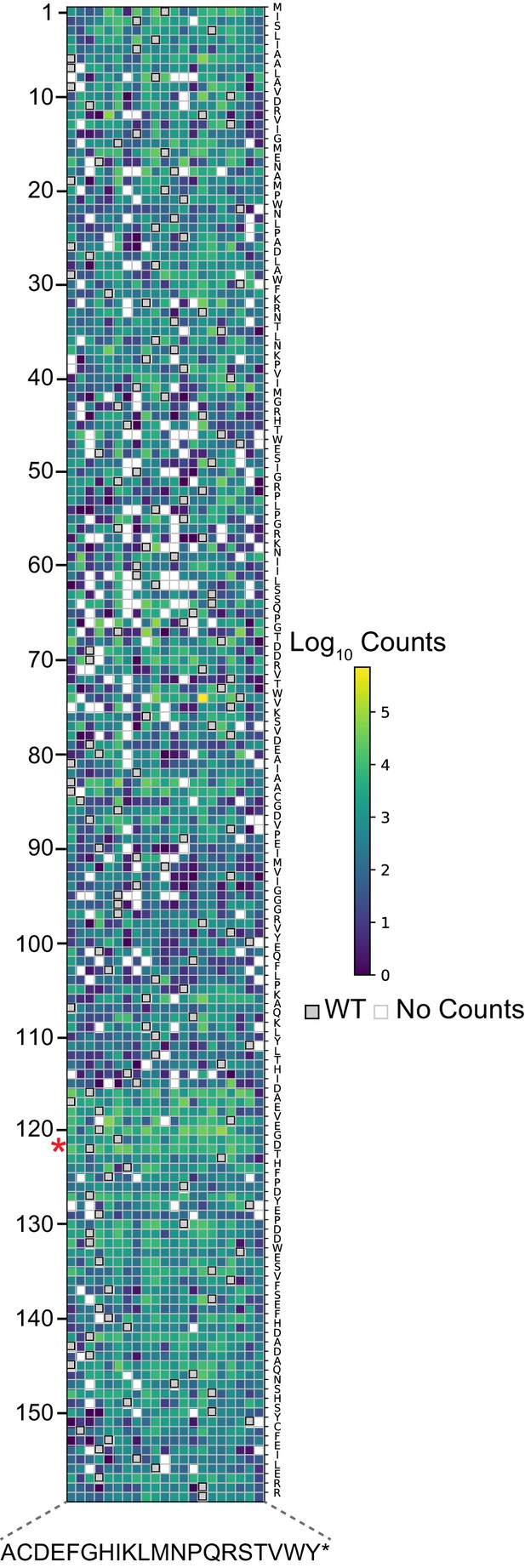
Deep mutational scanning library completeness – heatmap of counts for all mutants.
Log10(counts) of all possible mutations in DHFR domain of DL121 chimeric protein library at time point zero. The y axis corresponds to positions on E. coli DHFR domain as numbered in PDB ID: 1R × 2. A red star indicates the location of the LOV2 domain insertion. The x axis corresponds to possible mutations. Wild-type residues are shown in gray; positions with no counts are shown in white.
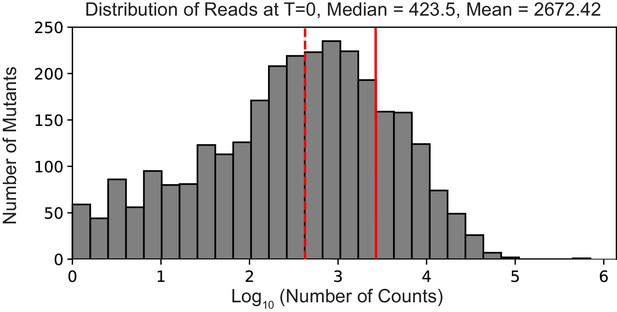
Deep mutational scanning library completeness – distribution of counts for all mutants.
A histogram of the number of counts per mutant at time point zero. The median and mean number of counts is shown as a dashed and solid red line, respectively.
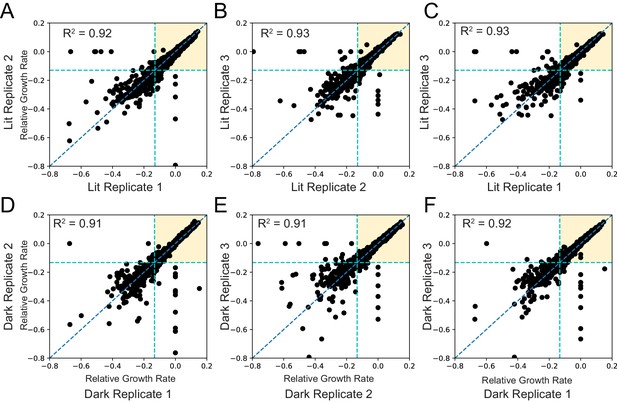
Reproducibility across biological replicates.
The relative growth rate (see Materials and methods) for each mutant is compared across all three lit (A-C) and dark (D-F) replicates. The line of best fit is indicated with a blue dashed line. The teal dashed lines represent the growth rate of DL121-D27N; mutants with a relative growth rate below this cutoff were considered near catalytically inactive and excluded from analysis of allostery.
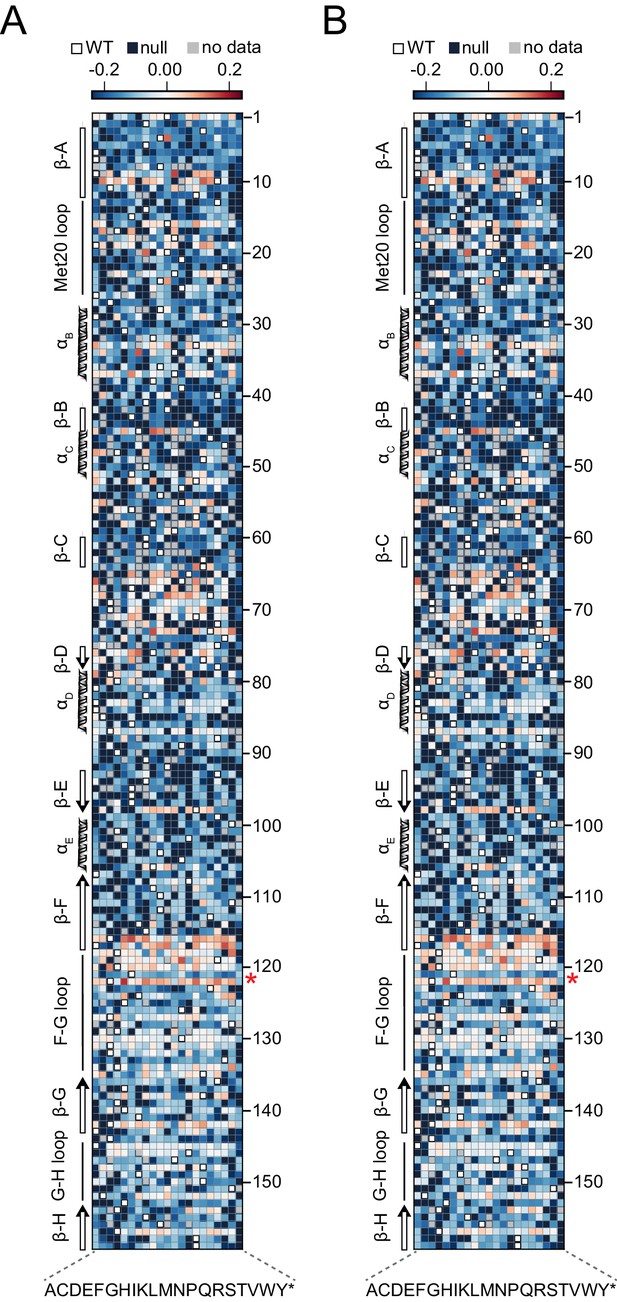
Heatmaps of relative growth rates.
(A) Relative growth rate in the dark. (B) Relative growth rate in the light. Blue and red indicate mutations with deleterious and beneficial effects on growth rate respectively. White squares with black outlines mark the WT residue at each position. Mutations missing from the library (‘no data’) are colored gray, and mutations that did not have sufficient counts for at least three time points (‘null data’, no relative growth rate could be fit) are colored navy.
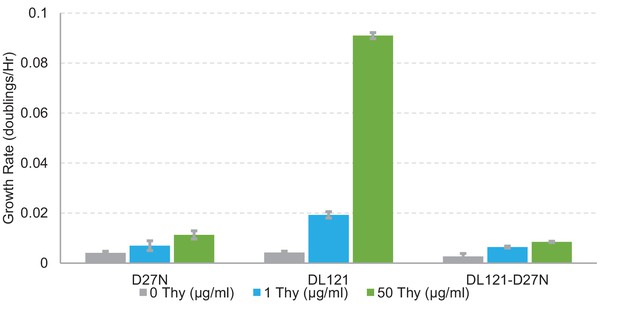
Growth rate measurements for DL121-D27N.
Comparison of growth rates as doublings per hour for three enzymes: nonchimeric E. coli DHFR with a D27N mutation (rendering it catalytically inactive), the unmutated fusion protein, DL121, and DL121 combined with the D27N mutation. All three mutants were grown in a 96-well plate in M9 media supplemented with either no thymidine, 1 μg/ml thymidine (the same media conditions as the experiments in this work), or 50 μg/ml thymidine at 30°C. Error bars represent standard deviation across six replicates.
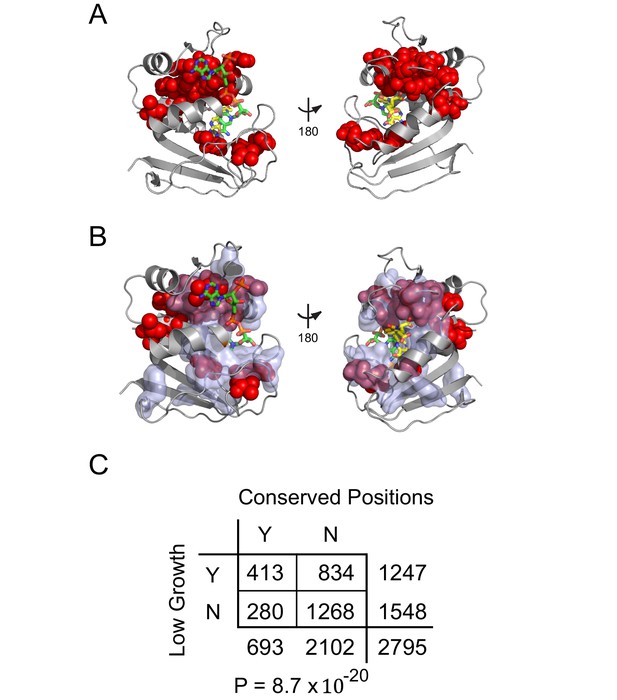
Relationship between catalytically inactivating mutations and evolutionarily conserved positions.
(A) Structural distribution of positions enriched for mutations with growth rates as low as or lower than DL121 D27N (indicated with red spheres). The DHFR backbone is in gray cartoon, the folate substrate in yellow sticks, and the NADP co-factor in green sticks. (B) Relationship of evolutionarily conserved positions (light blue surface) to positions enriched for growth-rate disrupting mutations (red spheres, same as in A). (C) A contingency table summarizes the overlap between conserved positions, and the mutations that yield low growth (growth rate <= DL121 D27N).
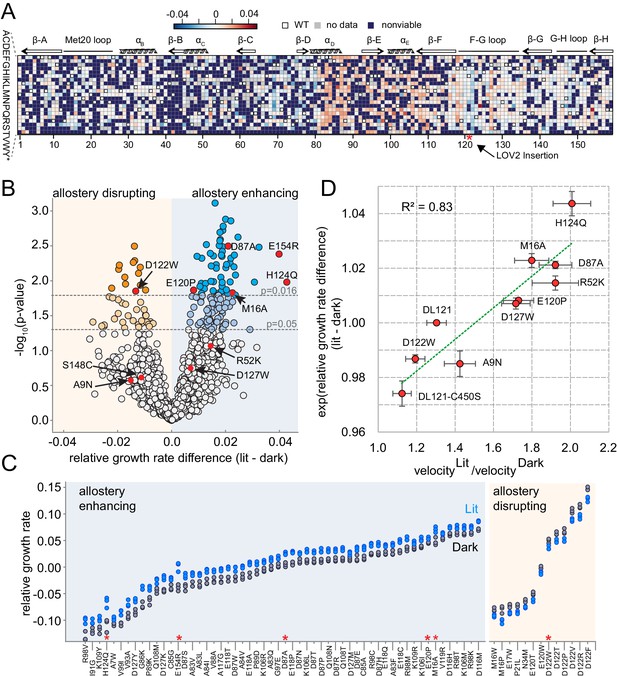
The effect of DL121 DHFR mutations on allostery.
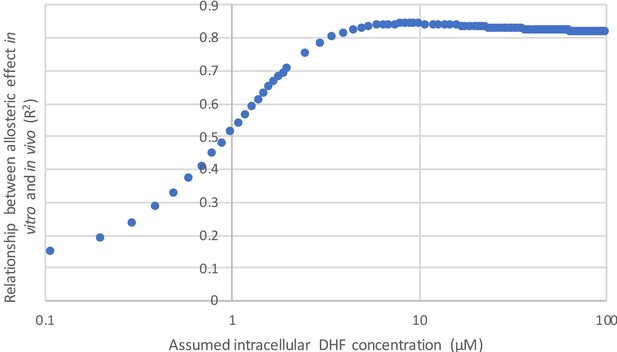
(A) Heatmap of mutational effects on allostery. Blue indicates allostery disrupting mutations, and red indicates allostery enhancing mutations. White squares with black outlines mark the WT residue at each position. Mutations missing from the library (‘no data’) are colored gray, and mutations that did not have sufficient sequencing counts for at least three time points (‘null data’) are colored navy. The LOV2 domain insertion site is indicated with a red star. (B) Volcano plot indicating the statistical significance of the light-dark growth rate difference (y-axis) as a function of relative growth rate difference (x-axis). p-Values were computed using a t-test across triplicate light and dark measurements. Individual points correspond to mutations; mutations on the left (yellow) side of the graph are allostery disrupting, while mutations on the right (blue) are allostery enhancing. Two cutoffs for statistical significance are indicated with dashed gray lines – both a standard value of p=0.05, and an adjusted p-value of 0.016, obtained by using Sequential Goodness of Fit (SGoF) to account for multiple hypothesis testing. Mutations selected for further in vitro experimental characterization are colored red and labeled. S148C and E154R did not yield sufficient quantities of active protein for further in vitro characterization. (C) Triplicate relative growth rate measurements under lit (blue) and dark (gray) conditions for all mutations with statistically significant allostery at the adjusted p-value (p<=0.016). The mutations are sorted by dark growth rate; mutations selected for in vitro characterization are marked with red asterisks. (D) Relationship between the allosteric effect as measured in vivo and in vitro. As we expect a log-linear relationship, we compare the ratio of velocity at 25 µM DHF (along x) to the exponent of the relative growth rate difference (along y). The relative growth rate difference under lit and dark conditions is the mean of triplicate measurements, error bars indicate SEM. All mutant effects on growth rate were measured in the same experiment (corresponding to a subset of the data in panel B) with the exception of DL121 C450S. The relative growth rate for this light-insensitive LOV2 mutant was measured in the ‘calibration curve’ experiment shown in Figure 2 (see also Materials and methods). The ratio between velocity in the light and velocity in the dark reflects the mean of triplicate measurements; error bars indicate SEM. The green line was fit by linear regression.
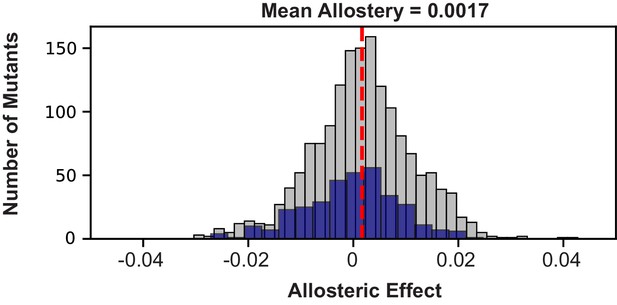
Distribution of mutational effects on allosteric regulation.
The allosteric effect of all viable mutants is shown in gray with the mean allosteric effect of 0.0017 shown as a red dotted line. The allosteric effect of viable mutants in the sector is shown overlaid in blue. The mean allosteric effect of sector positions is −0.0005. The cutoff for sector identity used is a p-value of 0.01 as calculated in Reynolds et al., 2011 (Rivoire et al., 2016).
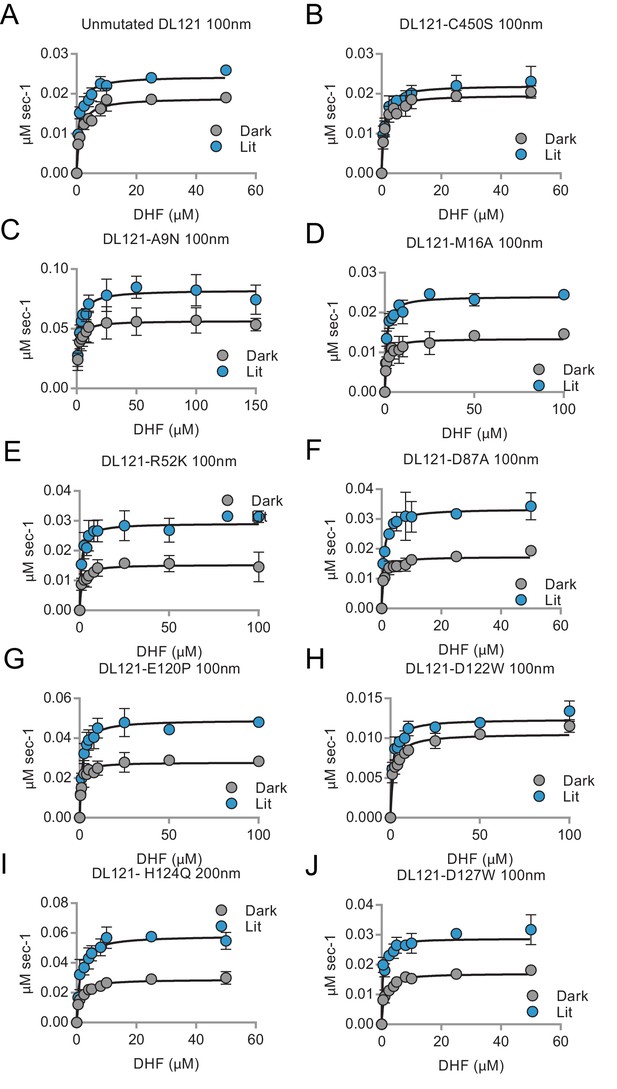
Steady state kinetics measurements for select mutants in the light and dark.
Initial velocity vs. substrate (dihydrofolate) concentration for the purified DL121 chimeric protein, allosterically inactivated DL121-C450S and eight point mutations to the DHFR domain of DL121. Lit (blue) and dark (gray) conditions are shown with error bars representing standard deviation across three replicates. The kcat, KM, catalytic efficiency and associated error are reported in Supplementary file 1a.
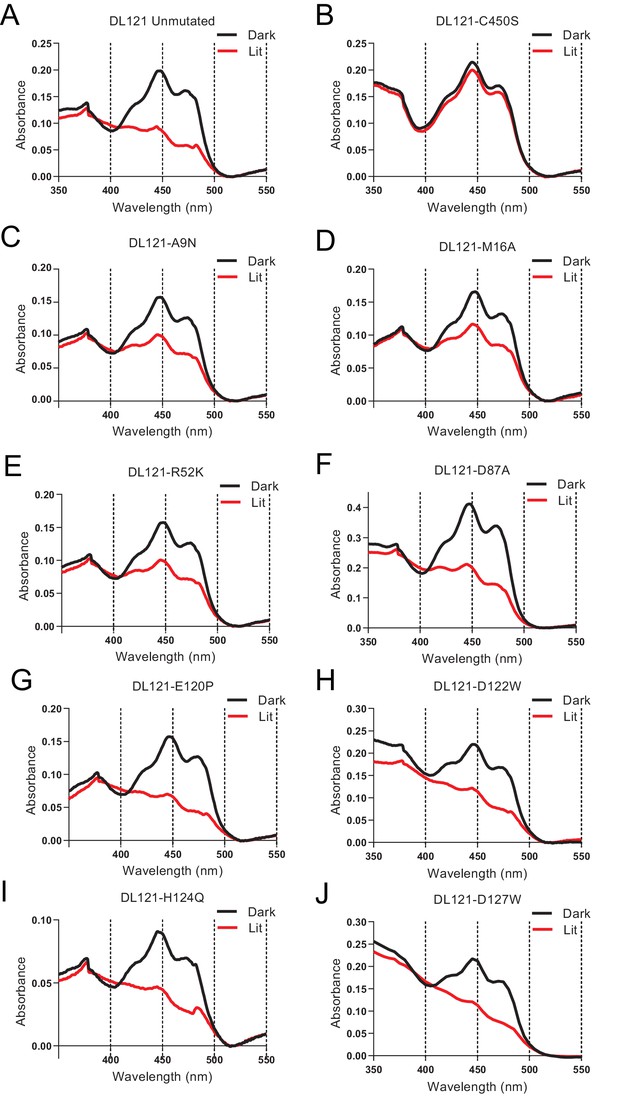
Spectroscopic characterization of LOV2 activation for select DL121 mutants.
The absorbance of purified DL121 chimeric protein, allosterically inactivated DL121-C450S and eight point mutations to the DHFR domain of DL121. Lit state absorbance spectra (red line) were measured after illumination for at least 2 min by full spectrum 125 watt 6400K fluorescent lamp (Hydrofarm Inc). Dark conditions are taken under the same conditions but using opaque tubes when the sample was placed under the lamp. With the exception of the DL121-C450S mutant, all show a characteristic spectral shift upon light stimulation consistent with an active LOV2 domain. Formation of a covalent FMN-thiol adduct in the LOV2 domain upon light exposure causes the 447 nm peak in the dark state to shift to 390 nm in the light.
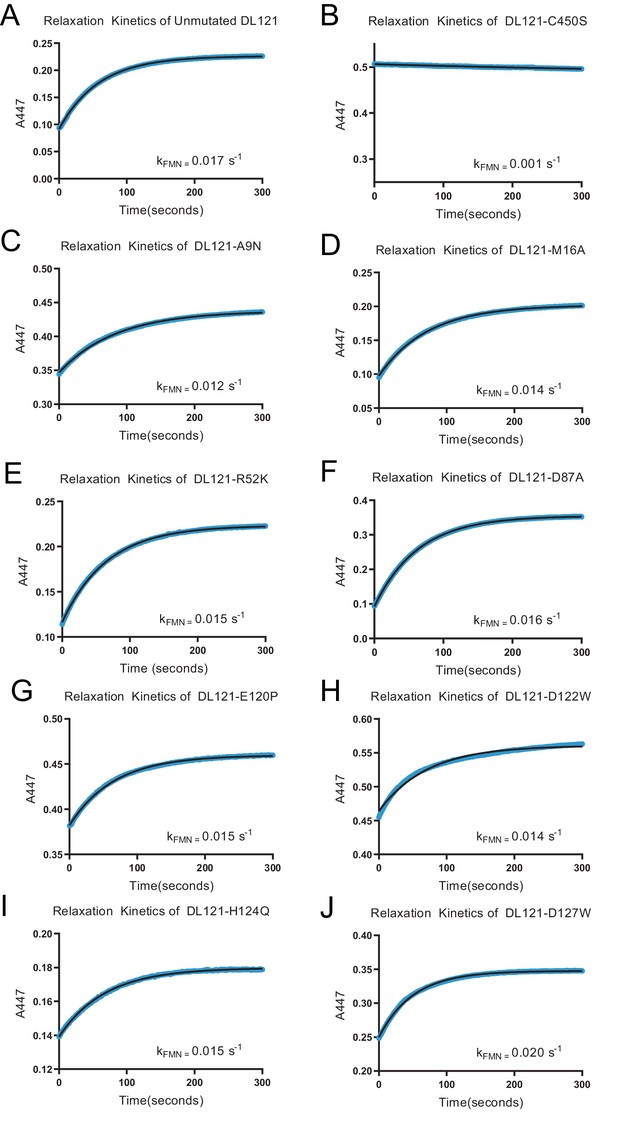
Relaxation rate of the LOV2 chromophore for select DL121 mutants.
The relaxation of the chromophore at 447 nm was observed for 5 min following illumination for at least 2 min by full spectrum 125 watt 6400K fluorescent lamp (Hydrofarm Inc). With the exception of the allosterically inactivated DL121-C450S all of the assayed LOV2 domains had exponential and reversible relaxation to the dark state near that of the unmutated DL121 (kFMN = 0.017 s−1), indicating an active light response in the protein.
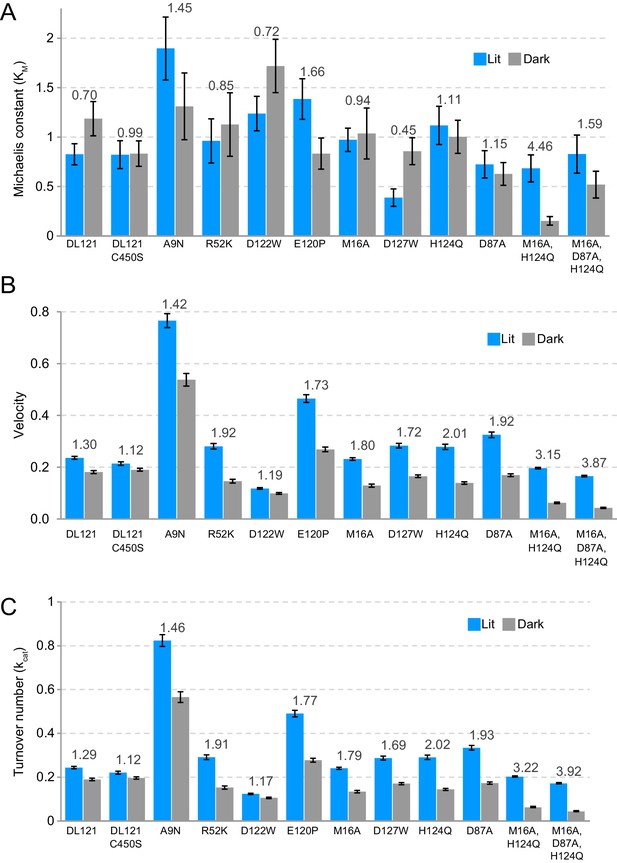
Steady state kinetics parameters under lit and dark conditions for select mutants of the DL121 fusion.
(A) The KM values (B) enzyme velocity and (C) kcat values are shown for both lit (blue bar) and dark (gray bar) conditions, error bars represent standard error across three replicates. Above each pair of bars the lit:dark ratio of the relevant catalytic parameter is shown. The Michaelis-Menten kinetics values are reported in Supplementary file 1a.
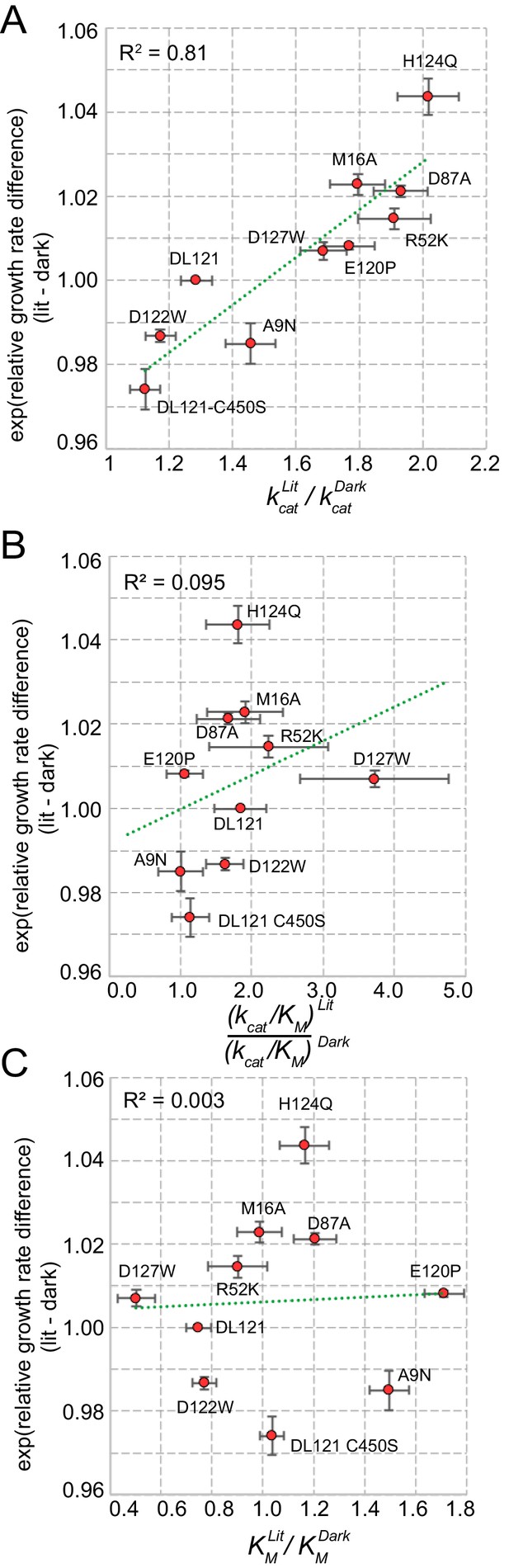
Correlation between in vivo allostery and in vitro steady state kinetics parameters for mutants of the DL121 fusion.
The relationship between the relative growth rate difference (in vivo) and the ratio of (A) catalytic turnover (kcat) (B) catalytic efficiency (kcat/KM) or (C) the Michaelis constant (KM). As we expect a log-linear relationship, we compare the ratio of catalytic constants to the exponent of the relative growth rate difference. The green dashed line is the linear regression with the coefficient of correlation (R2) shown. The low coefficient of correlation in comparisons (B-C) indicates that there is little relationship between the allosteric growth rate difference and both catalytic efficiency and the Michaelis constant ratios. The error bars represent standard error.
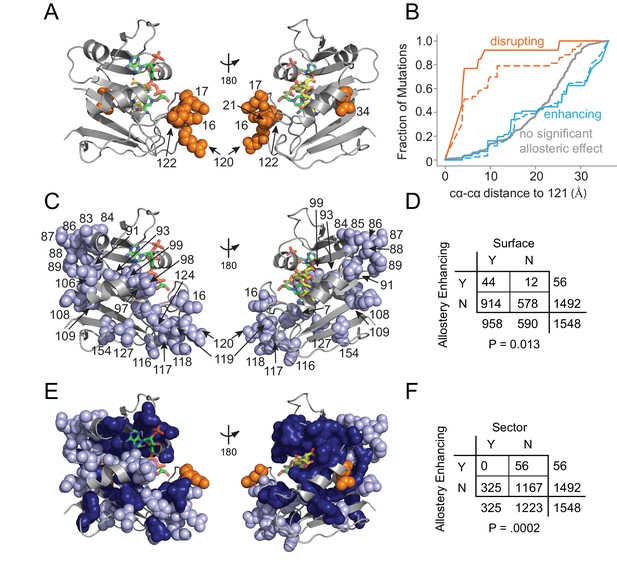
Structural distribution of allosteric mutations.
(A) Sites of allostery disrupting mutations (orange spheres). DHFR backbone is in gray cartoon, folate substrate in yellow sticks, and NADP co-factor in green sticks. (B) Fraction of mutations that enhance (blue), disrupt (orange), or do not significantly influence allostery (gray) as a function of distance to the LOV2 insertion site at DHFR position 121. Solid and dashed lines indicate mutations at either the p=0.016 and p=0.05 significance cutoffs for allostery, respectively. (C) Sites of allostery enhancing mutations (light blue spheres). (D) Contingency table summarizing the overlap between allostery enhancing mutations and mutations on the DHFR solvent accessible surface (considered as >25% relative solvent accessibility in the 1R × 2 PDB). (E) Sites of allostery enhancing (light blue spheres) and disrupting mutations (orange spheres) in the context of the sector (dark blue surface). (F) Contingency table summarizing the relationship between allostery enhancing mutations and sector mutations (sector defined at a p-value cutoff of 0.010). No allostery enhancing mutations occur within the sector.
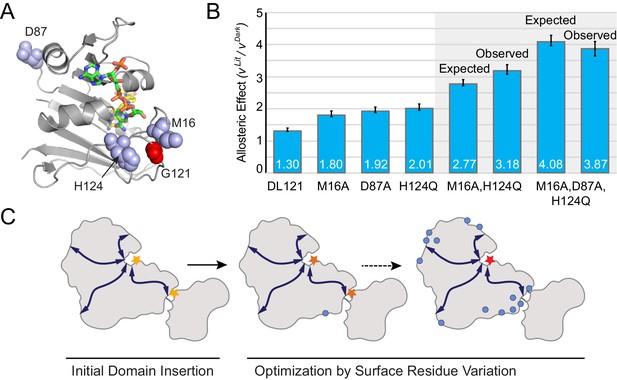
Combinatorial effect of allostery-enhancing mutations.
(A) Location of M16, D87, and H124 (blue spheres). The LOV2 insertion site, G121, is shown in red spheres. The DHFR backbone is in gray cartoon, the folate substrate in yellow sticks, and the NADP co-factor in green sticks. (B) The in vitro allosteric effect of the single, double and triple mutants. Included are the log-additive expectations (Expected) for the double and triple mutants given only the single mutation effects, and the experimentally measured effects (Observed). The ratio between velocity in the light and dark reflects the mean of triplicate measurements; error bars indicate SEM. There is not a statistically significant difference between the expected and observed allosteric effects (p=0.07 for M16A,H124Q, p=0.48 for M16A,D87A,H124Q; as computed by unpaired t-test). (C) Schematic whereby a novel domain insertion is iteratively optimized by surface residue variation.
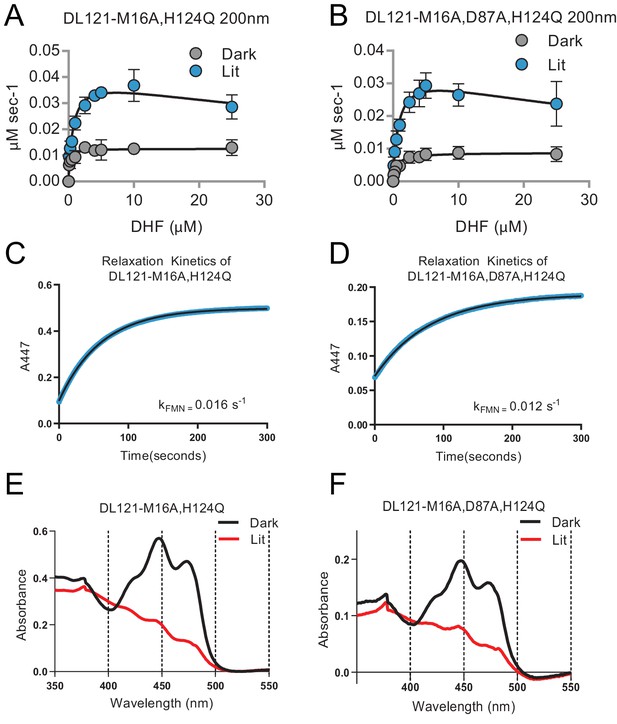
Characterization of the DL121- M16A,H124Q and DL121- M16A, D87A, H124Q mutants.
(A-B) Steady state kinetics measurements in the light and dark. Initial velocity vs. substrate (dihydrofolate) concentration is plotted, Lit (blue) and dark (gray) conditions are shown with error bars representing standard deviation across three replicates. For both the double and triple mutant, the lit states were better fit by a substrate inhibition model than a standard Michaelis-Menten model (p<0.05). The kcat, KM, catalytic efficiency and associated error are reported in Supplementary file 1a. (C-D) Relaxation rate of the LOV2 chromophore. The relaxation of the chromophore at 447 nm was observed for 5 min following illumination for at least 2 min by full spectrum 125 watt 6400K fluorescent lamp (Hydrofarm Inc). (E-F) Spectroscopic characterization of LOV2 activation. Lit state absorbance spectra (red line) were measured after illumination for at least 2 min by full spectrum 125 watt 6400K fluorescent lamp (Hydrofarm Inc). Dark conditions are taken under the same conditions but using opaque tubes when the sample was placed under the lamp. Both mutants show a characteristic spectral shift upon light stimulation consistent with an active LOV2 domain.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Escherichia coli) | DHFR-LOV2 121 | Reynolds et al. Cell 2011 [20] | Fusion of Escherichia coli DHFR and Avena sativa LOV2 | |
| Strain, strain background (Escherichia coli) | BL21(DE3) | New England Biolabs | NEB #: C2527H | Competent cells |
| Strain, strain background (Escherichia coli) | ER2566 ΔfolA ΔthyA | Dr. Steven Benkovic, described in [20, 26] | Competent cells | |
| Strain, strain background (Escherichia coli) | XL1-Blue | Agilent Technologies | Cat. #: 200249 | Competent cells |
| Recombinant DNA reagent | pACYC-Duet_DL121_WTTS(plasmid) | Reynolds et al. Cell 2011 [20] | Addgene ID 171954 | Contains chimeric DL121 with TYMS (selection vector) |
| Recombinant DNA reagent | pHIS8-3_DL121(plasmid) | Reynolds et al. Cell 2011 [20] | Addgene ID 171953 | Contains chimeric DL121 (expression vector) |
| Sequence-based reagent | DL121_pos1_fwd | This Paper | Mutagenic PCR primer | NNSATCAGTCTGATTGCGGCG |
| Sequence-based reagent | DL121_pos2_fwd | This Paper | Mutagenic PCR primer | NNSAGTCTGATTGCGGCGTTAG |
| Sequence-based reagent | DL121_pos3_fwd | This Paper | Mutagenic PCR primer | NNSCTGATTGCGGCGTTAGCG |
| Sequence-based reagent | DL121_pos4_fwd | This Paper | Mutagenic PCR primer | NNSATTGCGGCGTTAGCGGTA |
| Sequence-based reagent | DL121_pos5_fwd | This Paper | Mutagenic PCR primer | NNSGCGGCGTTAGCGGTAGAT |
| Sequence-based reagent | DL121_pos6_fwd | This Paper | Mutagenic PCR primer | NNSGCGTTAGCGGTAGATCGC |
| Sequence-based reagent | DL121_pos7_fwd | This Paper | Mutagenic PCR primer | NNSTTAGCGGTAGATCGCGTTATC |
| Sequence-based reagent | DL121_pos8_fwd | This Paper | Mutagenic PCR primer | NNSGCGGTAGATCGCGTTATCG |
| Sequence-based reagent | DL121_pos9_fwd | This Paper | Mutagenic PCR primer | NNSGTAGATCGCGTTATCGGCATG |
| Sequence-based reagent | DL121_pos10_fwd | This Paper | Mutagenic PCR primer | NNSGATCGCGTTATCGGCATGG |
| Sequence-based reagent | DL121_pos11_fwd | This Paper | Mutagenic PCR primer | NNSCGCGTTATCGGCATGGAAAA |
| Sequence-based reagent | DL121_pos12_fwd | This Paper | Mutagenic PCR primer | NNSGTTATCGGCATGGAAAACGC |
| Sequence-based reagent | DL121_pos13_fwd | This Paper | Mutagenic PCR primer | NNSATCGGCATGGAAAACGCC |
| Sequence-based reagent | DL121_pos14_fwd | This Paper | Mutagenic PCR primer | NNSGGCATGGAAAACGCCATG |
| Sequence-based reagent | DL121_pos15_fwd | This Paper | Mutagenic PCR primer | NNSATGGAAAACGCCATGCCG |
| Sequence-based reagent | DL121_pos16_fwd | This Paper | Mutagenic PCR primer | NNSGAAAACGCCATGCCGTGG |
| Sequence-based reagent | DL121_pos17_fwd | This Paper | Mutagenic PCR primer | NNSAACGCCATGCCGTGGAAC |
| Sequence-based reagent | DL121_pos18_fwd | This Paper | Mutagenic PCR primer | NNSGCCATGCCGTGGAACCTG |
| Sequence-based reagent | DL121_pos19_fwd | This Paper | Mutagenic PCR primer | NNSATGCCGTGGAACCTGCCT |
| Sequence-based reagent | DL121_pos20_fwd | This Paper | Mutagenic PCR primer | NNSCCGTGGAACCTGCCTGCC |
| Sequence-based reagent | DL121_pos21_fwd | This Paper | Mutagenic PCR primer | NNSTGGAACCTGCCTGCCGAT |
| Sequence-based reagent | DL121_pos22_fwd | This Paper | Mutagenic PCR primer | NNSAACCTGCCTGCCGATCTC |
| Sequence-based reagent | DL121_pos23_fwd | This Paper | Mutagenic PCR primer | NNSCTGCCTGCCGATCTCGCC |
| Sequence-based reagent | DL121_pos24_fwd | This Paper | Mutagenic PCR primer | NNSCCTGCCGATCTCGCCTGG |
| Sequence-based reagent | DL121_pos25_fwd | This Paper | Mutagenic PCR primer | NNSGCCGATCTCGCCTGGTTT |
| Sequence-based reagent | DL121_pos26_fwd | This Paper | Mutagenic PCR primer | NNSGATCTCGCCTGGTTTAAACGC |
| Sequence-based reagent | DL121_pos27_fwd | This Paper | Mutagenic PCR primer | NNSCTCGCCTGGTTTAAACGCAACA |
| Sequence-based reagent | DL121_pos28_fwd | This Paper | Mutagenic PCR primer | NNSGCCTGGTTTAAACGCAACAC |
| Sequence-based reagent | DL121_pos29_fwd | This Paper | Mutagenic PCR primer | NNSTGGTTTAAACGCAACACCTTAAATAAAC |
| Sequence-based reagent | DL121_pos30_fwd | This Paper | Mutagenic PCR primer | NNSTTTAAACGCAACACCTTAAATAAACCCG |
| Sequence-based reagent | DL121_pos31_fwd | This Paper | Mutagenic PCR primer | NNSAAACGCAACACCTTAAATAAACCCGTG |
| Sequence-based reagent | DL121_pos32_fwd | This Paper | Mutagenic PCR primer | NNSCGCAACACCTTAAATAAACCCGT |
| Sequence-based reagent | DL121_pos33_fwd | This Paper | Mutagenic PCR primer | NNSAACACCTTAAATAAACCCGTGATTATGG |
| Sequence-based reagent | DL121_pos34_fwd | This Paper | Mutagenic PCR primer | NNSACCTTAAATAAACCCGTGATTATGGG |
| Sequence-based reagent | DL121_pos35_fwd | This Paper | Mutagenic PCR primer | NNSTTAAATAAACCCGTGATTATGGGCC |
| Sequence-based reagent | DL121_pos36_fwd | This Paper | Mutagenic PCR primer | NNSAATAAACCCGTGATTATGGGCC |
| Sequence-based reagent | DL121_pos37_fwd | This Paper | Mutagenic PCR primer | NNSAAACCCGTGATTATGGGCC |
| Sequence-based reagent | DL121_pos38_fwd | This Paper | Mutagenic PCR primer | NNSCCCGTGATTATGGGCCGC |
| Sequence-based reagent | DL121_pos39_fwd | This Paper | Mutagenic PCR primer | NNSGTGATTATGGGCCGCCATAC |
| Sequence-based reagent | DL121_pos40_fwd | This Paper | Mutagenic PCR primer | NNSATTATGGGCCGCCATACCT |
| Sequence-based reagent | DL121_pos41_fwd | This Paper | Mutagenic PCR primer | NNSATGGGCCGCCATACCTGG |
| Sequence-based reagent | DL121_pos42_fwd | This Paper | Mutagenic PCR primer | NNSGGCCGCCATACCTGGGAA |
| Sequence-based reagent | DL121_pos43_fwd | This Paper | Mutagenic PCR primer | NNSCGCCATACCTGGGAATCG |
| Sequence-based reagent | DL121_pos44_fwd | This Paper | Mutagenic PCR primer | NNSCATACCTGGGAATCGATCGGT |
| Sequence-based reagent | DL121_pos45_fwd | This Paper | Mutagenic PCR primer | NNSACCTGGGAATCGATCGGT |
| Sequence-based reagent | DL121_pos46_fwd | This Paper | Mutagenic PCR primer | NNSTGGGAATCGATCGGTCGT |
| Sequence-based reagent | DL121_pos47_fwd | This Paper | Mutagenic PCR primer | NNSGAATCGATCGGTCGTCCG |
| Sequence-based reagent | DL121_pos48_fwd | This Paper | Mutagenic PCR primer | NNSTCGATCGGTCGTCCGTTG |
| Sequence-based reagent | DL121_pos49_fwd | This Paper | Mutagenic PCR primer | NNSATCGGTCGTCCGTTGCCA |
| Sequence-based reagent | DL121_pos50_fwd | This Paper | Mutagenic PCR primer | NNSGGTCGTCCGTTGCCAGGA |
| Sequence-based reagent | DL121_pos51_fwd | This Paper | Mutagenic PCR primer | NNSCGTCCGTTGCCAGGACGC |
| Sequence-based reagent | DL121_pos52_fwd | This Paper | Mutagenic PCR primer | NNSCCGTTGCCAGGACGCAAA |
| Sequence-based reagent | DL121_pos53_fwd | This Paper | Mutagenic PCR primer | NNSTTGCCAGGACGCAAAAATATTATCC |
| Sequence-based reagent | DL121_pos54_fwd | This Paper | Mutagenic PCR primer | NNSCCAGGACGCAAAAATATTATCCTGAG |
| Sequence-based reagent | DL121_pos55_fwd | This Paper | Mutagenic PCR primer | NNSGGACGCAAAAATATTATCCTGAGCTC |
| Sequence-based reagent | DL121_pos56_fwd | This Paper | Mutagenic PCR primer | NNSCGCAAAAATATTATCCTGAGCTCACAA |
| Sequence-based reagent | DL121_pos57_fwd | This Paper | Mutagenic PCR primer | NNSAAAAATATTATCCTGAGCTCACAACCGG |
| Sequence-based reagent | DL121_pos58_fwd | This Paper | Mutagenic PCR primer | NNSAATATTATCCTGAGCTCACAACCGGGTA |
| Sequence-based reagent | DL121_pos59_fwd | This Paper | Mutagenic PCR primer | NNSATTATCCTGAGCTCACAACCG |
| Sequence-based reagent | DL121_pos60_fwd | This Paper | Mutagenic PCR primer | NNSATCCTGAGCTCACAACCG |
| Sequence-based reagent | DL121_pos61_fwd | This Paper | Mutagenic PCR primer | NNSCTGAGCTCACAACCGGGT |
| Sequence-based reagent | DL121_pos62_fwd | This Paper | Mutagenic PCR primer | NNSAGCTCACAACCGGGTACG |
| Sequence-based reagent | DL121_pos63_fwd | This Paper | Mutagenic PCR primer | NNSTCACAACCGGGTACGGAC |
| Sequence-based reagent | DL121_pos64_fwd | This Paper | Mutagenic PCR primer | NNSCAACCGGGTACGGACGAT |
| Sequence-based reagent | DL121_pos65_fwd | This Paper | Mutagenic PCR primer | NNSCCGGGTACGGACGATCGC |
| Sequence-based reagent | DL121_pos66_fwd | This Paper | Mutagenic PCR primer | NNSGGTACGGACGATCGCGTA |
| Sequence-based reagent | DL121_pos67_fwd | This Paper | Mutagenic PCR primer | NNSACGGACGATCGCGTAACG |
| Sequence-based reagent | DL121_pos68_fwd | This Paper | Mutagenic PCR primer | NNSGACGATCGCGTAACGTGG |
| Sequence-based reagent | DL121_pos69_fwd | This Paper | Mutagenic PCR primer | NNSGATCGCGTAACGTGGGTG |
| Sequence-based reagent | DL121_pos70_fwd | This Paper | Mutagenic PCR primer | NNSCGCGTAACGTGGGTGAAG |
| Sequence-based reagent | DL121_pos71_fwd | This Paper | Mutagenic PCR primer | NNSGTAACGTGGGTGAAGTCGG |
| Sequence-based reagent | DL121_pos72_fwd | This Paper | Mutagenic PCR primer | NNSACGTGGGTGAAGTCGGTG |
| Sequence-based reagent | DL121_pos73_fwd | This Paper | Mutagenic PCR primer | NNSTGGGTGAAGTCGGTGGAT |
| Sequence-based reagent | DL121_pos74_fwd2 | This Paper | Mutagenic PCR primer | NNSGTGAAGTCGGTGGATGAAG |
| Sequence-based reagent | DL121_pos75_fwd | This Paper | Mutagenic PCR primer | NNSAAGTCGGTGGATGAAGCAATTG |
| Sequence-based reagent | DL121_pos76_fwd | This Paper | Mutagenic PCR primer | NNSTCGGTGGATGAAGCAATTGC |
| Sequence-based reagent | DL121_pos77_fwd | This Paper | Mutagenic PCR primer | NNSGTGGATGAAGCAATTGCGG |
| Sequence-based reagent | DL121_pos78_fwd | This Paper | Mutagenic PCR primer | NNSGATGAAGCAATTGCGGCG |
| Sequence-based reagent | DL121_pos79_fwd | This Paper | Mutagenic PCR primer | NNSGAAGCAATTGCGGCGTGT |
| Sequence-based reagent | DL121_pos80_fwd | This Paper | Mutagenic PCR primer | NNSGCAATTGCGGCGTGTGGT |
| Sequence-based reagent | DL121_pos81_fwd | This Paper | Mutagenic PCR primer | NNSATTGCGGCGTGTGGTGAC |
| Sequence-based reagent | DL121_pos82_fwd | This Paper | Mutagenic PCR primer | NNSGCGGCGTGTGGTGACGTAC |
| Sequence-based reagent | DL121_pos83_fwd | This Paper | Mutagenic PCR primer | NNSGCGTGTGGTGACGTACCA |
| Sequence-based reagent | DL121_pos84_fwd | This Paper | Mutagenic PCR primer | NNSTGTGGTGACGTACCAGAAATCAT |
| Sequence-based reagent | DL121_pos85_fwd | This Paper | Mutagenic PCR primer | NNSGGTGACGTACCAGAAATCATGG |
| Sequence-based reagent | DL121_pos86_fwd | This Paper | Mutagenic PCR primer | NNSGACGTACCAGAAATCATGGTGATTG |
| Sequence-based reagent | DL121_pos87_fwd | This Paper | Mutagenic PCR primer | NNSGTACCAGAAATCATGGTGATTGGC |
| Sequence-based reagent | DL121_pos88_fwd | This Paper | Mutagenic PCR primer | NNSCCAGAAATCATGGTGATTGGC |
| Sequence-based reagent | DL121_pos89_fwd | This Paper | Mutagenic PCR primer | NNSGAAATCATGGTGATTGGCGG |
| Sequence-based reagent | DL121_pos90_fwd | This Paper | Mutagenic PCR primer | NNSATCATGGTGATTGGCGGC |
| Sequence-based reagent | DL121_pos91_fwd | This Paper | Mutagenic PCR primer | NNSATGGTGATTGGCGGCGGC |
| Sequence-based reagent | DL121_pos92_fwd | This Paper | Mutagenic PCR primer | NNSGTGATTGGCGGCGGCCGC |
| Sequence-based reagent | DL121_pos93_fwd | This Paper | Mutagenic PCR primer | NNSATTGGCGGCGGCCGCGTT |
| Sequence-based reagent | DL121_pos94_fwd | This Paper | Mutagenic PCR primer | NNSGGCGGCGGCCGCGTTTAT |
| Sequence-based reagent | DL121_pos95_fwd | This Paper | Mutagenic PCR primer | NNSGGCGGCCGCGTTTATGAA |
| Sequence-based reagent | DL121_pos96_fwd | This Paper | Mutagenic PCR primer | NNSGGCCGCGTTTATGAACAGTT |
| Sequence-based reagent | DL121_pos97_fwd | This Paper | Mutagenic PCR primer | NNSCGCGTTTATGAACAGTTCTTGC |
| Sequence-based reagent | DL121_pos98_fwd | This Paper | Mutagenic PCR primer | NNSGTTTATGAACAGTTCTTGCCAAAAGCGC |
| Sequence-based reagent | DL121_pos99_fwd | This Paper | Mutagenic PCR primer | NNSTATGAACAGTTCTTGCCAAAAGCGCAAA |
| Sequence-based reagent | DL121_pos100_fwd | This Paper | Mutagenic PCR primer | NNSGAACAGTTCTTGCCAAAAGCGCAAAAGC |
| Sequence-based reagent | DL121_pos101_fwd | This Paper | Mutagenic PCR primer | NNSCAGTTCTTGCCAAAAGCGCAAAAGCTTT |
| Sequence-based reagent | DL121_pos102_fwd | This Paper | Mutagenic PCR primer | NNSTTCTTGCCAAAAGCGCAAAAG |
| Sequence-based reagent | DL121_pos103_fwd | This Paper | Mutagenic PCR primer | NNSTTGCCAAAAGCGCAAAAGC |
| Sequence-based reagent | DL121_pos104_fwd | This Paper | Mutagenic PCR primer | NNSCCAAAAGCGCAAAAGCTTTATCTG |
| Sequence-based reagent | DL121_pos105_fwd | This Paper | Mutagenic PCR primer | NNSAAAGCGCAAAAGCTTTATCTGACG |
| Sequence-based reagent | DL121_pos106_fwd | This Paper | Mutagenic PCR primer | NNSGCGCAAAAGCTTTATCTGACG |
| Sequence-based reagent | DL121_pos107_fwd | This Paper | Mutagenic PCR primer | NNSCAAAAGCTTTATCTGACGCATATCGAC |
| Sequence-based reagent | DL121_pos108_fwd | This Paper | Mutagenic PCR primer | NNSAAGCTTTATCTGACGCATATCGAC |
| Sequence-based reagent | DL121_pos109_fwd | This Paper | Mutagenic PCR primer | NNSCTTTATCTGACGCATATCGACGC |
| Sequence-based reagent | DL121_pos110_fwd | This Paper | Mutagenic PCR primer | NNSTATCTGACGCATATCGACGCA |
| Sequence-based reagent | DL121_pos111_fwd | This Paper | Mutagenic PCR primer | NNSCTGACGCATATCGACGCAG |
| Sequence-based reagent | DL121_pos112_fwd | This Paper | Mutagenic PCR primer | NNSACGCATATCGACGCAGAAGT |
| Sequence-based reagent | DL121_pos113_fwd | This Paper | Mutagenic PCR primer | NNSCATATCGACGCAGAAGTGGAAC |
| Sequence-based reagent | DL121_pos114_fwd | This Paper | Mutagenic PCR primer | NNSATCGACGCAGAAGTGGAACT |
| Sequence-based reagent | DL121_pos115_fwd | This Paper | Mutagenic PCR primer | NNSGACGCAGAAGTGGAACTGG |
| Sequence-based reagent | DL121_pos116_fwd | This Paper | Mutagenic PCR primer | NNSGCAGAAGTGGAACTGGCC |
| Sequence-based reagent | DL121_pos117_fwd | This Paper | Mutagenic PCR primer | NNSGAAGTGGAACTGGCCACC |
| Sequence-based reagent | DL121_pos118_fwd | This Paper | Mutagenic PCR primer | NNSGTGGAACTGGCCACCACT |
| Sequence-based reagent | DL121_pos119_fwd | This Paper | Mutagenic PCR primer | NNSGAACTGGCCACCACTCTAGA |
| Sequence-based reagent | DL121_pos120_fwd | This Paper | Mutagenic PCR primer | NNSCTGGCCACCACTCTAGAG |
| Sequence-based reagent | DL121_pos121_fwd | This Paper | Mutagenic PCR primer | NNSGACACCCATTTCCCGGATTAC |
| Sequence-based reagent | DL121_pos122_fwd | This Paper | Mutagenic PCR primer | NNSACCCATTTCCCGGATTACGA |
| Sequence-based reagent | DL121_pos123_fwd | This Paper | Mutagenic PCR primer | NNSCATTTCCCGGATTACGAGCC |
| Sequence-based reagent | DL121_pos124_fwd | This Paper | Mutagenic PCR primer | NNSTTCCCGGATTACGAGCCG |
| Sequence-based reagent | DL121_pos125_fwd | This Paper | Mutagenic PCR primer | NNSCCGGATTACGAGCCGGAT |
| Sequence-based reagent | DL121_pos126_fwd | This Paper | Mutagenic PCR primer | NNSGATTACGAGCCGGATGACTG |
| Sequence-based reagent | DL121_pos127_fwd | This Paper | Mutagenic PCR primer | NNSTACGAGCCGGATGACTGG |
| Sequence-based reagent | DL121_pos128_fwd | This Paper | Mutagenic PCR primer | NNSGAGCCGGATGACTGGGAA |
| Sequence-based reagent | DL121_pos129_fwd | This Paper | Mutagenic PCR primer | NNSCCGGATGACTGGGAATCG |
| Sequence-based reagent | DL121_pos130_fwd | This Paper | Mutagenic PCR primer | NNSGATGACTGGGAATCGGTATTCAG |
| Sequence-based reagent | DL121_pos131_fwd | This Paper | Mutagenic PCR primer | NNSGACTGGGAATCGGTATTCAGC |
| Sequence-based reagent | DL121_pos132_fwd | This Paper | Mutagenic PCR primer | NNSTGGGAATCGGTATTCAGCGAATT |
| Sequence-based reagent | DL121_pos133_fwd | This Paper | Mutagenic PCR primer | NNSGAATCGGTATTCAGCGAATTCCAC |
| Sequence-based reagent | DL121_pos134_fwd | This Paper | Mutagenic PCR primer | NNSTCGGTATTCAGCGAATTCCAC |
| Sequence-based reagent | DL121_pos135_fwd | This Paper | Mutagenic PCR primer | NNSGTATTCAGCGAATTCCACGATG |
| Sequence-based reagent | DL121_pos136_fwd | This Paper | Mutagenic PCR primer | NNSTTCAGCGAATTCCACGATGC |
| Sequence-based reagent | DL121_pos137_fwd | This Paper | Mutagenic PCR primer | NNSAGCGAATTCCACGATGCTG |
| Sequence-based reagent | DL121_pos138_fwd | This Paper | Mutagenic PCR primer | NNSGAATTCCACGATGCTGATGC |
| Sequence-based reagent | DL121_pos139_fwd | This Paper | Mutagenic PCR primer | NNSTTCCACGATGCTGATGCG |
| Sequence-based reagent | DL121_pos140_fwd | This Paper | Mutagenic PCR primer | NNSCACGATGCTGATGCGCAG |
| Sequence-based reagent | DL121_pos141_fwd | This Paper | Mutagenic PCR primer | NNSGATGCTGATGCGCAGAACT |
| Sequence-based reagent | DL121_pos142_fwd | This Paper | Mutagenic PCR primer | NNSGCTGATGCGCAGAACTCTC |
| Sequence-based reagent | DL121_pos143_fwd | This Paper | Mutagenic PCR primer | NNSGATGCGCAGAACTCTCACAG |
| Sequence-based reagent | DL121_pos144_fwd | This Paper | Mutagenic PCR primer | NNSGCGCAGAACTCTCACAGC |
| Sequence-based reagent | DL121_pos145_fwd | This Paper | Mutagenic PCR primer | NNSCAGAACTCTCACAGCTATTGCTTTG |
| Sequence-based reagent | DL121_pos146_fwd | This Paper | Mutagenic PCR primer | NNSAACTCTCACAGCTATTGCTTTGAGATT |
| Sequence-based reagent | DL121_pos147_fwd | This Paper | Mutagenic PCR primer | NNSTCTCACAGCTATTGCTTTGAGATTCT |
| Sequence-based reagent | DL121_pos148_fwd | This Paper | Mutagenic PCR primer | NNSCACAGCTATTGCTTTGAGATTCTGG |
| Sequence-based reagent | DL121_pos149_fwd | This Paper | Mutagenic PCR primer | NNSAGCTATTGCTTTGAGATTCTGGAG |
| Sequence-based reagent | DL121_pos150_fwd | This Paper | Mutagenic PCR primer | NNSTATTGCTTTGAGATTCTGGAGCG |
| Sequence-based reagent | DL121_pos151_fwd | This Paper | Mutagenic PCR primer | NNSTGCTTTGAGATTCTGGAGCG |
| Sequence-based reagent | DL121_pos152_fwd | This Paper | Mutagenic PCR primer | NNSTTTGAGATTCTGGAGCGGC |
| Sequence-based reagent | DL121_pos153_fwd | This Paper | Mutagenic PCR primer | NNSGAGATTCTGGAGCGGCGG |
| Sequence-based reagent | DL121_pos154_fwd | This Paper | Mutagenic PCR primer | NNSATTCTGGAGCGGCGGTAA |
| Sequence-based reagent | DL121_pos155_fwd | This Paper | Mutagenic PCR primer | NNSCTGGAGCGGCGGTAACAT |
| Sequence-based reagent | DL121_pos156_fwd | This Paper | Mutagenic PCR primer | NNSGAGCGGCGGTAACATCCG |
| Sequence-based reagent | DL121_pos157_fwd | This Paper | Mutagenic PCR primer | NNSCGGCGGTAACATCCGTCG |
| Sequence-based reagent | DL121_pos158_fwd | This Paper | Mutagenic PCR primer | NNSCGGTAACATCCGTCGACAAG |
| Sequence-based reagent | DL121_pos159_fwd | This Paper | Mutagenic PCR primer | NNSTAACATCCGTCGACAAGCTTG |
| Sequence-based reagent | DL121_pos1_rev | This Paper | Mutagenic PCR primer | CGGATCCTGGCTGTGGTG |
| Sequence-based reagent | DL121_pos2_rev | This Paper | Mutagenic PCR primer | CATCGGATCCTGGCTGTG |
| Sequence-based reagent | DL121_pos3_rev | This Paper | Mutagenic PCR primer | GATCATCGGATCCTGGCTG |
| Sequence-based reagent | DL121_pos4_rev | This Paper | Mutagenic PCR primer | ACTGATCATCGGATCCTGG |
| Sequence-based reagent | DL121_pos5_rev | This Paper | Mutagenic PCR primer | CAGACTGATCATCGGATCCTG |
| Sequence-based reagent | DL121_pos6_rev | This Paper | Mutagenic PCR primer | AATCAGACTGATCATCGGATCCTG |
| Sequence-based reagent | DL121_pos7_rev | This Paper | Mutagenic PCR primer | CGCAATCAGACTGATCATCGG |
| Sequence-based reagent | DL121_pos8_rev | This Paper | Mutagenic PCR primer | CGCCGCAATCAGACTGATC |
| Sequence-based reagent | DL121_pos9_rev | This Paper | Mutagenic PCR primer | TAACGCCGCAATCAGACTGA |
| Sequence-based reagent | DL121_pos10_rev | This Paper | Mutagenic PCR primer | CGCTAACGCCGCAATCAG |
| Sequence-based reagent | DL121_pos11_rev | This Paper | Mutagenic PCR primer | TACCGCTAACGCCGCAAT |
| Sequence-based reagent | DL121_pos12_rev | This Paper | Mutagenic PCR primer | ATCTACCGCTAACGCCGC |
| Sequence-based reagent | DL121_pos13_rev | This Paper | Mutagenic PCR primer | GCGATCTACCGCTAACGC |
| Sequence-based reagent | DL121_pos14_rev | This Paper | Mutagenic PCR primer | AACGCGATCTACCGCTAAC |
| Sequence-based reagent | DL121_pos15_rev | This Paper | Mutagenic PCR primer | GATAACGCGATCTACCGCTAAC |
| Sequence-based reagent | DL121_pos16_rev | This Paper | Mutagenic PCR primer | GCCGATAACGCGATCTACC |
| Sequence-based reagent | DL121_pos17_rev | This Paper | Mutagenic PCR primer | CATGCCGATAACGCGATCTAC |
| Sequence-based reagent | DL121_pos18_rev | This Paper | Mutagenic PCR primer | TTCCATGCCGATAACGCG |
| Sequence-based reagent | DL121_pos19_rev | This Paper | Mutagenic PCR primer | GTTTTCCATGCCGATAACGC |
| Sequence-based reagent | DL121_pos20_rev | This Paper | Mutagenic PCR primer | GGCGTTTTCCATGCCGATAACG |
| Sequence-based reagent | DL121_pos21_rev | This Paper | Mutagenic PCR primer | CATGGCGTTTTCCATGCC |
| Sequence-based reagent | DL121_pos22_rev | This Paper | Mutagenic PCR primer | CGGCATGGCGTTTTCCAT |
| Sequence-based reagent | DL121_pos23_rev | This Paper | Mutagenic PCR primer | CCACGGCATGGCGTTTTC |
| Sequence-based reagent | DL121_pos24_rev | This Paper | Mutagenic PCR primer | GTTCCACGGCATGGCGTT |
| Sequence-based reagent | DL121_pos25_rev | This Paper | Mutagenic PCR primer | CAGGTTCCACGGCATGGC |
| Sequence-based reagent | DL121_pos26_rev | This Paper | Mutagenic PCR primer | AGGCAGGTTCCACGGCAT |
| Sequence-based reagent | DL121_pos27_rev | This Paper | Mutagenic PCR primer | GGCAGGCAGGTTCCACGG |
| Sequence-based reagent | DL121_pos28_rev | This Paper | Mutagenic PCR primer | ATCGGCAGGCAGGTTCCA |
| Sequence-based reagent | DL121_pos29_rev | This Paper | Mutagenic PCR primer | GAGATCGGCAGGCAGGTT |
| Sequence-based reagent | DL121_pos30_rev | This Paper | Mutagenic PCR primer | GGCGAGATCGGCAGGCAG |
| Sequence-based reagent | DL121_pos31_rev | This Paper | Mutagenic PCR primer | CCAGGCGAGATCGGCAGG |
| Sequence-based reagent | DL121_pos32_rev | This Paper | Mutagenic PCR primer | AAACCAGGCGAGATCGGC |
| Sequence-based reagent | DL121_pos33_rev | This Paper | Mutagenic PCR primer | TTTAAACCAGGCGAGATCGG |
| Sequence-based reagent | DL121_pos34_rev | This Paper | Mutagenic PCR primer | GCGTTTAAACCAGGCGAGAT |
| Sequence-based reagent | DL121_pos35_rev | This Paper | Mutagenic PCR primer | GTTGCGTTTAAACCAGGCGA |
| Sequence-based reagent | DL121_pos36_rev | This Paper | Mutagenic PCR primer | GGTGTTGCGTTTAAACCAGG |
| Sequence-based reagent | DL121_pos37_rev | This Paper | Mutagenic PCR primer | TAAGGTGTTGCGTTTAAACCAGG |
| Sequence-based reagent | DL121_pos38_rev | This Paper | Mutagenic PCR primer | ATTTAAGGTGTTGCGTTTAAACCAGG |
| Sequence-based reagent | DL121_pos39_rev | This Paper | Mutagenic PCR primer | TTTATTTAAGGTGTTGCGTTTAAACCAG |
| Sequence-based reagent | DL121_pos40_rev | This Paper | Mutagenic PCR primer | GGGTTTATTTAAGGTGTTGCGTTTAAAC |
| Sequence-based reagent | DL121_pos41_rev | This Paper | Mutagenic PCR primer | CACGGGTTTATTTAAGGTGTTGCGT |
| Sequence-based reagent | DL121_pos42_rev | This Paper | Mutagenic PCR primer | AATCACGGGTTTATTTAAGGTGTTGC |
| Sequence-based reagent | DL121_pos43_rev | This Paper | Mutagenic PCR primer | CATAATCACGGGTTTATTTAAGGTGTTG |
| Sequence-based reagent | DL121_pos44_rev | This Paper | Mutagenic PCR primer | GCCCATAATCACGGGTTTATTTAAGG |
| Sequence-based reagent | DL121_pos45_rev | This Paper | Mutagenic PCR primer | GCGGCCCATAATCACGGG |
| Sequence-based reagent | DL121_pos46_rev | This Paper | Mutagenic PCR primer | ATGGCGGCCCATAATCAC |
| Sequence-based reagent | DL121_pos47_rev | This Paper | Mutagenic PCR primer | GGTATGGCGGCCCATAATC |
| Sequence-based reagent | DL121_pos48_rev | This Paper | Mutagenic PCR primer | CCAGGTATGGCGGCCCATA |
| Sequence-based reagent | DL121_pos49_rev | This Paper | Mutagenic PCR primer | TTCCCAGGTATGGCGGCC |
| Sequence-based reagent | DL121_pos50_rev | This Paper | Mutagenic PCR primer | CGATTCCCAGGTATGGCG |
| Sequence-based reagent | DL121_pos51_rev | This Paper | Mutagenic PCR primer | GATCGATTCCCAGGTATGGCG |
| Sequence-based reagent | DL121_pos52_rev | This Paper | Mutagenic PCR primer | ACCGATCGATTCCCAGGTATG |
| Sequence-based reagent | DL121_pos53_rev | This Paper | Mutagenic PCR primer | ACGACCGATCGATTCCCA |
| Sequence-based reagent | DL121_pos54_rev | This Paper | Mutagenic PCR primer | CGGACGACCGATCGATTC |
| Sequence-based reagent | DL121_pos55_rev | This Paper | Mutagenic PCR primer | CAACGGACGACCGATCGA |
| Sequence-based reagent | DL121_pos56_rev | This Paper | Mutagenic PCR primer | TGGCAACGGACGACCGAT |
| Sequence-based reagent | DL121_pos57_rev | This Paper | Mutagenic PCR primer | TCCTGGCAACGGACGACC |
| Sequence-based reagent | DL121_pos58_rev | This Paper | Mutagenic PCR primer | GCGTCCTGGCAACGGACG |
| Sequence-based reagent | DL121_pos59_rev | This Paper | Mutagenic PCR primer | TTTGCGTCCTGGCAACGG |
| Sequence-based reagent | DL121_pos60_rev | This Paper | Mutagenic PCR primer | ATTTTTGCGTCCTGGCAAC |
| Sequence-based reagent | DL121_pos61_rev | This Paper | Mutagenic PCR primer | AATATTTTTGCGTCCTGGCAAC |
| Sequence-based reagent | DL121_pos62_rev | This Paper | Mutagenic PCR primer | GATAATATTTTTGCGTCCTGGCAAC |
| Sequence-based reagent | DL121_pos63_rev | This Paper | Mutagenic PCR primer | CAGGATAATATTTTTGCGTCCTGGC |
| Sequence-based reagent | DL121_pos64_rev | This Paper | Mutagenic PCR primer | GCTCAGGATAATATTTTTGCGTCCTG |
| Sequence-based reagent | DL121_pos65_rev | This Paper | Mutagenic PCR primer | TGAGCTCAGGATAATATTTTTGCGTCCT |
| Sequence-based reagent | DL121_pos66_rev | This Paper | Mutagenic PCR primer | TTGTGAGCTCAGGATAATATTTTTGCG |
| Sequence-based reagent | DL121_pos67_rev | This Paper | Mutagenic PCR primer | CGGTTGTGAGCTCAGGATAATATTTTTG |
| Sequence-based reagent | DL121_pos68_rev | This Paper | Mutagenic PCR primer | ACCCGGTTGTGAGCTCAG |
| Sequence-based reagent | DL121_pos69_rev | This Paper | Mutagenic PCR primer | CGTACCCGGTTGTGAGCT |
| Sequence-based reagent | DL121_pos70_rev | This Paper | Mutagenic PCR primer | GTCCGTACCCGGTTGTGA |
| Sequence-based reagent | DL121_pos71_rev | This Paper | Mutagenic PCR primer | ATCGTCCGTACCCGGTTG |
| Sequence-based reagent | DL121_pos72_rev | This Paper | Mutagenic PCR primer | GCGATCGTCCGTACCCGG |
| Sequence-based reagent | DL121_pos73_rev | This Paper | Mutagenic PCR primer | TACGCGATCGTCCGTACC |
| Sequence-based reagent | DL121_pos74_rev2 | This Paper | Mutagenic PCR primer | CGTTACGCGATCGTCC |
| Sequence-based reagent | DL121_pos75_rev | This Paper | Mutagenic PCR primer | CCACGTTACGCGATCGTC |
| Sequence-based reagent | DL121_pos76_rev | This Paper | Mutagenic PCR primer | CACCCACGTTACGCGATC |
| Sequence-based reagent | DL121_pos77_rev | This Paper | Mutagenic PCR primer | CTTCACCCACGTTACGCG |
| Sequence-based reagent | DL121_pos78_rev | This Paper | Mutagenic PCR primer | CGACTTCACCCACGTTACG |
| Sequence-based reagent | DL121_pos79_rev | This Paper | Mutagenic PCR primer | CACCGACTTCACCCACGT |
| Sequence-based reagent | DL121_pos80_rev | This Paper | Mutagenic PCR primer | ATCCACCGACTTCACCCA |
| Sequence-based reagent | DL121_pos81_rev | This Paper | Mutagenic PCR primer | TTCATCCACCGACTTCACC |
| Sequence-based reagent | DL121_pos82_rev | This Paper | Mutagenic PCR primer | TGCTTCATCCACCGACTTCACC |
| Sequence-based reagent | DL121_pos83_rev | This Paper | Mutagenic PCR primer | AATTGCTTCATCCACCGACTTC |
| Sequence-based reagent | DL121_pos84_rev | This Paper | Mutagenic PCR primer | CGCAATTGCTTCATCCACC |
| Sequence-based reagent | DL121_pos85_rev | This Paper | Mutagenic PCR primer | CGCCGCAATTGCTTCATC |
| Sequence-based reagent | DL121_pos86_rev | This Paper | Mutagenic PCR primer | ACACGCCGCAATTGCTTC |
| Sequence-based reagent | DL121_pos87_rev | This Paper | Mutagenic PCR primer | ACCACACGCCGCAATTGC |
| Sequence-based reagent | DL121_pos88_rev | This Paper | Mutagenic PCR primer | GTCACCACACGCCGCAAT |
| Sequence-based reagent | DL121_pos89_rev2 | This Paper | Mutagenic PCR primer | TACGTCACCACACGCC |
| Sequence-based reagent | DL121_pos90_rev | This Paper | Mutagenic PCR primer | TGGTACGTCACCACACGC |
| Sequence-based reagent | DL121_pos91_rev | This Paper | Mutagenic PCR primer | TTCTGGTACGTCACCACACGC |
| Sequence-based reagent | DL121_pos92_rev | This Paper | Mutagenic PCR primer | GATTTCTGGTACGTCACCACACGCC |
| Sequence-based reagent | DL121_pos93_rev | This Paper | Mutagenic PCR primer | CATGATTTCTGGTACGTCACCACACGC |
| Sequence-based reagent | DL121_pos94_rev | This Paper | Mutagenic PCR primer | CACCATGATTTCTGGTACGTCACCACA |
| Sequence-based reagent | DL121_pos95_rev | This Paper | Mutagenic PCR primer | AATCACCATGATTTCTGGTACGTCA |
| Sequence-based reagent | DL121_pos96_rev | This Paper | Mutagenic PCR primer | GCCAATCACCATGATTTCTGGTAC |
| Sequence-based reagent | DL121_pos97_rev | This Paper | Mutagenic PCR primer | GCCGCCAATCACCATGATTT |
| Sequence-based reagent | DL121_pos98_rev | This Paper | Mutagenic PCR primer | GCCGCCGCCAATCACCATG |
| Sequence-based reagent | DL121_pos99_rev | This Paper | Mutagenic PCR primer | GCGGCCGCCGCCAATCAC |
| Sequence-based reagent | DL121_pos100_rev | This Paper | Mutagenic PCR primer | AACGCGGCCGCCGCCAAT |
| Sequence-based reagent | DL121_pos101_rev | This Paper | Mutagenic PCR primer | ATAAACGCGGCCGCCGCC |
| Sequence-based reagent | DL121_pos102_rev | This Paper | Mutagenic PCR primer | TTCATAAACGCGGCCGCC |
| Sequence-based reagent | DL121_pos103_rev | This Paper | Mutagenic PCR primer | CTGTTCATAAACGCGGCC |
| Sequence-based reagent | DL121_pos104_rev | This Paper | Mutagenic PCR primer | GAACTGTTCATAAACGCGGC |
| Sequence-based reagent | DL121_pos105_rev | This Paper | Mutagenic PCR primer | CAAGAACTGTTCATAAACGCGG |
| Sequence-based reagent | DL121_pos106_rev | This Paper | Mutagenic PCR primer | TGGCAAGAACTGTTCATAAACGC |
| Sequence-based reagent | DL121_pos107_rev | This Paper | Mutagenic PCR primer | TTTTGGCAAGAACTGTTCATAAACG |
| Sequence-based reagent | DL121_pos108_rev | This Paper | Mutagenic PCR primer | CGCTTTTGGCAAGAACTGTTCATAAA |
| Sequence-based reagent | DL121_pos109_rev | This Paper | Mutagenic PCR primer | TTGCGCTTTTGGCAAGAACT |
| Sequence-based reagent | DL121_pos110_rev | This Paper | Mutagenic PCR primer | CTTTTGCGCTTTTGGCAAGAAC |
| Sequence-based reagent | DL121_pos111_rev | This Paper | Mutagenic PCR primer | AAGCTTTTGCGCTTTTGGC |
| Sequence-based reagent | DL121_pos112_rev | This Paper | Mutagenic PCR primer | ATAAAGCTTTTGCGCTTTTGGCA |
| Sequence-based reagent | DL121_pos113_rev | This Paper | Mutagenic PCR primer | CAGATAAAGCTTTTGCGCTTTTGG |
| Sequence-based reagent | DL121_pos114_rev | This Paper | Mutagenic PCR primer | CGTCAGATAAAGCTTTTGCGCTTT |
| Sequence-based reagent | DL121_pos115_rev | This Paper | Mutagenic PCR primer | ATGCGTCAGATAAAGCTTTTGCG |
| Sequence-based reagent | DL121_pos116_rev | This Paper | Mutagenic PCR primer | GATATGCGTCAGATAAAGCTTTTGC |
| Sequence-based reagent | DL121_pos117_rev | This Paper | Mutagenic PCR primer | GTCGATATGCGTCAGATAAAGCTTTTG |
| Sequence-based reagent | DL121_pos118_rev | This Paper | Mutagenic PCR primer | TGCGTCGATATGCGTCAGATAAA |
| Sequence-based reagent | DL121_pos119_rev | This Paper | Mutagenic PCR primer | TTCTGCGTCGATATGCGTCA |
| Sequence-based reagent | DL121_pos120_rev | This Paper | Mutagenic PCR primer | CACTTCTGCGTCGATATGCG |
| Sequence-based reagent | DL121_pos121_rev | This Paper | Mutagenic PCR primer | GTCGATGTTCTCGGCGGT |
| Sequence-based reagent | DL121_pos122_rev | This Paper | Mutagenic PCR primer | GCCGTCGATGTTCTCGGC |
| Sequence-based reagent | DL121_pos123_rev | This Paper | Mutagenic PCR primer | GTCGCCGTCGATGTTCTC |
| Sequence-based reagent | DL121_pos124_rev | This Paper | Mutagenic PCR primer | GGTGTCGCCGTCGATGTT |
| Sequence-based reagent | DL121_pos125_rev | This Paper | Mutagenic PCR primer | ATGGGTGTCGCCGTCGAT |
| Sequence-based reagent | DL121_pos126_rev | This Paper | Mutagenic PCR primer | GAAATGGGTGTCGCCGTC |
| Sequence-based reagent | DL121_pos127_rev | This Paper | Mutagenic PCR primer | CGGGAAATGGGTGTCGCC |
| Sequence-based reagent | DL121_pos128_rev | This Paper | Mutagenic PCR primer | ATCCGGGAAATGGGTGTC |
| Sequence-based reagent | DL121_pos129_rev | This Paper | Mutagenic PCR primer | GTAATCCGGGAAATGGGTGTC |
| Sequence-based reagent | DL121_pos130_rev | This Paper | Mutagenic PCR primer | CTCGTAATCCGGGAAATGGG |
| Sequence-based reagent | DL121_pos131_rev | This Paper | Mutagenic PCR primer | CGGCTCGTAATCCGGGAA |
| Sequence-based reagent | DL121_pos132_rev | This Paper | Mutagenic PCR primer | ATCCGGCTCGTAATCCGG |
| Sequence-based reagent | DL121_pos133_rev | This Paper | Mutagenic PCR primer | GTCATCCGGCTCGTAATCC |
| Sequence-based reagent | DL121_pos134_rev | This Paper | Mutagenic PCR primer | CCAGTCATCCGGCTCGTA |
| Sequence-based reagent | DL121_pos135_rev | This Paper | Mutagenic PCR primer | TTCCCAGTCATCCGGCTC |
| Sequence-based reagent | DL121_pos136_rev | This Paper | Mutagenic PCR primer | CGATTCCCAGTCATCCGG |
| Sequence-based reagent | DL121_pos137_rev | This Paper | Mutagenic PCR primer | TACCGATTCCCAGTCATCCG |
| Sequence-based reagent | DL121_pos138_rev | This Paper | Mutagenic PCR primer | GAATACCGATTCCCAGTCATCC |
| Sequence-based reagent | DL121_pos139_rev | This Paper | Mutagenic PCR primer | GCTGAATACCGATTCCCAGTC |
| Sequence-based reagent | DL121_pos140_rev | This Paper | Mutagenic PCR primer | TTCGCTGAATACCGATTCCCA |
| Sequence-based reagent | DL121_pos141_rev | This Paper | Mutagenic PCR primer | GAATTCGCTGAATACCGATTCCC |
| Sequence-based reagent | DL121_pos142_rev | This Paper | Mutagenic PCR primer | GTGGAATTCGCTGAATACCGATTC |
| Sequence-based reagent | DL121_pos143_rev | This Paper | Mutagenic PCR primer | ATCGTGGAATTCGCTGAATACC |
| Sequence-based reagent | DL121_pos144_rev | This Paper | Mutagenic PCR primer | AGCATCGTGGAATTCGCTG |
| Sequence-based reagent | DL121_pos145_rev | This Paper | Mutagenic PCR primer | ATCAGCATCGTGGAATTCGC |
| Sequence-based reagent | DL121_pos146_rev | This Paper | Mutagenic PCR primer | CGCATCAGCATCGTGGAATT |
| Sequence-based reagent | DL121_pos147_rev | This Paper | Mutagenic PCR primer | CTGCGCATCAGCATCGTG |
| Sequence-based reagent | DL121_pos148_rev | This Paper | Mutagenic PCR primer | GTTCTGCGCATCAGCATC |
| Sequence-based reagent | DL121_pos149_rev | This Paper | Mutagenic PCR primer | AGAGTTCTGCGCATCAGC |
| Sequence-based reagent | DL121_pos150_rev | This Paper | Mutagenic PCR primer | GTGAGAGTTCTGCGCATCAG |
| Sequence-based reagent | DL121_pos151_rev | This Paper | Mutagenic PCR primer | GCTGTGAGAGTTCTGCGC |
| Sequence-based reagent | DL121_pos152_rev | This Paper | Mutagenic PCR primer | ATAGCTGTGAGAGTTCTGCG |
| Sequence-based reagent | DL121_pos153_rev | This Paper | Mutagenic PCR primer | GCAATAGCTGTGAGAGTTCTGC |
| Sequence-based reagent | DL121_pos154_rev | This Paper | Mutagenic PCR primer | AAAGCAATAGCTGTGAGAGTTCTG |
| Sequence-based reagent | DL121_pos155_rev | This Paper | Mutagenic PCR primer | CTCAAAGCAATAGCTGTGAGAGTTC |
| Sequence-based reagent | DL121_pos156_rev | This Paper | Mutagenic PCR primer | AATCTCAAAGCAATAGCTGTGAGAGTT |
| Sequence-based reagent | DL121_pos157_rev | This Paper | Mutagenic PCR primer | CAGAATCTCAAAGCAATAGCTGTGAG |
| Sequence-based reagent | DL121_pos158_rev | This Paper | Mutagenic PCR primer | CTCCAGAATCTCAAAGCAATAGCTG |
| Sequence-based reagent | DL121_pos159_rev | This Paper | Mutagenic PCR primer | CCGCTCCAGAATCTCAAAGC |
| Sequence-based reagent | DL121_E154R_F | This Paper | Mutagenic PCR primer | ctctcacagctattgctttaggattctggagcggcggtaa |
| Sequence-based reagent | DL121_E154R_R | This Paper | Mutagenic PCR primer | ttaccgccgctccagaatcctaaagcaatagctgtgagag |
| Sequence-based reagent | DL121_D122W_F | This Paper | Mutagenic PCR primer | gtaatccgggaaatgggtccagccgtcgatgttctcggc |
| Sequence-based reagent | DL121_D122W_R | This Paper | Mutagenic PCR primer | gccgagaacatcgacggctggacccatttcccggattac |
| Sequence-based reagent | DL121_D127W_F | This Paper | Mutagenic PCR primer | cagtcatccggctcgtaccacgggaaatgggtgtcgc |
| Sequence-based reagent | DL121_D127W_R | This Paper | Mutagenic PCR primer | gcgacacccatttcccgtggtacgagccggatgactg |
| Sequence-based reagent | DL121_M16A_F | This Paper | Mutagenic PCR primer | cggcatggcgttttccgcgccgataacgcgatct |
| Sequence-based reagent | DL121_M16A_R | This Paper | Mutagenic PCR primer | agatcgcgttatcggcgcggaaaacgccatgccg |
| Sequence-based reagent | DL121_A9N_F | This Paper | Mutagenic PCR primer | catgccgataacgcgatctacatttaacgccgcaatcagactgatc |
| Sequence-based reagent | DL121_A9N_R | This Paper | Mutagenic PCR primer | gatcagtctgattgcggcgttaaatgtagatcgcgttatcggcatg |
| Sequence-based reagent | DL121_R52K_F | This Paper | Mutagenic PCR primer | tcctggcaacggcttaccgatcgattcccaggtatggc |
| Sequence-based reagent | DL121_R52K_R | This Paper | Mutagenic PCR primer | gccatacctgggaatcgatcggtaagccgttgccagga |
| Sequence-based reagent | DL121_E120P_F | This Paper | Mutagenic PCR primer | ctagagtggtggccagtggcacttctgcgtcgatat |
| Sequence-based reagent | DL121_E120P_R | This Paper | Mutagenic PCR primer | atatcgacgcagaagtgccactggccaccactctag |
| Sequence-based reagent | DL121_S148C_F | This Paper | Mutagenic PCR primer | aagcaatagctgtgacagttctgcgcatcagcatc |
| Sequence-based reagent | DL121_S148C_R | This Paper | Mutagenic PCR primer | gatgctgatgcgcagaactgtcacagctattgctt |
| Sequence-based reagent | DL121_H124Q_F | This Paper | Mutagenic PCR primer | tcgtaatccgggaactgggtgtcgccgtc |
| Sequence-based reagent | DL121_H12RQ_R | This Paper | Mutagenic PCR primer | gacggcgacacccagttcccggattacga |
| Sequence-based reagent | DL121_D27N_F | This Paper | Mutagenic PCR primer | aaaccaggcgagattggcaggcaggttcc |
| Sequence-based reagent | DL121_D27N_R | This Paper | Mutagenic PCR primer | ggaacctgcctgccaatctcgcctggttt |
| Sequence-based reagent | DL121_D87A_F | This Paper | Mutagenic PCR primer | catgatttctggtacggcaccacacgccgcaat |
| Sequence-based reagent | DL121_D87A_R | This Paper | Mutagenic PCR primer | attgcggcgtgtggtgccgtaccagaaatcatg |
| Sequence-based reagent | Thrombin_to_TEV_F | This Paper | Mutagenic PCR primer | cttccagggtcatgggatgatgatcagtctgattgc |
| Sequence-based reagent | Thrombin_to_TEV_R | This Paper | Mutagenic PCR primer | tacaggttctcaccaccgtggtggtggtg |
| Sequence-based reagent | DL121_SL1V2_F | This Paper | Round one Amplicon PCR primer | cactctttccctacacgacgctcttccgatctnnnnatcaccatcatcaccacagc |
| Sequence-based reagent | DL121_SL1V2_R | This Paper | Round one Amplicon PCR primer | tgactggagttcagacgtgtgctcttccgatctnnnnaccgatcgattcccaggta |
| Sequence-based reagent | DL121_SL2V2_F | This Paper | Round one Amplicon PCR primer | cactctttccctacacgacgctcttccgatctnnnngcaacaccttaaataaacccg |
| Sequence-based reagent | DL121_SL2V2_R | This Paper | Round one Amplicon PCR primer | tgactggagttcagacgtgtgctcttccgatctnnnngatttctggtacgtcaccaca |
| Sequence-based reagent | DL121_SL3V2_F | This Paper | Round one Amplicon PCR primer | cactctttccctacacgacgctcttccgatctnnnngtaacgtgggtgaagtcg |
| Sequence-based reagent | DL121_SL3V2_R | This Paper | Round one Amplicon PCR primer | tgactggagttcagacgtgtgctcttccgatctnnnnctcgatgcgctctagagtg |
| Sequence-based reagent | DL121_SL4V2_F | This Paper | Round one Amplicon PCR primer | cactctttccctacacgacgctcttccgatctnnnnaagaagaccgccgagaacat |
| Sequence-based reagent | DL121_SL4V2_R | This Paper | Round one Amplicon PCR primer | tgactggagttcagacgtgtgctcttccgatctnnnncttaagcattatgcggccg |
| Sequence-based reagent | DL121_CLV3_F | This Paper | Round one Amplicon PCR primer | cactctttccctacacgacgctcttccgatctnnnngacacccatttcccggattacgagc |
| Sequence-based reagent | DL_WTTS_R3 | This Paper | Round one Amplicon PCR primer | tgactggagttcagacgtgtgctcttccgatctnnnngccgtgtacaatacgattactttctg |
| Sequence-based reagent | D501 | Illumina/Reynolds et al. Cell 2011 [20] | Round two Amplicon PCR primer | aatgatacggcgaccaccgagatctacactatagcctacactctttccctacacgac |
| Sequence-based reagent | D502 | Illumina/Reynolds et al. Cell 2011 [20] | Round two Amplicon PCR primer | aatgatacggcgaccaccgagatctacacatagaggcacactctttccctacacgac |
| Sequence-based reagent | D503 | Illumina/Reynolds et al. Cell 2011 [20] | Round two Amplicon PCR primer | aatgatacggcgaccaccgagatctacaccctatcctacactctttccctacacgac |
| Sequence-based reagent | D504 | Illumina/Reynolds et al. Cell 2011 [20] | Round two Amplicon PCR primer | aatgatacggcgaccaccgagatctacacggctctgaacactctttccctacacgac |
| Sequence-based reagent | D505 | Illumina/Reynolds et al. Cell 2011 [20] | Round two Amplicon PCR primer | aatgatacggcgaccaccgagatctacacaggcgaagacactctttccctacacgac |
| Sequence-based reagent | D506 | Illumina/Reynolds et al. Cell 2011 [20] | Round two Amplicon PCR primer | aatgatacggcgaccaccgagatctacactaatcttaacactctttccctacacgac |
| Sequence-based reagent | D507 | Illumina/Reynolds et al. Cell 2011 [20] | Round two Amplicon PCR primer | aatgatacggcgaccaccgagatctacaccaggacgtacactctttccctacacgac |
| Sequence-based reagent | D508 | Illumina/Reynolds et al. Cell 2011 [20] | Round two Amplicon PCR primer | aatgatacggcgaccaccgagatctacacgtactgacacactctttccctacacgac |
| Sequence-based reagent | D701 | Illumina/Reynolds et al. Cell 2011 [20] | Round two Amplicon PCR primer | caagcagaagacggcatacgagatcgagtaatgtgactggagttcagacgtg |
| Sequence-based reagent | D702 | Illumina/Reynolds et al. Cell 2011 [20] | Round two Amplicon PCR primer | caagcagaagacggcatacgagattctccggagtgactggagttcagacgtg |
| Sequence-based reagent | D703 | Illumina/Reynolds et al. Cell 2011 [20] | Round two Amplicon PCR primer | caagcagaagacggcatacgagataatgagcggtgactggagttcagacgtg |
| Sequence-based reagent | D704 | Illumina/Reynolds et al. Cell 2011 [20] | Round two Amplicon PCR primer | caagcagaagacggcatacgagatggaatctcgtgactggagttcagacgtg |
| Sequence-based reagent | D705 | Illumina/Reynolds et al. Cell 2011 [20] | Round two Amplicon PCR primer | caagcagaagacggcatacgagatttctgaatgtgactggagttcagacgtg |
| Sequence-based reagent | D706 | Illumina/Reynolds et al. Cell 2011 [20] | Round two Amplicon PCR primer | caagcagaagacggcatacgagatacgaattcgtgactggagttcagacgtg |
| Sequence-based reagent | D707 | Illumina/Reynolds et al. Cell 2011 [20] | Round two Amplicon PCR primer | caagcagaagacggcatacgagatagcttcaggtgactggagttcagacgtg |
| Sequence-based reagent | D708 | Illumina/Reynolds et al. Cell 2011 [20] | Round two Amplicon PCR primer | caagcagaagacggcatacgagatgcgcattagtgactggagttcagacgtg |
| Sequence-based reagent | D709 | Illumina/Reynolds et al. Cell 2011 [20] | Round two Amplicon PCR primer | caagcagaagacggcatacgagatcatagccggtgactggagttcagacgtg |
| Sequence-based reagent | D710 | Illumina/Reynolds et al. Cell 2011 [20] | Round two Amplicon PCR primer | caagcagaagacggcatacgagatttcgcggagtgactggagttcagacgtg |
| Sequence-based reagent | D711 | Illumina/Reynolds et al. Cell 2011 [20] | Round two Amplicon PCR primer | caagcagaagacggcatacgagatgcgcgagagtgactggagttcagacgtg |
| Sequence-based reagent | D712 | Illumina/Reynolds et al. Cell 2011 [20] | Round two Amplicon PCR primer | caagcagaagacggcatacgagatctatcgctgtgactggagttcagacgtg |
| Commercial assay or kit | QuikChange II site-directed mutagenesis kit | Agilent | Cat. #: 200523 | |
| Software, algorithm | usearch v11.0.667 | Edgar Bioinformatics 2010 (PMID:20709691) | Merge read pairs | https://www.drive5.com/usearch/ |
Additional files
-
Supplementary file 1
Supplementary tables.
(a) Steady state kinetic parameters for select point mutants of the DL121 fusion. The parameter kcat is reported in units of s−1, Km is in units of µM. Error is calculated as standard error of the mean over three replicates. Related to Figure 4 of the main text. (b) Fisher Exact Test p-values for the null hypothesis that the sector and inactivating mutants are independent properties. Inactivating mutations are defined as those that yield relative growth rates at or below the growth rate for DL121-D27N. Over a range of sector definitions, the null hypothesis is rejected at a confidence level of 0.05 or better, shown in red. Sector definitions were taken from Reynolds et al., 2011 (Rivoire et al., 2016). (c) Fisher Exact Test p-values for the null hypothesis that conserved positions and inactivating mutants are independent. Calculations were made over a range of conservation definitions chosen to result in an equal number positions as the sector positions in Supplementary file 1b 23, 36, 40, and 49 positions respectively. In all cases, the null hypothesis is rejected at a confidence level of 0.05 or better (red), and inactivating mutations are enriched at conserved positions beyond expectation due to random chance. Conservation values are calculated as in Reynolds et al., 2011 (Rivoire et al., 2016), and reflect the Kullback-Leibler relative entropy of amino acid frequencies at each DHFR position. (d) Fisher Exact Test p-values for the null hypothesis that the sector and allosteric mutations are independent. We compared over four sector cutoffs (as defined in Rivoire et al., 2016) and at two cutoffs for allostery significance (a standard p-value of 0.05, and an adjusted p-value of 0.016). The multiple hypothesis testing adjusted p-value was obtained by Sequential Goodness of Fit (SGoF, Carvajal-Rodriguez and de Uña-Alvarez, 2011). The top table shows the association between sector positions and allostery enhancing mutations; the bottom table computes the associate between sector positions and allostery disrupting mutations. In nearly all cases, the null hypothesis is rejected at a confidence level of 0.05 or better, shown in red. (e) Fisher Exact Test p-values for the null hypothesis that the solvent accessible DHFR surface and allosteric mutations are independent. At two cutoffs for allostery (a standard p-value of 0.05, and an adjusted p-value of 0.016), the null hypothesis is rejected at a confidence level of 0.05 or better, shown in red. (f) Statistical association of allosteric mutations and surface positions that are either within or contacting the sector. Contacting was defined as two atoms within the sum of their Pauling radii plus 20%. A surface site contacts the sector if the peptide bond atoms of the surface site contact any atoms in the sector position. P-values were computed by Fisher exact test with the null hypothesis that the sector and allosteric mutations are independent. Cutoffs for sector definition as defined in Rivoire et al., 2016 are shown as well as mutants determined to effect allostery either at a 95% confidence interval (p<0.05) or at the multiple hypothesis testing adjusted p-value (p<0.016). The null hypothesis that there is no relationship between allosteric mutations and sector or sector contacting positions on the surface of DHFR of the DL121 chimera is rejected at a confidence level of 0.05 or better over a range of cutoffs, shown in red. Allostery enhancing mutations are depleted from sector connected surface sites, while allostery disrupting mutations are enriched (in comparison with random expectation). (g) Statistical association of allosteric mutations and surface positions that are contacting the sector. In contrast to Supplementary file 1f, surface positions within the sector are excluded. Fisher Exact Test p-values were calculated for the null hypothesis that the sector and allosteric mutations are independent. Cutoffs for sector definition as defined in Rivoire et al., 2016 are shown as well as mutants determined to effect allostery either at a 95% confidence interval (p<0.05) or at the multiple hypothesis testing adjusted p-value (p<0.016). At most cutoff combinations, there is not a statistically significant association between sector connected surface sites and either mutations that enhance (top panel) or disrupt allostery (bottom panel).
- https://cdn.elifesciences.org/articles/68346/elife-68346-supp1-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/68346/elife-68346-transrepform-v2.docx