Cis-regulatory variants affect gene expression dynamics in yeast
Figures
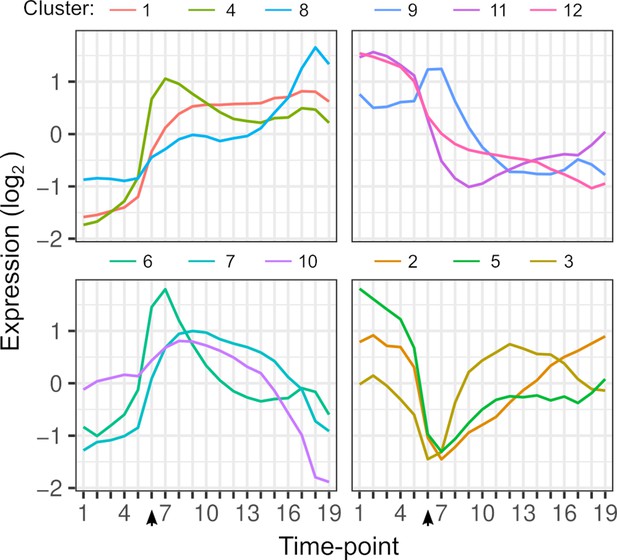
Gene expression dynamics.
Each line shows the average expression of genes in each k-means cluster over timepoints. Clustering is based on the expression of 4703 genes from each hybrid. The arrow indicates the timepoint when glucose was depleted.
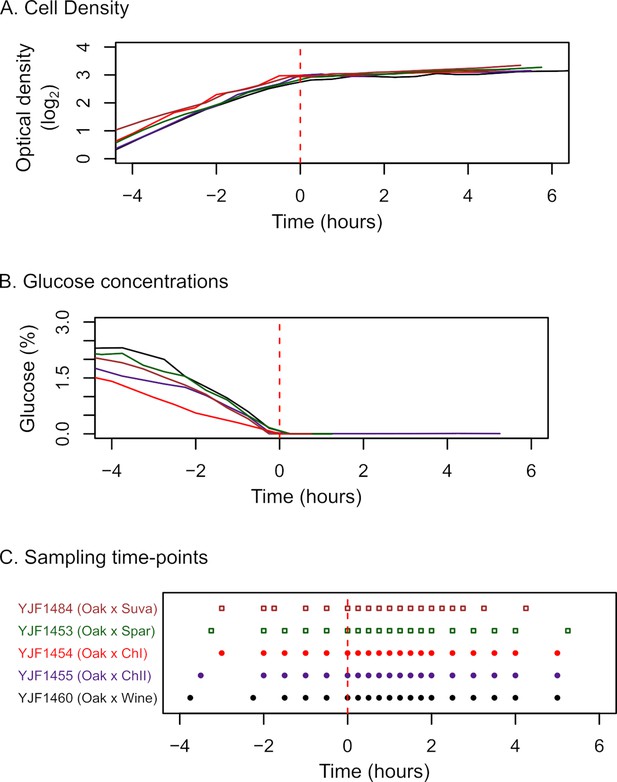
Sampling scheme for gene expression dynamics during the diauxic shift.
Cell density (A) and glucose concentrations (B) were used to choose sampling timepoints for gene expression measurements (C). Time of glucose depletion is marked as 0 hr by a red dashed line. Sample timepoints were taken every 15 min for 2 hr after glucose depletion. Intra-specific hybrid samples are circles, inter-specific hybrid samples are squares.
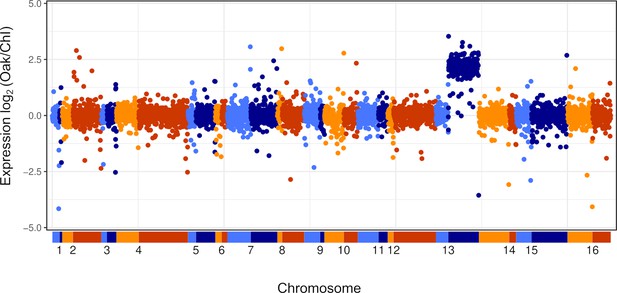
Chromosome 13R aneuploidy in YJM1454 (Oak × ChI).
Expression of the Oak relative to the ChI allele for all genes ordered by position along each chromosome, where expression is the average across all timepoints. Points are color coded by chromosome (alternating blue and orange, bottom), with light colors for the left arm and dark colors for the right arm.
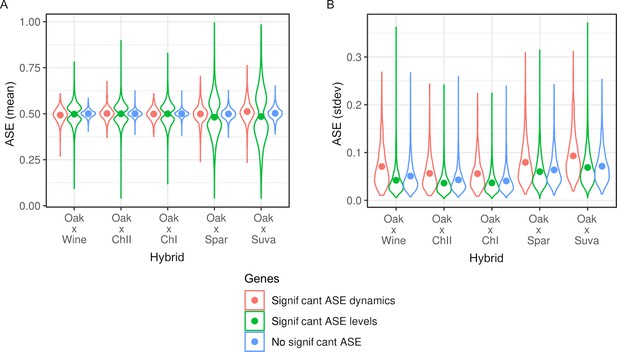
Characteristics of differentially expressed genes.
(A) Average allele-specific expression (ASE) expression levels and (B) standard deviation in ASE expression over time for genes with significant ASE dynamics (red), ASE levels (green), and no ASE (blue) for each of the five hybrids. Points show the mean, and density is indicated by the width of the color-coded regions.
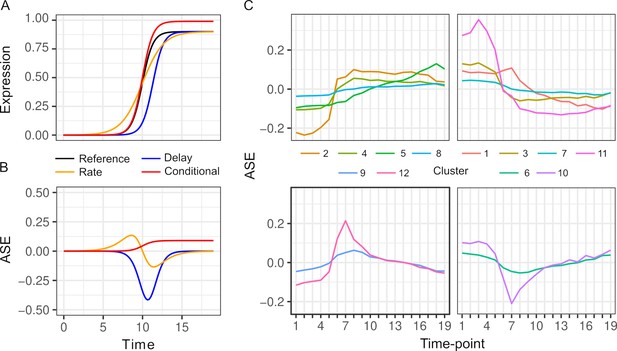
Patterns of allele-specific expression (ASE) dynamics.
(A) Three hypothetical types of differences in expression dynamics in comparison to a common reference (black) are shown by a time delay (blue), rate change (orange), and condition-specific expression difference (red). (B) ASE based on the three types of differences in comparison to the common reference. (C) Average ASE across 19 timepoints of k-means clustering of 6135 genes with significant ASE dynamics. Clusters 6, 9, 10, and 12 (bottom panels) show maximum deviation during the diauxic shift, whereas the others generally show increasing or decreasing ASE differences over time consistent with condition-specific ASE.
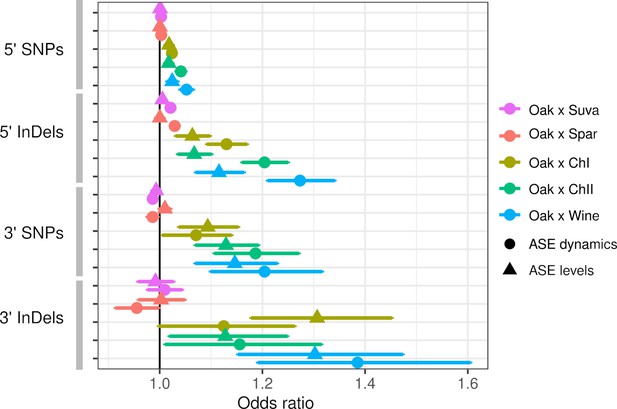
Allele-specific expression (ASE) is associated with intergenic single-nucleotide polymorphisms (SNPs) and insertions/deletions (InDels).
The odds ratio (OR) and 95% confidence interval for associations between the number of SNPs or InDels and significant ASE levels (triangles) and dynamics (circles). The OR of each hybrid is shown separately for upstream (5′) and downstream (3′) intergenic variants.
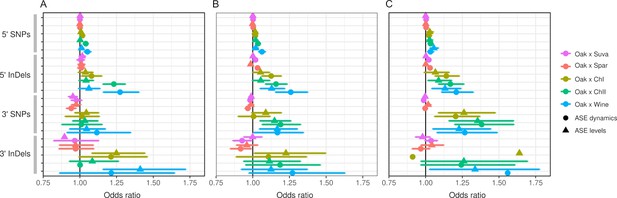
Allele-specific expression (ASE) associations with single-nucleotide polymorphisms (SNPs) and insertions/deletions (InDels) for genes with small (A), medium (B), and large (C) expression differences.
Genes were split into three equal-sized groups based on the sum of squared deviations from an allele frequency of 0.5. Each panel shows the odds ratio and 95% confidence interval for associations between the number of variants (SNPs or InDels) and significant ASE dynamics (circles) or levels (triangles).
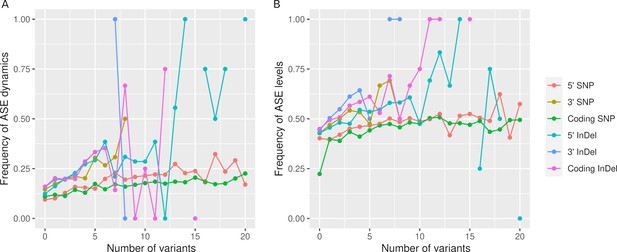
The frequency of significant allele-specific expression (ASE) dynamics (A) and ASE levels (B) as a function of the number of variants.
The data for all three intra-specific hybrids is shown together.
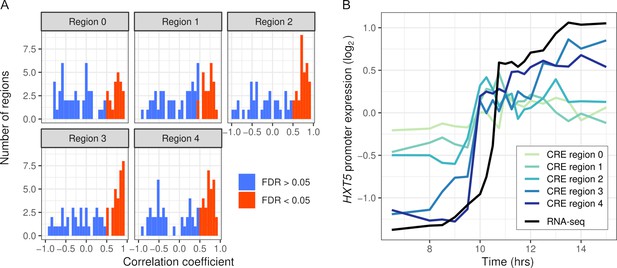
Intra-specific cis-regulatory elements (CREs) recapitulate endogenous expression dynamics.
(A) Histogram of the correlations between CRE expression and the endogenous (RNA-seq) expression of 69 genes. Correlations are shown separately for CRE regions 0–4, ordered proximal to distal of the transcription start site with shifts of 30 bp. CREs with a significant (false discovery rate < 0.05) correlation are shown in red, the rest in blue. (B) CRE expression of five regions upstream of HXT5 as well as its endogenous expression from RNA-seq.
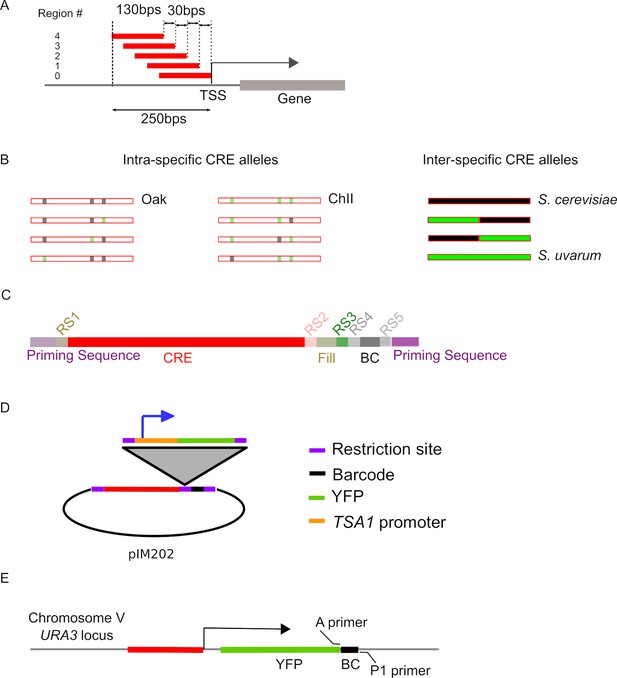
Design and cloning of cis-regulatory element (CRE-seq) libraries.
(A) CRE sequences were designed using 130 bp regions with 30 bp steps that covered the 250 bp upstream of the transcription start site (TSS). (B) Intra-specific CREs include the Oak (YJF153, black) and ChII (SX6, green) alleles, as well as each variant substituted into the Oak and ChII background. Inter-specific CREs include the S. cerevisiae (black) and S. uvarum (green) alleles as well as two chimeric alleles designed with recombination in the center of CREs. (C) Synthesized 200 bp sequences contained priming sequences to amplify the library, restriction sites (RS) for cloning, fill sequences to compensate for any difference length due to insertion deletion polymorphisms (InDels), barcodes (BC), and CREs. (D) Synthesized sequences were cloned into pIM202, then YFP with the TSA1 core promoter was inserted between the CRE and barcode. This eliminated the fill sequence and placed the BC in the 3' UTR of YFP. (E) The reporter gene (YFP) with the barcoded CREs was integrated at the URA3 locus. The A and P1 primers were used for barcode sequencing of extracted RNA and DNA.
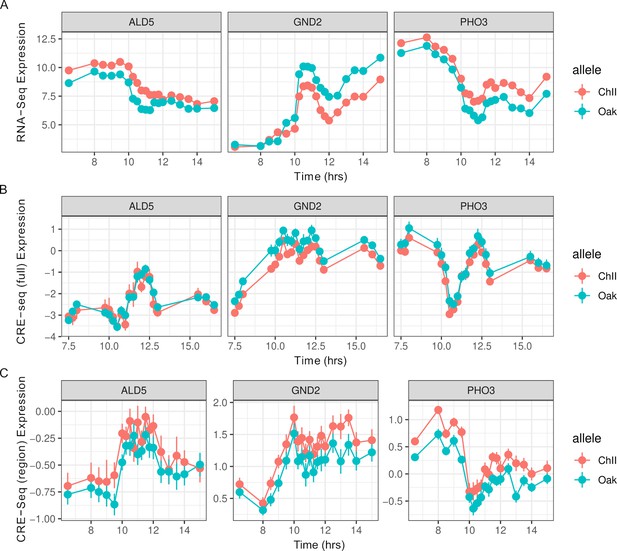
Short cis-regulatory elements (CREs) recapitulate longer CRE expression.
Gene expression dynamics of three genes (ALD5, GND2, PHO3) measured by RNA-seq (A) in comparison to CRE-seq expression from full-length promoters (B) and CRE-seq expression from short 130 bp promoter regions (C). Points and lines show the mean and standard error from replicate barcodes of the Oak and ChII alleles. Expression is shown on a log2 scale.
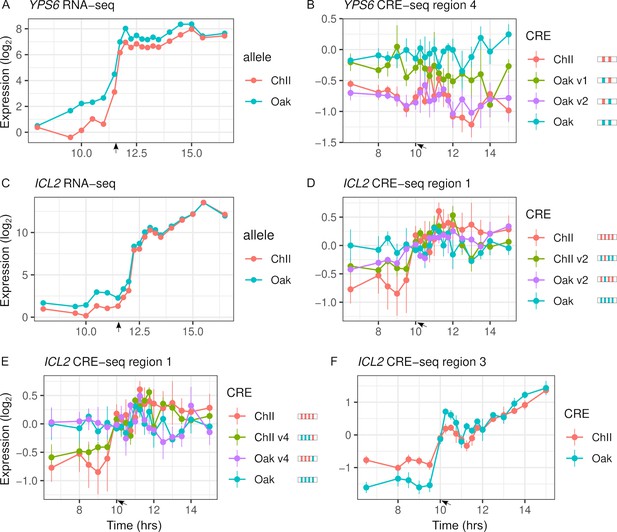
Intra-specific cis-regulatory elements (CREs) show differences in expression levels and dynamics.
(A) YPS6 shows allele differences in endogenous expression levels and dynamics. (B) CRE region 4 of the YPS6 promoter shows allele differences in expression levels, but is not correlated with endogenous expression patterns. Substituting the Oak insertions/deletions (InDel) into the ChII allele (Oak v1) increases expression levels, but substituting the Oak single-nucleotide polymorphism (SNP) into the ChII allele (Oak v2) has no effect. (C) ICL2 shows allele differences in endogenous expression dynamics. Of the four SNPs and one InDel that differentiate the region 1 CRE alleles, two SNPs (v2 and v4) alter expression dynamics in both the Oak and ChII background (D, E). (F) CRE region 3 of ICL2 has a single InDel between the Oak and ChII alleles and also shows allele differences in expression dynamics. For panels (B), (D), and (E), CRE alleles are shown by rectangles with colored ticks to indicate the Oak and ChII variants. Bars indicate standard errors. Arrows indicate the approximate time of glucose depletion.
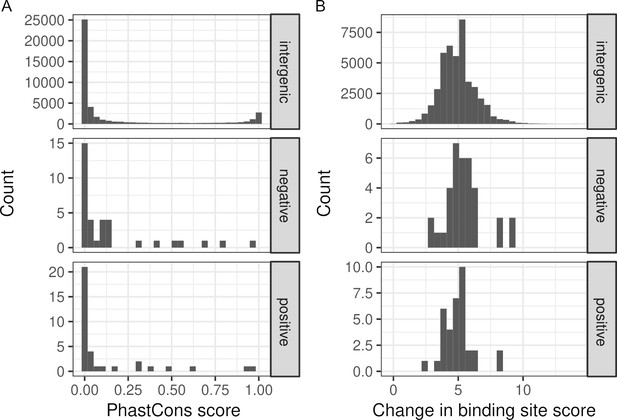
Binding site and conservation scores of variants.
(A) Histograms of PhastCons conservation scores (0, least conserved; 1, most conserved). (B) Histograms of the maximum change in transcription factor binding scores cause by a variant across 196 binding site models. The distribution is shown for variants associated with cis-regulatory element (CRE)-expression dynamics (positive, n = 35) or not (negative, n = 35), and all other intergenic variants in the Oak × ChII hybrid (n = 44,514).
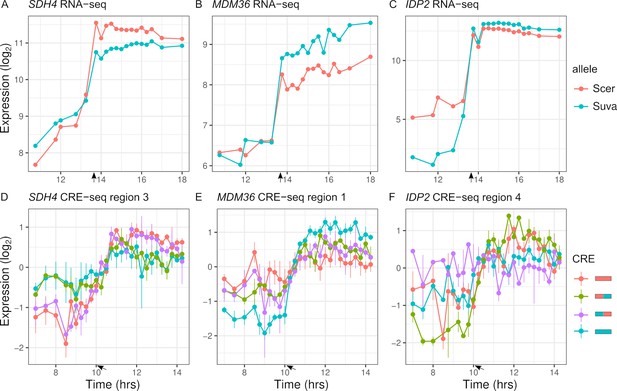
Inter-specific cis-regulatory elements (CREs) show differences in expression dynamics.
(A–C) Endogenous expression of S. cerevisiae (Scer) and S. uvarum (Suva) alleles of SDH4, MDM36, and IDP2. (D–F) CRE-seq expression of region 3 (SDH4), region 1 (MDM36), and region 4 (IDP2) for parental S. cerevisiae (red) and S. uvarum (blue) CRE alleles and both chimeric CREs. SDH4 shows expression divergence maps to the proximal promoter region, MDM36 and IDP2 show chimera expression that differ from both parents, with the MDM36 chimeras being between the two parents and IDP2 chimeras being outside the two parents. Bars indicate standard errors. Arrows indicate the approximate time of glucose depletion.
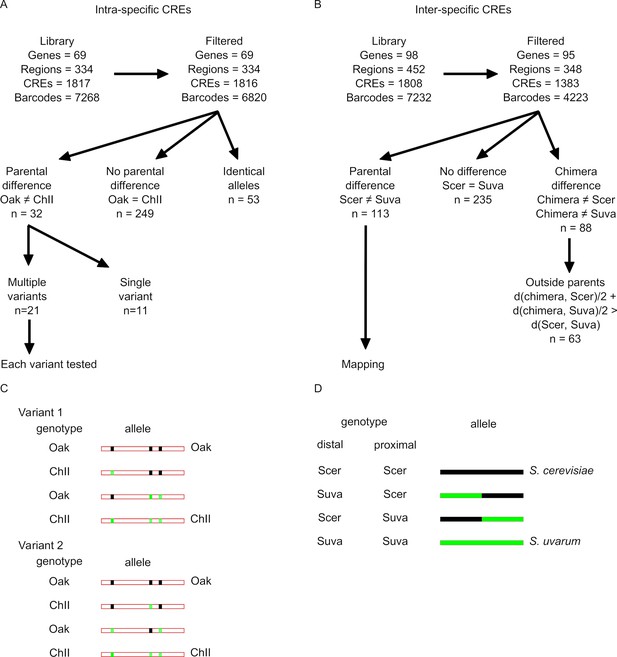
Identification of significant differences for the intra-specific and inter-specific cis-regulatory element sequence (CRE-seq) libraries.
The number of genes, promoter regions, CREs (including variants and chimeras), and barcodes is shown for the intra-specific (A) and inter-specific (B) library. The same numbers are shown after filtering to remove low-abundance barcodes and CREs without replicates. For the intra-specific data, differences between the parental (Oak and ChII) alleles were identified for all CRE regions that were not identical between Oak and ChII. Variants within these CRE region were tested when there was more than one difference between the Oak and ChII alleles. Examples of how two variants were tested are shown by the variant genotype and the allele background in which it occurs (C). For the inter-specific library, differences between the parental S. cerevisiae (Scer) and S. uvarum (Suva) alleles were identified, and subsequently mapped using the genotype of the proximal or distal region of the parental and chimeric alleles (D). Chimeras with expression outside the parental range were identified by those where a chimera differed from both parental alleles and where the average distance of the chimera to each parent is greater than the distance between the two parents.
Tables
Number of genes with allele-specific expression.
| Intra-specific hybrids‡ | Inter-specific hybrids‡ | ||||
|---|---|---|---|---|---|
| S. cerevisiae (Oak × Wine) | S. cerevisiae (Oak × China II) | S. cerevisiae (Oak × China I)† | S. cerevisiae × S. paradoxus | S. cerevisiae × S. uvarum | |
| Group* | YJF1460 | YJF1455 | YJF14542 | YJF1453 | YJF1484 |
| Dynamics | 671 | 699 | 911 | 2055 | 1827 |
| Levels | 1964 | 2088 | 2260 | 2930 | 3237 |
| Both | 371 | 375 | 501 | 1260 | 1253 |
-
*Genes with significant (false discovery rate < 0.01) allele-specific differences in dynamics, levels, or both dynamics and levels.
†The total number of genes is 4703 except for 358 genes on chromosome 13R of the China I hybrid that were removed.
-
‡Oak is most closely related to the Wine strain, followed by China II, China I, S. paradoxus, and S. uvarum.
CRE regions and variants affecting gene expression.
| Library | Type | Genes | Regions | SNPs | InDels |
|---|---|---|---|---|---|
| Intra-specific | Levels | 1/59 | 1/240 | 0/1 | 1/1 |
| Intra-specific | Dynamics | 22/50 | 31/201 | 30/57 | 5/13 |
| Inter-specific | Levels | 2/86 | 2/317 | -/68 | -/12 |
| Inter-specific | Dynamics | 59/72 | 113/257 | -/3560 | -/479 |
-
Genes, regions, SNPs, and InDels are the number significant out of the number tested. Individual SNPs and InDels were not tested for the inter-specific library.
CRE: cis-regulatory element; SNPs: single-nucleotide polymorphisms; InDels: insertions/deletions.
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Saccharomyces cerevisiae) | ALD5 | Saccharomyces Genome Database | SGD:S000000875 | |
| Gene (Saccharomyces cerevisiae) | GND2 | Saccharomyces Genome Database | SGD:S000003488 | |
| Gene (Saccharomyces cerevisiae) | PHO3 | Saccharomyces Genome Database | SGD:S000000296 | |
| Strain, strain background (Saccharomyces cerevisiae) | Oak; YJF153 | PMID:12702333 | YPS163 background | |
| Strain, strain background (Saccharomyces cerevisiae) | Wine; YJF1442 | PMID:16103919 | UCD2120 background | |
| Strain, strain background (Saccharomyces cerevisiae) | ChI; YJF1373 | PMID:22913817 | HN6 background | |
| Strain, strain background (Saccharomyces cerevisiae) | ChII; YJF1375 | PMID:22913817 | SX6 background | |
| Strain, strain background (Saccharomyces paradoxus) | YJF694 | PMID:19212322 | N17 background | |
| Strain, strain background (Saccharomyces uvarum) | YJF1450 | PMID:22384314 | CBS7001 background | |
| Recombinant DNA reagent | pIM202 | PMID:23921661 | CRE cloning vector | |
| Sequence-based reagent | S288c genome | PMID:22384314 | ||
| Sequence-based reagent | N17 genome | PMID:22384314 | ||
| Sequence-based reagent | CBS 7001 genome | PMID:22384314 | ||
| Sequence-based reagent | Oak genome | This paper | YJF153.fasta; YJF153.gff | https://doi.org/10.17605/OSF.IO/Y5748 |
| Sequence-based reagent | Wine genome | This paper | BC217.fasta; BC217.gff | https://doi.org/10.17605/OSF.IO/Y5748 |
| Sequence-based reagent | ChI genome | This paper | HN6.fasta; HN6.gff | https://doi.org/10.17605/OSF.IO/Y5748 |
| Sequence-based reagent | ChII genome | This paper | SX6.fasta; SX6.gff | https://doi.org/10.17605/OSF.IO/Y5748 |
| Sequence-based reagent | Intra-specific CRE-library | This paper | CRE_Libraries.YJF1455.csv | https://doi.org/10.17605/OSF.IO/Y5748 |
| Sequence-based reagent | Inter-specific CRE-library | This paper | CRE_Libraries.YJF1484.csv | https://doi.org/10.17605/OSF.IO/Y5748 |
| Commercial assay or kit | Dynabeads mRNA Direct kit | Invitrogen | Invitrogen:61011 | |
| Commercial assay or kit | YeaStar DNA kit | Zymo Research | Zymo:D2002 | |
| Commercial assay or kit | YeaStar RNA kit | Zymo Research | Zymo:R1002 | |
| Commercial assay or kit | Dynabeads mRNA Direct kit | Zymo Research | Zymo:D2002 | |
| Commercial assay or kit | Glucose (GO) Assay Kit | Sigma | Sigma:GAGO20 | |
| Software, algorithm | BWA v0.7.5 | PMID:19451168 | RRID:SCR_010910 | https://github.com/lh3/bwa |
| Software, algorithm | PicardTools v1.114 | Broad Institute | RRID:SCR_006525 | https://github.com/broadinstitute/picard |
| Software, algorithm | GATK HaplotypeCaller v3.3–0 | Broad Institute | RRID:SCR_001876 | https://github.com/broadinstitute/gatk/ |
| Software, algorithm | liftOver | UCSC Genome Browser | RRID:SCR_018160 | https://genome-store.ucsc.edu/ |
| Software, algorithm | Fastx-toolkit | Hannon Lab | RRID:SCR_005534 | https://github.com/agordon/fastx_toolkit |
| Software, algorithm | Bowtie2 v2.1.0 | PMID:22388286 | RRID:SCR_016368 | https://github.com/BenLangmead/bowtie2 |
| Software, algorithm | Htseq-count | PMID:25260700 | RRID:SCR_011867 | https://github.com/htseq/htseq |
| Software, algorithm | DESeq2 | PMID:25516281 | RRID:SCR_015687 | https://doi.org/10.18129/B9.bioc.DESeq2 |
| Software, algorithm | Patser | PMID:10487864 | http://stormo.wustl.edu/software.html | |
| Software, algorithm | Custom R scripts | This paper | https://doi.org/10.17605/OSF.IO/Y5748 |
Additional files
-
Supplementary file 1
Supplementary tables.
Table S1: strains used in this study. Table S2: k-means clusters of gene expression dynamics. Table S3: k-means clusters of allelic differences in expression. Table S4: logistic regression of allele-specific expression (ASE) dynamics and levels. Table S5: average number of single-nucleotide polymorphism (SNP) and insertion/deletion (InDel) differences within hybrids. Table S6: logistic regression with binding site and conservation scores. Table S7: genome assemblies used to identify variants.
- https://cdn.elifesciences.org/articles/68469/elife-68469-supp1-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/68469/elife-68469-transrepform-v2.pdf





