Live-cell single-molecule tracking highlights requirements for stable Smc5/6 chromatin association in vivo
Figures
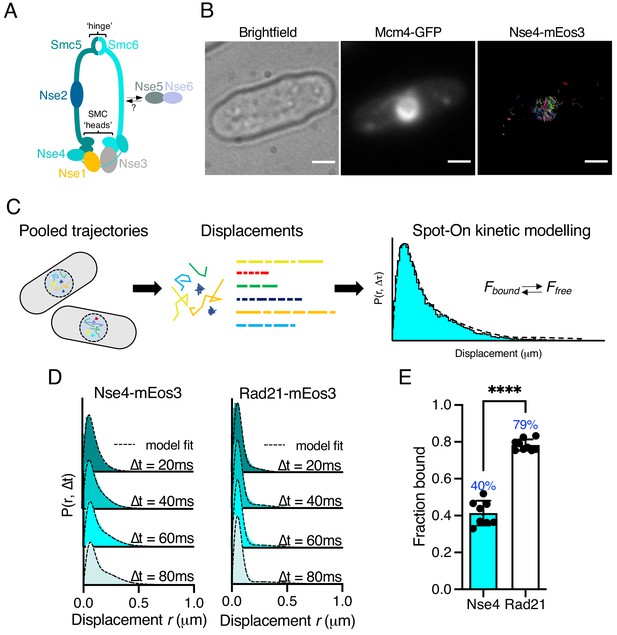
Single-particle tracking of Smc5/6 to monitor chromatin association in live cells.
(A) Schematic representation of the Smc5/6 complex in fission yeast. (B) Nse4–mEos3 tracking shows nuclear localisation of trajectories. SPT trajectories demonstrated confinement within nuclear region (right) that colocalised with the nuclear replication protein Mcm4 fused to GFP. Scale bar = 2 μm. (C) Overview of approach to quantifying chromatin association using SPT data and Spot-On kinetic modelling. (D) Probability density function (PDF) histograms and Spot-On model fitting (dashed line) for Nse4–mEos3 (Smc5/6) and Rad21–mEos3 (cohesin) single-molecule displacements at different time intervals. Displacements are from three pooled independent experiments, each with three technical repeats. (E) Fraction-bound values derived from Spot-On model fitting. Mean (±S.D). Black dots indicate Spot-On Fbound values derived from each technical repeat from three independent experiments. Percentages in blue denote fraction-bound value from fitting pooled data in (D). ****p<0.0001.
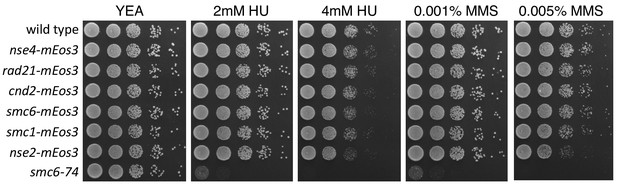
Phenotypic characterisation of mEos3-tagged SMC subunits.
Spot assay of S. pombe strains expressing different SMC components fused to the mEos3 fluorescent tag. Cells were spotted onto YEA plates containing either hydroxyurea (HU) or methyl methanesulfonate (MMS) to assay for sensitivity to replication stress. Plates were incubated at 30°C for 3 days.
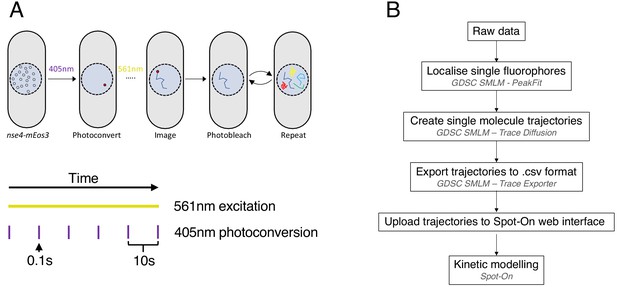
Outline of the single-particle tracking technique.
(A) Diagram of SPT experimental approach. (Top) Single mEos3 fluorophores fused to SMC components are stochastically photoconverted and imaged using 405 nm and 561 nm laser light, respectively. In each frame, the position of the fluorophore is recorded in each frame it is detected allowing for the creation of a trajectory. Multiple molecules are imaged in each cell over the course of an experiment. (Bottom) Laser illumination scheme for each experiment. mEos3 is photoconverted using 0.1 s pulses of 405 nm laser every 10 s and photoconverted species are imaged by continuous 561 nm illumination. (B) Raw data processing pipeline for SPT experiments. For specific details, see Materials and methods.
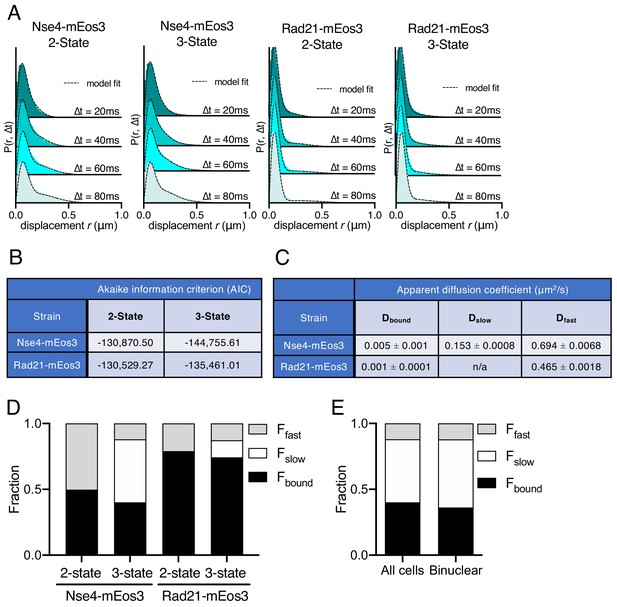
Smc5/6 behaviour fits a three-state model.
(A) Probability density function (PDF) histograms of single-molecule displacements for Nse4–mEos3 and Rad21-mEos3 over multiple Δt fit with either a two-state or three-state Spot-On model. Data are pooled from three independent experiments, each with three technical repeats. Dashed line indicates model derived from cumulative distribution function (CDF) fitting in Spot-On. (B) Akaike information criterion (AIC) scores from Spot-On model fitting in (A). Nse4–mEos3 three-state fitting showed a large difference in AIC scores compared to three-state fitting. This indicates the data are best described by a three-state model. The difference in AIC scores for Rad21–mEos3 was much smaller and thus a two-state model was used. (C) Apparent diffusion coefficients of Spot-On sub-populations of Nse4–mEos3 (three state) and Rad21–mEos3 (two state). (D) Fractions of the total population of molecules observed residing in each kinetic state extracted from Spot-On model fitting data in (A). (E) Comparison of the fractions of Nse4–mEos3 molecules observed residing in each kinetic state extracted from all cells (mostly G2) in the wild-type data or only binuclear cells (S-phase, n = 75).
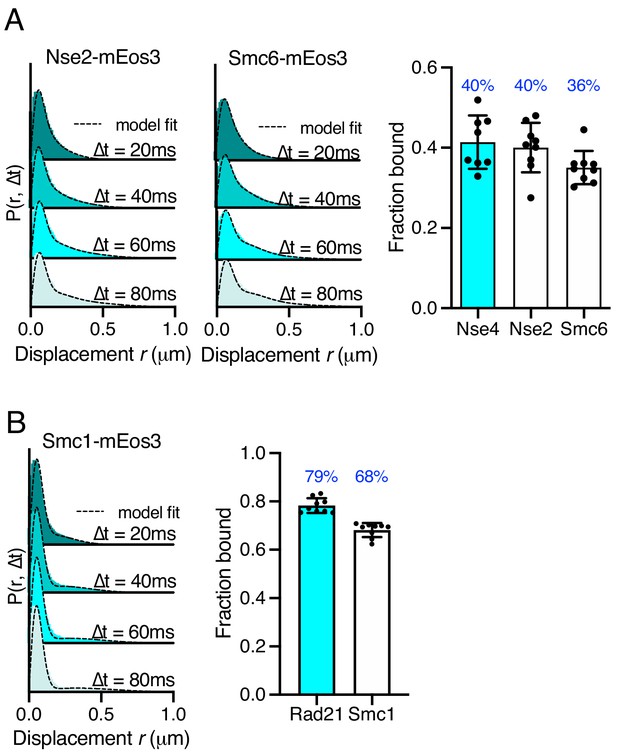
Single-particle tracking of SMC complex subunits.
(A) Probability density function (PDF) histograms of single-molecule displacements (left) and fraction-bound values calculated from Spot-On model fitting (right) of alternative Smc5/6 subunits tagged with mEos3 compared to Nse4–mEos3. Mean (±S.D). Black dots values derived from independent technical repeats, percentages in blue denote fraction-bound value from fitting pooled data from all repeats. (B) PDF histograms of single-molecule displacements (left) and fraction-bound values calculated from Spot-On model fitting (right) of alternative cohesin subunit, Smc1, tagged with mEos3 compared to Rad21–mEos3. Mean (±S.D). Black dots values derived from independent technical repeats; percentages in blue denote fraction-bound value from fitting pooled data from all repeats.
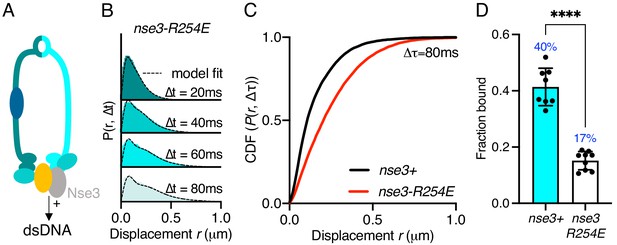
Stable Smc5/6 chromatin association requires dsDNA binding activity.
(A) Schematic representation of the region of known dsDNA interaction in S. pombe Smc5/6. (B) Probability density function histogram of pooled Nse4–mEos3 single-molecule in nse3-R254E background and Spot-On model fitting (dashed line). The resulting fraction of bound molecules compared to wild-type data in Figure 1D. Bar chart shows mean ± S.E.M. Black dots denote independent repeats. ***p=0.0003. (C) Cumulative distribution function (CDF) of pooled Δt = 80 ms data from (B). (D) Fbound values derived from Spot-On model fitting of Nse4–mEos3 in nse3-R254E background. Black dots denote each technical repeat from three independent experiments. Percentages in blue denote fraction-bound value from fitting pooled data in (B). Mean (±S.D). ****p<0.0001.
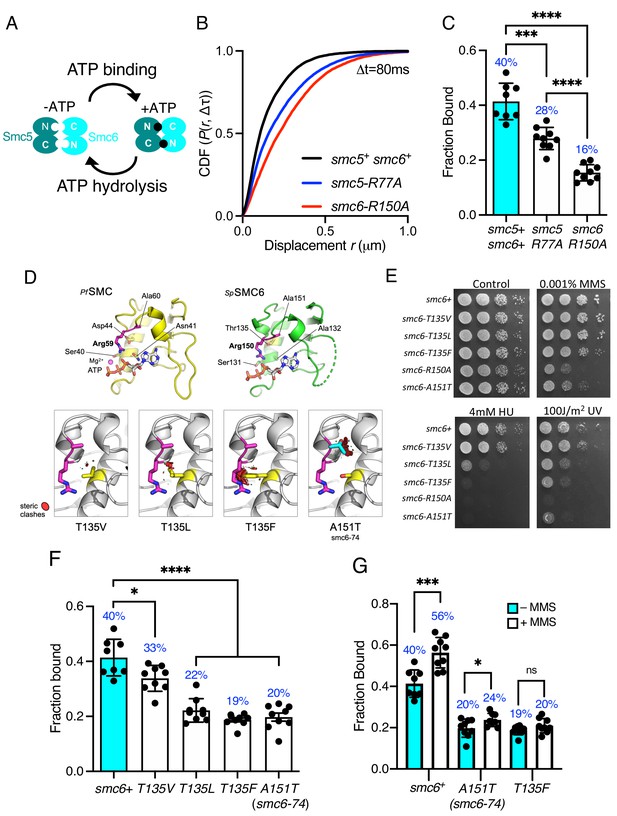
Smc5/6 ATPase activity regulates chromatin association.
(A) Schematic representation of SMC head engagement upon ATP binding. (B) CDF of pooled Δt = 80 ms single-molecule displacements of Nse4–mEos3 in smc5+ smc6+, smc6-R150A, and smc5-R77A genetic backgrounds. (C) Comparison of the fraction of bound molecules from Nse4–mEos3 sptPALM experiments in asynchronous smc6-R150A and smc5-R77A genetic backgrounds to wild=type data from Figure 1D. Black dots denote each technical repeat from three independent experiments. Percentages in blue denote fraction-bound value from fitting pooled data. Mean (±S.D). ***p=0.0001, ****p<0.0001. (D) Secondary structure molecular cartoons of homology models for the head domains of S. pombe Smc6, highlighting the arginine finger and its interaction with ATP. The X-ray crystal structure for the head domain of Pyrococcus furiosus SMC in complex with ATP served as a reference, providing the expected position of bound ATP the homology model. Key amino acids are shown in ‘stick representation’. The lower panel shows the predicted increase in severity of steric clashes made with the arginine finger through introduction of each of the indicated mutations. (E) Yeast spot assay of S. pombe strains harbouring different smc6 ATPase mutations grown at 30°C for 3 days. (F) Fraction-bound values in each of the smc6-T135 mutant backgrounds compared to wild-type data from Figure 1D and smc6-74 (A151T). Black dots denote each technical repeat from three independent experiments. Percentages in blue denote fraction-bound value from fitting pooled data. Mean (±S.D). *p=0.0158, ****p<0.0001. (G) Fraction-bound values derived from SPT analysis of MMS-treated (0.03%, 5 hr) cells compared to asynchronous untreated data in (F). *p=0.0495, ***p=0.0005.
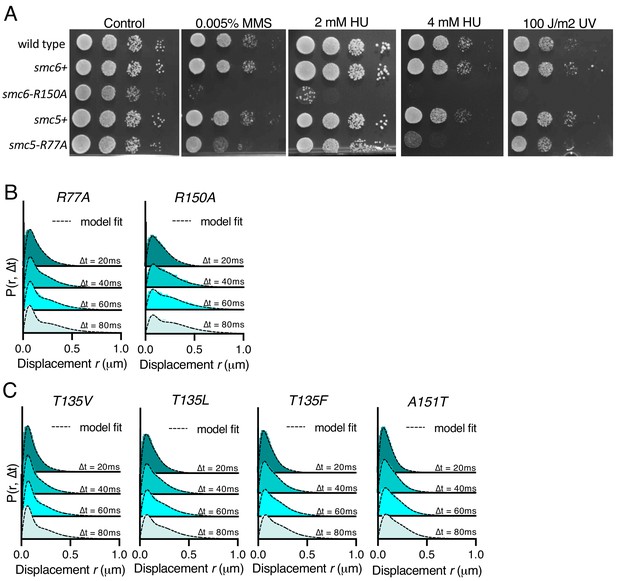
Characterisation of mEos3-tagged Smc5/6 ATPase mutants.
(A) Spot assay of S. pombe strains harbouring arginine finger mutations in either smc5 or smc6. Plates were incubated at 30°C for 3 days. (B) Probability density function (PDF) histograms of single-molecule displacements for multiple Δt of Nse4–mEos3 for smc5 (R77A) or smc6 (R150A) arginine finger mutants (see Figure 3C and F for fraction bound). Dashed line indicates model derived from cumulative distribution function (CDF) fitting in Spot-On. Data are pooled from three individual experiments, each with three technical repeats. (C) PDF histograms of single-molecule displacements for multiple Δt of Nse4–mEos3 in the indicated mutants (see Figure 4E for fraction bound). Dashed line indicates model derived from CDF fitting in Spot-On. Data are pooled from three individual experiments, each with three technical repeats.
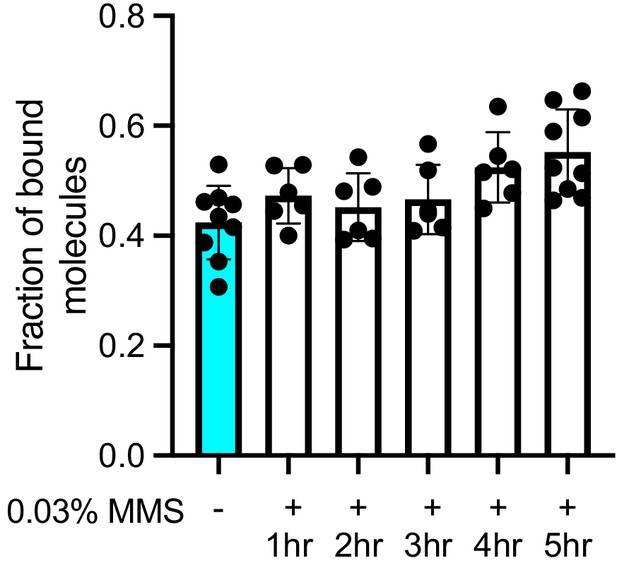
Spot-On analysis of Nse4 chromatin association during 5 hr methyl methanesulfonate (MMS) treatment.
Fraction of bound Nse4–mEos3 extracted from Spot-On analysis of MMS-treated cells. Cultures were incubated in the presence of 0.03% MMS over the course of 5 hr, and cells were isolated and imaged at each hour time point. Maximal chromatin association was detected between 4 and 5 hr. Mean ± S.D. Black dots denote independent technical repeats.
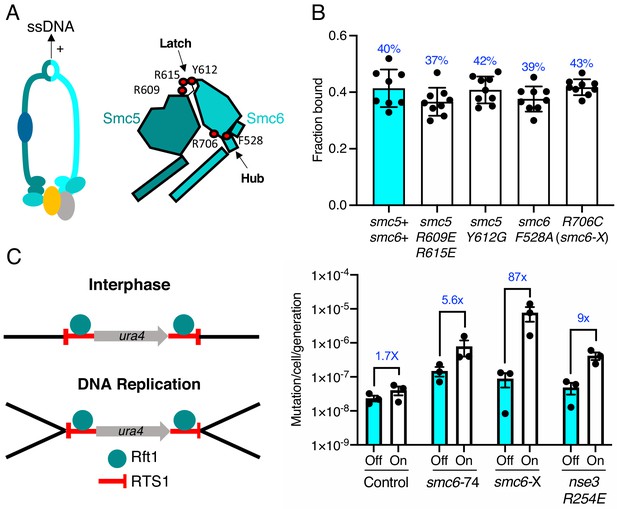
ssDNA interactions are required to prevent gross chromosomal re-arrangements but dispensable for stable Smc5/6 chromatin association.
(A) Left: Schematic representation of the hinge region known to interact with ssDNA interaction in Smc5/6. Right: Schematic diagram of the S. pombe hinge region adapted from Alt et al., 2017. Residues implicated in ssDNA interaction are highlighted with red filled circles. (B) Fraction-bound values of Nse4–mEos3 derived from SPT experiments in Smc5/6 hinge mutant backgrounds compared to wild-type data from Figure 1D. Mean ±S.D. Black dots denote independent repeats and percentages in blue denote fraction-bound value from fitting pooled data from all repeats. (C) Diagram of the site-specific replication stall system RTS1-ura4-RTS1 (Lambert et al., 2005), which consists of two inverted RTS1 sequences integrated on either sides of the ura4 gene. Rtf1 binds the RTS1 sequence and stalls incoming replication forks coming from both centromeric and telomeric sides. Rtf1 is expressed under the control of the nmt41 promoter which is ‘off’ in the presence of thiamine and ‘on’ upon thiamine removal. (D) Induction of rtf1 in cells harbouring RuraR construct induces ura4 marker loss as assayed by 5-fluoroorotic acid (5-FOA) resistance. Cells growing in the presence (Off, arrest repressed) or absence (On, arrest induced) of thiamine were analysed by fluctuation analysis. Mean ± S.E.M. Black dots denote independent repeats.
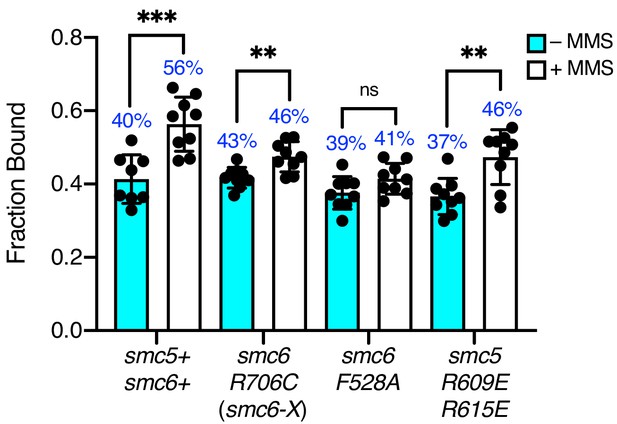
Spot-On analysis of MMS-treated ssDNA interaction mutants.
Fraction of bound Nse4–mEos3 extracted from Spot-On analysis of Nse4–mEos3 SPT in smc6 hinge mutants treated with 0.03% MMS for 5 hr. Compared to asynchronous untreated datasets from Figure 4B. Mean ± S.D. Black dots denote independent technical repeats, percentages denote fraction-bound value from fitting pooled data from all repeats. **p<0.005 (smc6-X = 0.0034, smc5-RR = 0.0025), ***p=0.0005.
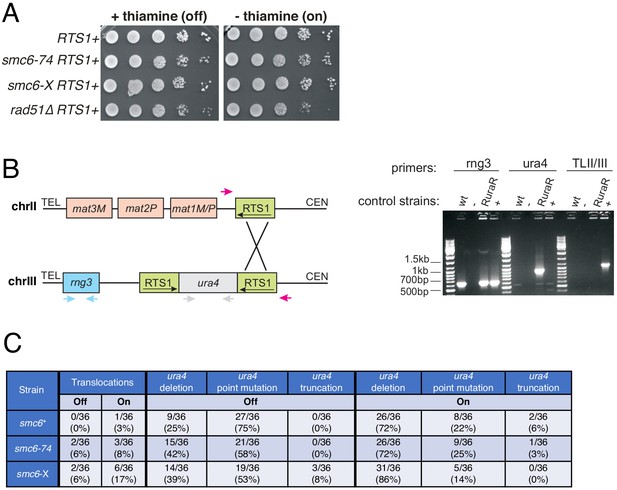
Analysis of the consequences of site-specific replication fork stalling on cell viability and gross chromosomal re-arrangements.
(A) S. pombe strains harbouring the site-specific replication stall system RuraR. Replication fork stalling at RTS1 is induced in the absence of thiamine (on). Plates were incubated at 30°C for 3 days. Unlike the HR-defective rad51Δ strain, smc6 hypomorphs do not lose viability on induction of replication stalling at RTS1. (B) PCR-based assay for translocation between RTS1 at RuraR and the native RTS1 at the mating type locus in ura4− colonies generated in the ura4 loss of gene function assay (Lambert et al., 2005). Left: Schematic to show the three primer pairs used. One pair (red arrows) amplifies the junction resulting from ectopic recombination between chromosome II and III (TLII/III). The second pair (grey arrows) amplifies the ura4 locus to distinguish point mutations, truncations (internal deletions), and full-length deletions. rng3 (blue arrows), an essential gene located between RuraR and the telomere, is amplified as positive control. Right: Example of control PCRs (top) and PCRs of 5′-fluoroorotic acid (5-FOA) resistant/ura4− colonies (bottom). The rng3 product is amplified in all strains, but not in the negative control (‘-'). ura4 is amplified only in a RuraR strain, but not in wild type (wt) (harbours full deletion of ura4, ura4-D18), the translocation positive control (‘+', gift from S. Lambert [Lambert et al., 2005]) or the negative control. Translocation between chromosome II and III can only be detected in the positive control. (C) PCR assay results for ura- colonies of smc6+, smc6-74 and smc6-X derived from the RuraR ura4 loss assay carried out in the presence (RuraR arrest ‘Off’) or absence (RuraR arrest ‘On’) of thiamine.
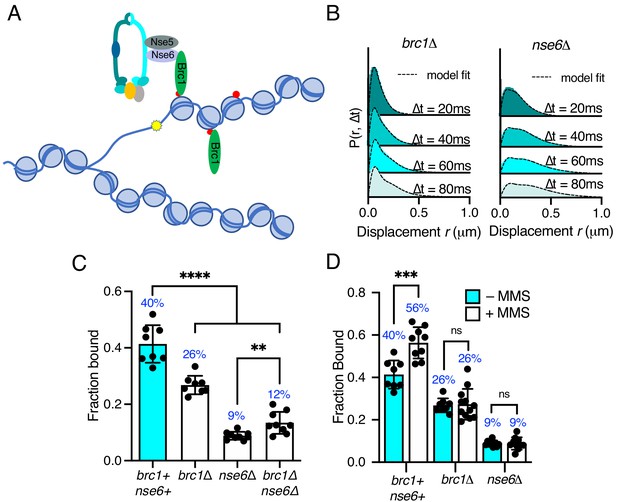
Differential requirements of Nse6 and Brc1 for Smc5/6 chromatin association.
(A) Schematic diagram of Smc5/6 recruitment to γ-H2A (red dots: H2A phosphorylation) at stalled replication forks. Brc1 binds to γ-H2A and recruits Smc5/6 via an interaction with Nse6. Yellow star indicates a DNA lesion. (B) Displacement PDF histograms from asynchronous cells expressing Nse4–mEos3 in brc1Δ and nse6Δ genetic backgrounds. Data are from three pooled independent experiments, each with three technical repeats. Spot-On model fit is denoted by dashed line. (C) Comparison of Nse4–mEos3 Fbound values derived from Spot-On fitting of SPT displacement histograms in wild type, brc1Δ, nse6Δ, and brc1Δ nse6Δ genetic backgrounds. Mean ± S.D. Black dot values derived from independent technical repeats; percentages in blue denote fraction-bound value from fitting pooled data from all repeats. ****p<0.0001, **p=0.0043. (D) FBound fraction values from brc1Δ and nse6Δ cells in (C) compared to parallel experiments where cells were treated with 0.03% MMS for 5 hr. ***p<0.005, ns = not significant.
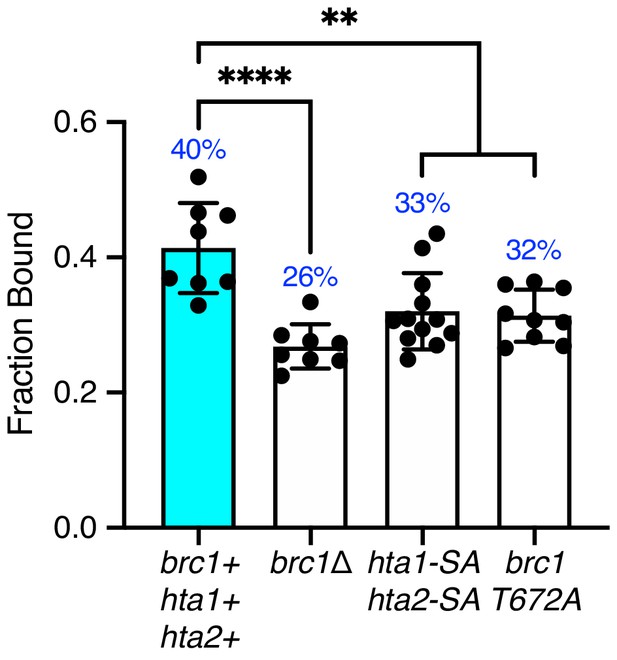
Spot-On analysis of Nse4 chromatin association in histone phosphorylation site and brc1γ-H2A interaction mutants.
Fraction of bound Nse4–mEos3 extracted from Spot-On analysis of Nse4–mEos3 SPT in hta1-S128A hta2-S129A and brc1-T672A mutants compared to wild-type and brc1Δ data sets from Figure 5C. Mean ± S.D. Black dots denote independent technical repeats; percentages denote fraction-bound value from fitting pooled data from all repeats. **p<0.005 (hta1-SA hta2-SA = 0.0034, smc5-RR = 0.0016), ****p<0.0001.
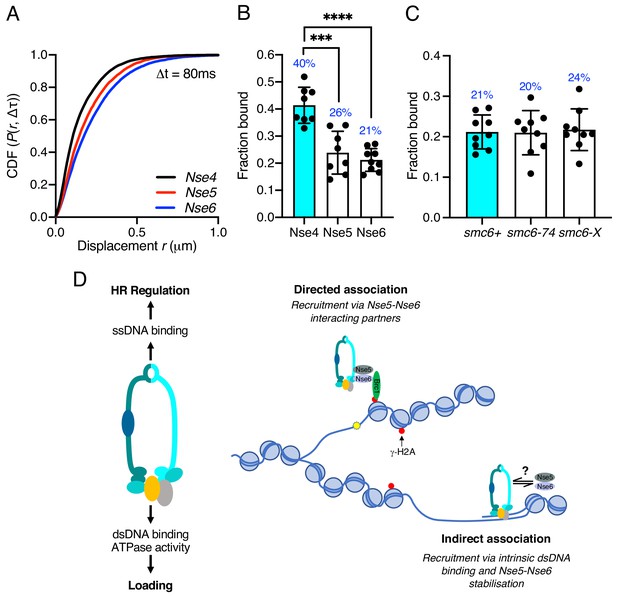
Nse5–Nse6 chromatin association is distinct from other Smc5/6 subunits.
(A) CDF histogram of pooled single-molecule displacements at Δt = 80 ms time interval of Nse4–mEos3, Nse5–mEos3, and Nse6–mEos3. (B) Fraction of bound molecules extracted from Spot-On model fits from experiment in (A). Mean ± S.D. Black dots denote independent technical repeats, percentages denote fraction-bound value from fitting pooled data from all repeats. ***p=0.0003, ****p<0.0001. (C) Fraction of bound molecules extracted from Spot-On model fits from SPT Nse6–mEos3 in smc6-74 or smc6-X genetic backgrounds compared to wild-type data in (B). (D) Schematic diagram of Smc5/6 DNA interactions and their roles (left) and proposed model of Smc5/6 chromatin association (right). Loading requires dsDNA binding by Nse3 and Smc5 and Smc6 ATPase activity. ssDNA binding at the hinge is not required for loading but is required for subsequent functions to regulate homologous recombination, suppress non-allelic recombination and gross chromosomal rearrangements (GCRs). Smc5/6 association with chromatin is dependent on Nse5 and Nse6 and either directed (e.g. Brc1-dependent recruitment to γ-H2A) (top) or non-directed via dsDNA binding and subsequent loading (bottom). Nse5/6 is required in both instances and may act either to directly load Smc5/6 or may stabilise its association after initial loading by dsDNA interaction.
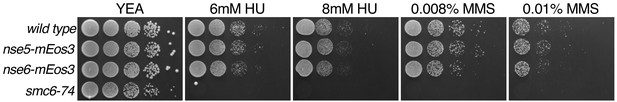
Phenotypic characterisation of mEos3 tagged Nse5 and Nse6 subunits.
Spot assay of S. pombe strains expressing either Nse5 or Nse6 Smc5/6 components fused to the mEos3 fluorescent tag. Cells were spotted onto YEA plates containing either hydroxyurea (HU) or methyl methanesulfonate (MMS) to assay for sensitivity to replication stress. Plates were incubated at 30°C for 3 days.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Chemical compound, drug | Methylmethane sulfonate | Sigma–Aldrich | 129925–25G | |
| Chemical compound, drug | Hydroxyurea | Sigma–Aldrich | H8627-100G | |
| Chemical compound, drug | 5-Fluoroorotic acid | Formedium | 5FOA10 | |
| Other | Agarose, Type I-A, low EEO | Sigma–Aldrich | A0169-25G | |
| Other | Circular coverslips: #1.5H, ∅25 mm | Thorlabs | CG15XH | |
| Other | UV-Ozone cleaning system | Novascan | PSD-UV | |
| Software, algorithm | GDSC SMLM | Fiji plugin update site | GDSC SMLM2 | Underlying source-code is freely avaliable at https://github.com/aherbert/gdsc-smlm |
| Software, algorithm | Prism 9 | Graphpad software |
Additional files
-
Supplementary file 1
Fluctuation experiment data tables.
Data from individual experimental repeats of ura4 loss assay in Figure 4C.
- https://cdn.elifesciences.org/articles/68579/elife-68579-supp1-v2.docx
-
Supplementary file 2
Strain table.
Strains used during this study.
- https://cdn.elifesciences.org/articles/68579/elife-68579-supp2-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/68579/elife-68579-transrepform-v2.pdf





