Duox-generated reactive oxygen species activate ATR/Chk1 to induce G2 arrest in Drosophila tracheoblasts
Figures
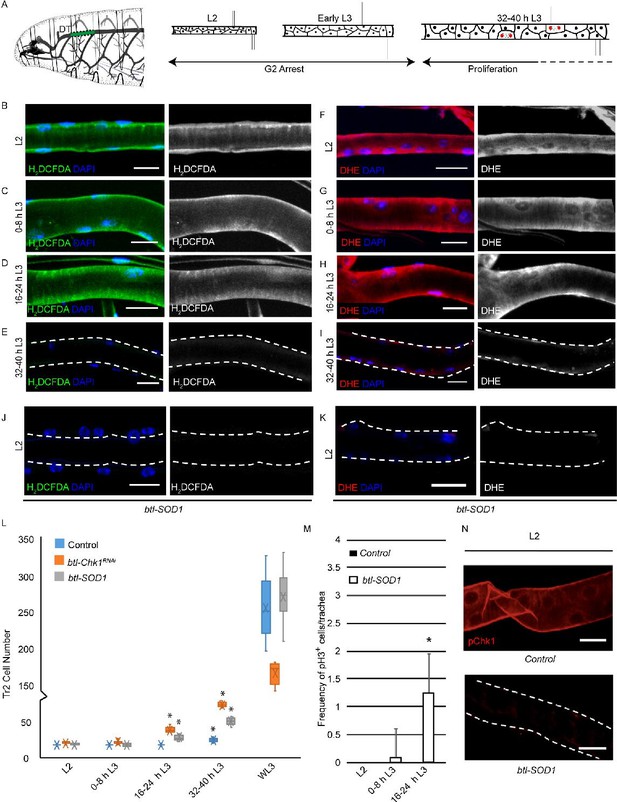
High levels of reactive oxygen species (ROS) are required for checkpoint kinase 1 (Chk1) activation and G2 arrest in tracheoblasts.
(A) A diagram of the third instar larva showing the dorsal trunk (DT) of the second thoracic metamere (Tr2, colored in green and marked by dashed line). The diagram also shows the timecourse of G2 arrest and cell division in Tr2. The cells in Tr2 DT remain geographically isolated from tracheal cells in other branches during larval life. (B–E) Levels of the ROS reporter 2',7'-dichlorodihydrofluorescein diacetate (H2DCFDA) in Tr2 DT during larval stages. Shown in the figures are H2DCFDA staining in L2 (B), 0–8 hr L3 (C), 16–24 hr L3 (D), and 32–40 hr L3 (E) in wild type (btl-Gal4) animals. (F–I) Levels of the ROS reporter dihydroethidium (DHE) in Tr2 DT during larval stages. Shown in the figures are DHE staining in L2 (F), 0–8 hr L3 (G), 16–24 hr L3 (H), and 32–40 hr L3 (I) in wild type (btl-Gal4) animals. (J, K) Effect of btl-Gal4-dependent overexpression of superoxide dismutase 1 (SOD1) on levels of ROS reporters in Tr2 DT. (J) H2DCFDA staining in btl-SOD1 (btl-GAL4/UAS-SOD1)-expressing larvae (n ≥ 6 tracheae per condition per timepoint). (K) DHE staining in btl-SOD1 larvae (n ≥ 6 tracheae per condition per timepoint). (L) Effect of SOD1 overexpression on cell numbers in Tr2 DT at different larval stages. Graph shows numbers of Tr2 tracheoblasts in wild type (btl-Gal4), btl-SOD1 (btl-GAL4/UAS-SOD1), and btl-Chk1RNAi (btl-GAL4/UAS-Chk1RNAi) larvae at L2, 0–8 hr L3, 16–24 hr L3, 32–40 hr L3, and wandering L3 (WL3) (n ≥ 7 tracheae per condition per timepoint). (M) Effect of SOD1 overexpression on mitotic indices in Tr2 DT (see text). Graph shows mitotic indices in Tr2 DT in wild type and btl-SOD1 (btl-GAL4/UAS-SOD1)-expressing larvae at L2, 0–8 hr L3 and 16–24 hr L3 (mean values ± standard deviation, n ≥ 7 tracheae per condition per timepoint). (N) Effect of SOD1 overexpression on Chk1 phosphorylation in Tr2 tracheoblasts. Shown in the figure is phosphorylated Chk1 (pChk1, phospho-Chk1Ser345) immunostaining (red) in Tr2 DT in wild type (btl-GAL4) and btl-SOD1 (btl-GAL4/UAS-SOD1) larvae at L2. Scale bars = 10 µm. Dashed lines outline the cuticular lumen of the tracheal tube here and elsewhere and are shifted outward to include the epithelial lining when they overlap with the signal. Student’s t-test: *p<0.00001.
-
Figure 1—source data 1
Cell Frequencies in SOD1 overexpressing animals.
- https://cdn.elifesciences.org/articles/68636/elife-68636-fig1-data1-v3.xlsx
-
Figure 1—source data 2
Mitotic indices in SOD1 overexpressing animals.
- https://cdn.elifesciences.org/articles/68636/elife-68636-fig1-data2-v3.xlsx
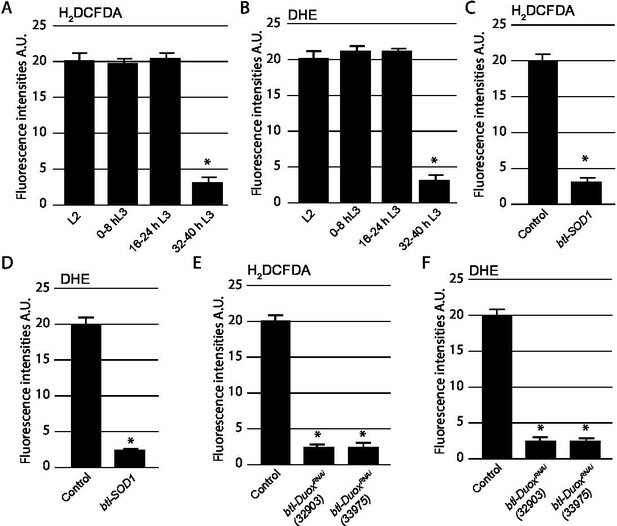
Quantification of reactive oxygen species (ROS) levels in Tr2 tracheoblasts.
(A–F) Quantification of fluorescence intensities of redox-sensitive fluorescent dyes 2',7'-dichlorodihydrofluorescein diacetate (H2DCFDA) and dihydroethidium (DHE) in Tr2 tracheoblasts. (A) Quantification of fluorescence intensities of H2DCFDA at L2, 0–8 hr L3, 16–24 hr L3, and 32–40 hr L3 in wild type (btl-Gal4) larvae. (B) Quantification of fluorescence intensities of DHE at L2, 0–8 hr L3, 16–24 hr L3, and 32–40 hr L3 in wild type (btl-Gal4) larvae. (C) Quantification of fluorescence intensities of H2DCFDA in wild type (btl-Gal4) and btl-Sod1(btl-GAL4/UAS-Sod1)-expressing tracheae at L2. (D) Quantification of fluorescence intensities of DHE in wild type (btl-Gal4) and btl-Sod1(btl-GAL4/UAS-Sod1)-expressing tracheae at L2. (E) Quantification of fluorescence intensities of H2DCFDA in wild type (btl-Gal4) and btl-DuoxRNAi (btl-GAL4/+; UAS-DuoxRNAi (32903)/+) and (btl-GAL4/+; UAS-DuoxRNAi (33975)/+)--expressing tracheae at L2. (F) Quantification of fluorescence intensities of DHE in wild type (btl-Gal4) and btl-DuoxRNAi (btl-GAL4/+; UAS-DuoxRNAi (32903)/+) and (btl-GAL4/+; UAS-DuoxRNAi (33975)/+)-expressing tracheae at L2. (mean values ± standard deviation, n = 5 tracheae per condition per timepoint). A.U: arbitrary units. Note that L2 tracheae were used as controls in all experiments and representative images have been quantified here (H2DCFDA n=8, DHE n=9): H2DCFDA Intensity value: 20266.0747 ±1199.05881 A.U. and DHE Intensity value 20006.8505±1023.51711 A.U. A.U: arbitrary units. Student’s t-test: *p<0.00001.
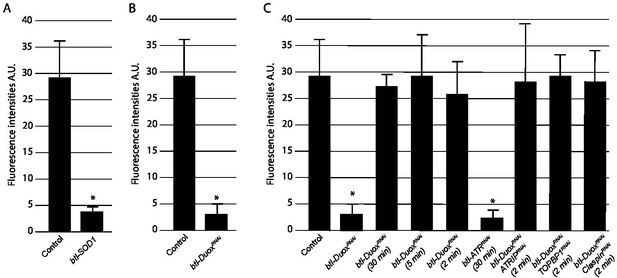
Quantification of phosphorylated checkpoint kinase 1 (pChk1) levels in Tr2 tracheoblasts.
(A–C) Quantification of fluorescence intensities after pChk1 immunostaining and tyramide-based amplification. (A) Quantification of fluorescence intensities after pChk1 immunostaining in Tr2 tracheoblasts in wild type (btl-Gal4) and btl-Sod1 (btl-GAL4/UAS-Sod1)-expressing animals at L2. (B) Quantification of fluorescence intensities after pChk1 immunostaining in Tr2 tracheoblasts in wild type (btl-Gal4) and btl-DuoxRNAi (btl-GAL4/+; UAS-DuoxRNAi (32903)/+)-expressing animals at L2. (C) Quantification of fluorescence intensities after pChk1 immunostaining in wild type (btl-Gal4), btl-DuoxRNAi(btl-GAL4/+; UAS-DuoxRNAi(32903)/+) and btl-DuoxRNAi(btl-GAL4/+; UAS-DuoxRNAi(32903)/+) animals treated with 100 µM H2O2 for 30 min, 5 min, and 2 min, btl-ATRRNAi(btl-GAL4/UAS-ATRRNAi) animals treated with 100 µM H2O2 for 30 min and btl-DuoxRNAi, ATRIPRNAi(btl-GAL4/UAS-ATRIPRNAi; UAS-DuoxRNAi(32903)/+),btl-DuoxRNAi, TOPBP1RNAi(btl-GAL4/+; UAS-DuoxRNAi(32903)/UAS-TOPBP1RNAi), btl-DuoxRNAi, ClaspinRNAi(btl-GAL4/+; UAS-DuoxRNAi(32903)/UAS-ClaspinRNAi) animals treated with 100 µM H2O2 for 2 min at L2. (mean values ± standard deviation, n = 5 tracheae per condition per timepoint). Note that L2 tracheae were used as controls in all experiments and representative images have been quantified here (n=9, pChk1 intensity value 29441.4784 ±6706.78889 A.U. ). A.U: arbitrary units. Student’s t-test: *p<0.00001.
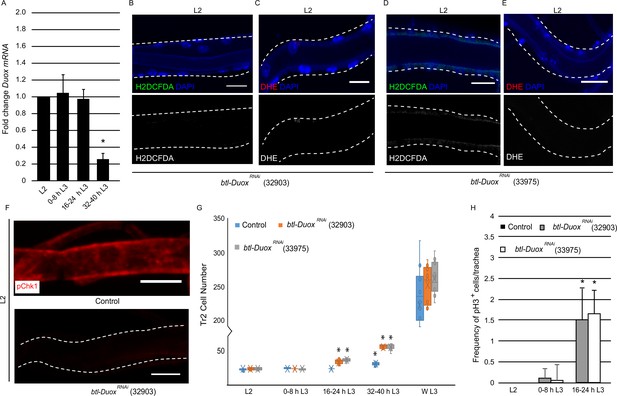
High reactive oxygen species (ROS) in tracheoblasts is dependent on Duox expression.
(A) Quantitative PCR analysis of Duox mRNA levels in micro-dissected Tr2 dorsal trunk (DT) fragments at different stages. Graph shows fold change in Duox mRNA in Tr2 DT fragments from wild type (btl-GAL4) larvae at L2, 0–8 hr L3, 16–24 hr L3, and 32–40 hr L3. Fold change has been represented with respect to L2 (n = 3 experiments, n ≥ 15 Tr2 DT fragments/stage/experiment, mean ± standard deviation, p<0.0001). (B–E) Effect of the knockdown of Duox expression on the levels of ROS reporters in Tr2 DT in L2. Shown here are the results of the expression of two different Duox RNAi lines (32903 and 33975). (B, D) 2',7'-Dichlorodihydrofluorescein diacetate (H2DCFDA) staining and (C, E) dihydroethidium (DHE) staining in Tr2 DT in btl-DuoxRNAi. ([B, C] btl-GAL4/+; UAS-DuoxRNAi (32903)/+ and [D, E] btl-GAL4/+; UAS-DuoxRNAi (33975)/+) larvae (n ≥ 6 tracheae per condition per timepoint). (F) Effect of reduction of Duox expression on levels of phosphorylated checkpoint kinase 1 (pChk1) in Tr2 DT in L2. pChk1 immunostaining (red) in Tr2 DT in wild type (btl-Gal4) and btl-DuoxRNAi (btl-GAL4/+; UAS- DuoxRNAi (32903)/+) larvae. (G) Effect of the knockdown of Duox expression on cell numbers in Tr2 DT at different larval stages. Graph shows cell numbers of Tr2 tracheoblasts in wild type (btl-Gal4) and btl-DuoxRNAi (btl-GAL4/+; UAS-DuoxRNAi (32903)/+ and btl-GAL4/+; UAS-DuoxRNAi (33975)/+) larvae at L2, 0–8 hr L3, 16–24 hr L3, 32–40 hr L3, and WL3 (n ≥ 7 tracheae per condition per timepoint). (H) Effect of the knockdown of Duox expression on mitotic indices in Tr2 DT. Graph shows mitotic indices in Tr2 DT in wild type (btl-Gal4) and btl-DuoxRNAi (btl-GAL4/+; UAS-DuoxRNAi (32903)/+ and btl-GAL4/+; UAS-DuoxRNAi (33975)/+) larvae at L2, 0–8 hr L3 and 16–24 hr L3 (mean values ± standard deviation, n ≥ 7 tracheae per condition per timepoint). Scale bars = 10 µm. Student’s t-test: *p<0.0001
-
Figure 2—source data 1
Cell frequencies in Duox RNAi expressing animals.
- https://cdn.elifesciences.org/articles/68636/elife-68636-fig2-data1-v3.xlsx
-
Figure 2—source data 2
Mitotic indices in Duox RNAi expressing animals.
- https://cdn.elifesciences.org/articles/68636/elife-68636-fig2-data2-v3.xlsx
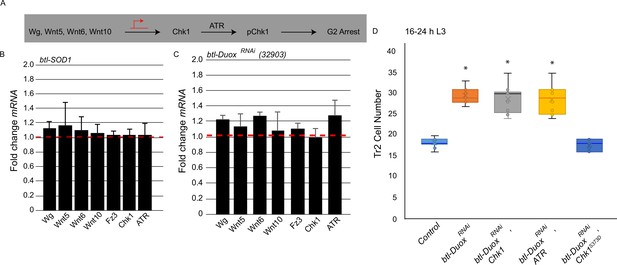
Reactive oxygen species (ROS) dependence identifies a novel pathway for the regulation of ataxia telangiectasia mutated-related kinase/checkpoint kinase 1 (ATR/Chk1) in tracheoblasts.
(A) Model for G2 arrest mechanism in Tr2 tracheoblasts based on previous studies. Earlier work has shown that four Wnt ligands (Wg, Wnt5, Wnt6, Wnt10) act synergistically to upregulate Chk1 mRNA levels in arrested tracheoblasts. High levels of Chk1 expression are necessary for G2 arrest and Chk1 overexpression can rescue defects in Wnt signaling (Kizhedathu et al., 2020). (B, C) Effect of superoxide dismutase 1 (SOD1) overexpression and Dual oxidase (Duox) knockdown on expression of Wnts and Wnt-target genes. Quantitative PCR analysis of levels of Wg, Wnt5, Wnt6, Wnt10, Fz3, Chk1, and ATR mRNA in micro-dissected Tr2 DT fragments at L2. Graph shows fold change in Wg, Wnt5, Wnt6, Wnt10, Fz3, Chk1, and ATR mRNA levels in Tr2 dorsal trunk (DT) fragments expressing (B) btl-SOD1 (btl-GAL4/UAS-SOD1) and (C) btl-DuoxRNAi (btl-GAL4/+; UAS-DuoxRNAi (32903)/+). Fold change has been represented with respect to wild type (btl-Gal4, shown by dashed red line, n = 3 experiments, n ≥ 15 Tr2 DT fragments/stage/experiment, mean ± standard deviation). (D) Effect of overexpression of a phosphomimic variant of Chk1 in btl-DuoxRNAi larvae at 16–24 hr L3. Graph shows numbers of Tr2 tracheoblasts in wild type (btl-Gal4), btl-DuoxRNAi (btl-GAL4/+; UAS-DuoxRNAi (32903)/+), btl-DuoxRNAi, Chk1 (btl-GAL4/+; UAS-DuoxRNAi (32903)/ UAS-Chk1), btl-DuoxRNAi, ATR (btl-GAL4/+; UAS-DuoxRNAi (32903)/ UAS-ATR) and btl-DuoxRNAi, Chk1S373D (btl-GAL4/+; UAS-DuoxRNAi (32903)/ UAS-Chk1S373D) larvae at 16–24 hr L3 (n ≥ 7 tracheae per condition per timepoint). Student’s t-test: *p<0.00001.
-
Figure 3—source data 1
Cell frequencies in Chk1S373D expressing animals.
- https://cdn.elifesciences.org/articles/68636/elife-68636-fig3-data1-v3.xlsx
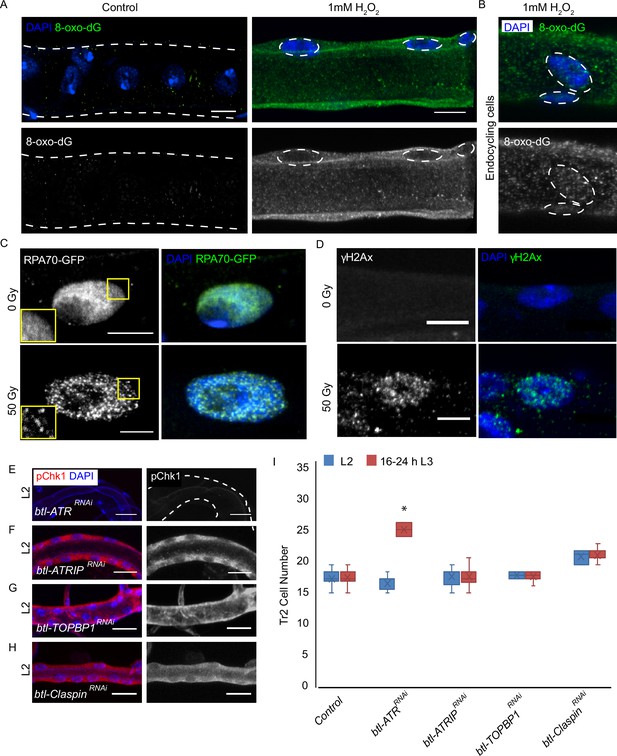
ATRIP, TOPBP1, and claspin are not required for reactive oxygen species (ROS)-mediated checkpoint kinase 1 (Chk1) activation in tracheoblasts.
(A–D) Detailed analysis of DNA damage in Tr2 dorsal trunk (DT). Shown here are findings from three different reporters of genotoxic stress. (A) 8-Oxo-2'-deoxyguanosine (8-Oxo-dG) immunostaining in wild type (btl-GAL4) Tr2 DT in untreated tracheae (left panel) and tracheae exposed to 1 mM H2O2 for 30 min ex vivo (right panel) at L2. (B) 8-Oxo-dG immunostaining in wild type (btl-GAL4) endocycling cells of the tracheae exposed to 1 mM H2O2 for 30 min ex vivo at L2. (C) GFP immunostaining in non-irradiated and γ-irradiated larvae expressing RPA70-GFP. Shown in the figure are GFP immunostaining in non-irradiated larvae (top panel) and larvae exposed to 50 Gy of γ-radiation (bottom panel) at L2. (D) γ-H2AXSer139 immunostaining in Tr2 DT in wild type (btl-GAL4) non-irradiated larvae (top panel) and larvae irradiated with 50 Gy of γ-radiation (bottom panel) at L2. (E–H) Analysis of the contribution of components of the DNA damage-dependent activation of ATR/Chk1 to Chk1 activation in Tr2 DT. Effects of the knockdown of ATR, ATRIP, TOPBP1, and Claspin on phosphorylated checkpoint kinase 1 (pChk1) levels in Tr2 DT at L2. pChk1 immunostaining (red) in Tr2 DT in (E) btl-ATRRNAi (btl-GAL4/UAS-ATRRNAi), (F) btl-ATRIPRNAi (btl-GAL4/UAS-ATRIPRNAi), (G) btl-TOPBP1RNAi (btl-GAL4/+; UAS-TOPBP1RNAi/+), and (H) btl-ClaspinRNAi (btl-GAL4/+; UAS-ClaspinRNAi/+) larvae at L2. (I) Effects of knockdown of ATR, ATRIP, TOPBP1, and Claspin on cell numbers in Tr2 DT. Graph shows numbers of Tr2 tracheoblasts in wild type (btl-Gal4), btl-ATRRNAi (btl-GAL4/UAS-ATRRNAi), btl-ATRIPRNAi (btl-GAL4/UAS-ATRIPRNAi), btl-TOPBP1RNAi (btl-GAL4/+; UAS-TOPBP1RNAi/+), and btl-ClaspinRNAi (btl-GAL4/+; UAS-ClaspinRNAi/+) at L2 and 16–24 hr L3 (mean values ± standard deviation, n ≥ 7 tracheae per condition per timepoint). Scale bars = 5 µm (A–D), 10 µm (E–H). Student’s t-test: *p<0.00001.
-
Figure 4—source data 1
Cell frequencies in ATR RNAi, ATRIP RNAi, TOPBP1 RNAi and Claspin RNAi expressing animals.
- https://cdn.elifesciences.org/articles/68636/elife-68636-fig4-data1-v3.xlsx
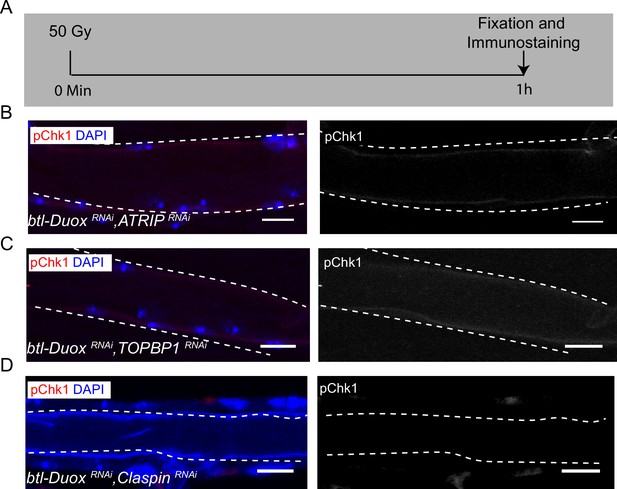
ATRIP, TOPBP1, and claspin are required for DNA damage-dependent activation ofATR/Chk1.
Effect of knockdown of Duox and ATRIP or TOPBP1 or Claspin on phosphorylated checkpoint kinase 1 (pChk1) levels in Tr2 dorsal trunk (DT) in larvae exposed to 50 Gy of γ-radiation at L2. (A) Schematic describing the protocol for exposure to γ-radiation and immunostaining. (B–D) pChk1 immunostaining in Tr2 DT in (B) btl-DuoxRNAi, ATRIPRNAi (btl-GAL4/UAS-ATRIPRNAi; UAS-DuoxRNAi (32903)/+), (C) btl-DuoxRNAi, TOPBP1RNAi (btl-GAL4/+; UAS-DuoxRNAi(32903)/UAS-TOPBP1RNAi), and (D) btl-DuoxRNAi, ClaspinRNAi (btl-GAL4/+; UAS-DuoxRNAi(32903)/UAS-ClaspinRNAi) larvae at 1 hr post exposure to 50 Gy of γ-radiation at L2 (n ≥ 6 tracheae per condition). Scale bars = 10 µm.
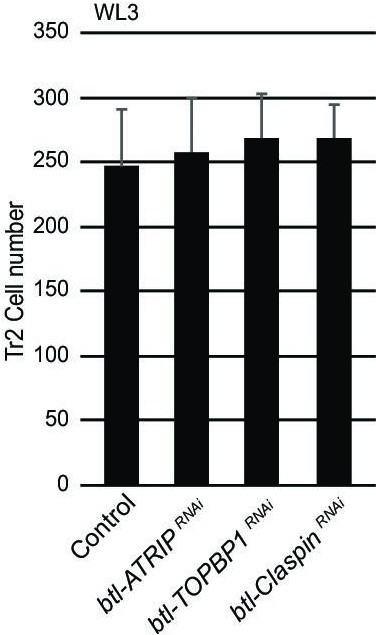
Loss of ATRIP, TOPBP1, and claspin does not affect cell numbers at WL3.
Effect of reduction of ATRIP, TOPBP1, or Claspin on cell numbers in Tr2 dorsal trunk (DT) at WL3. Graph shows cell numbers at WL3 in control (btl-GAL4), btl-ATRIPRNAi (btl-GAL4/UAS-ATRIPRNAi), btl-TOPBP1RNAi (btl-GAL4/+; UAS-TOPBP1RNAi/+), and btl-ClaspinRNAi (btl-GAL4/+; UAS-ClaspinRNAi/+) (mean values ± standard deviation, n ≥ 7 tracheae).
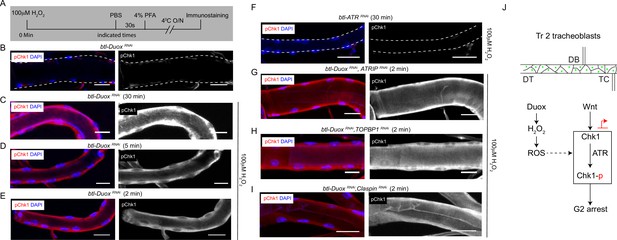
Incubation with H2O2 can restore phosphorylated checkpoint kinase 1 (pChk1) levels in Dual oxidase (Duox)-deficient tracheoblasts.
(A–E) Kinetics of Chk1 phosphorylation upon exposure to H2O2 ex vivo. (A) Regimen for H2O2 treatment and analysis of pChk1 in Tr2 dorsal trunk (DT) in L2. pChk1 immunostaining (red) in Tr2 DT in (B) untreated btl-DuoxRNAi (btl-GAL4/+; UAS-DuoxRNAi (32903)/+)-expressing tracheae and treated with 100 µM H2O2 for (C) 30 min, (D) 5 min, and (E) 2 min. (F) Effect of knockdown of ATR on Chk1 activation in Tr2 DT upon exposure to H2O2 ex vivo. pChk1 immunostaining (red) in Tr2 DT in btl-ATRRNAi (btl-GAL4/UAS-ATRRNAi) tracheae treated with 100 µM H2O2 for 30 min. (G–I) Effect of knockdown of Duox and ATRIP or TOPBP1 or Claspin on pChk1 levels in Tr2 DT in tracheae exposed to 100 µM H2O2 at L2. pChk1 immunostaining (red) in Tr2 DT in (G) btl-DuoxRNAi, ATRIPRNAi (btl-GAL4/ UAS-ATRIPRNAi; UAS-DuoxRNAi (32903)/+), (H) btl-DuoxRNAi, TOPBP1RNAi (btl-GAL4/+; UAS-DuoxRNAi(32903)/UAS-TOPBP1RNAi) and (I) btl-DuoxRNAi, ClaspinRNAi (btl-GAL4/+; UAS-DuoxRNAi(32903)/UAS-ClaspinRNAi) tracheae treated with 100 µM H2O2 for 2 min at L2. (J) Model for the regulation of ATR/Chk1 activation in Tr2 DT. We propose that H2O2 can induce ATR-dependent phosphorylation and activation of Chk1 in the absence of detectable DNA damage, leading to G2 arrest in Tr2 tracheoblasts. Scale bars = 10 µm.
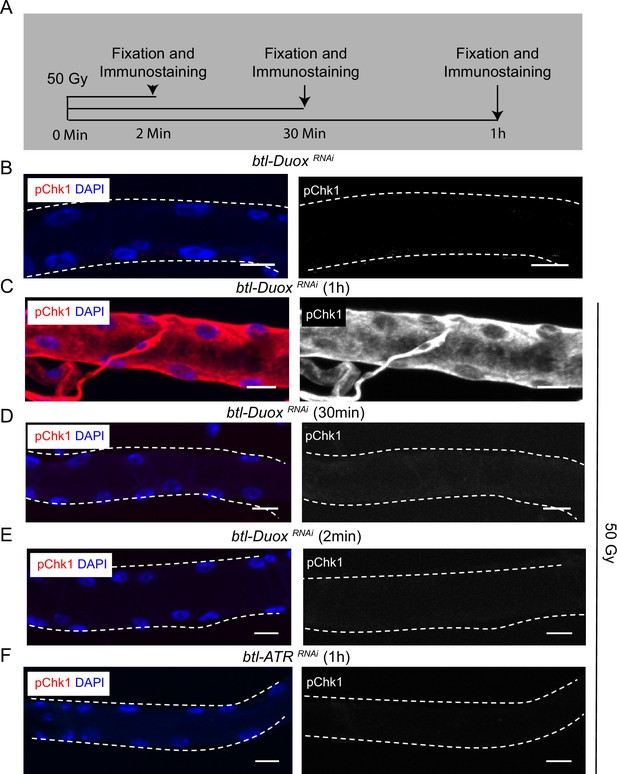
Exposure to γ-radiation can restore phosphorylated checkpoint kinase 1 (pChk1) levels in dual oxidase (Duox)-deficient tracheoblasts.
(A–E) Kinetics of Chk1 phosphorylation on exposure to γ-radiation. (A) Regimen for γ-irradiation and analysis of pChk1. pChk1 immunostaining (red) in Tr2 dorsal trunk (DT) in btl-DuoxRNAi (btl-GAL4/+; UAS-DuoxRNAi (32903/+)) in non-irradiated larvae (B) and larvae exposed to with 50 Gy of γ-radiation after (C) 1 hr, (D) 30 min, and (E) 2 min post irradiation at L2. (F) Effect of knockdown of ATR on Chk1 activation in Tr2 DT in larvae exposed to γ-radiation. pChk1 immunostaining (red) in Tr2 DT in btl-ATRRNAi (btl-GAL4/UAS-ATRRNAi) larvae exposed to 50 Gy of γ-radiation at 1 hr post exposure at L2 (n ≥ 6 tracheae per condition) Scale bars = 10 µm.
Tables
| Reagent type(species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (Drosophila melanogaster) | btl-GAL4 | Shiga et al., 1996 | FLYB: FBtp0001208 | This line was a gift from Dr. Shigeo Hayashi |
| Genetic reagent (D. melanogaster) | UAS-Chk1RNAi | VDRC | 110076 | |
| Genetic reagent (D. melanogaster) | UAS-Chk1S373D | This study | Please see Materials and methods for a detailed description. (Can be obtained through NCBS Fly Facility: https://bangalorefly.ncbs.res.in/) | |
| Genetic reagent (D. melanogaster) | RPA-70GFP | Blythe and Wieschaus, 2015 | This line was a gift from Dr. Eric F Wieschaus | |
| Genetic reagent (D. melanogaster) | UAS-DuoxRNAi | BDSC | RRID:BDSC_33975 and RRID:BDSC_32903 | |
| Genetic reagent (D. melanogaster) | UAS-ATR | Bayer et al., 2018 | This line was a gift from Dr. Anja C Nagel | |
| Antibody | Phospho-Chk1 (Ser345) (rabbit monoclonal) antibody | CST | Cat #2348 (RRID:AB_331212) | (1:200) |
| Antibody | Anti-8-hydroxy-2’-deoxyguanosine antibody (mouse monoclonal) antibody | Abcam | Cat #ab48508 (RRID:AB_867461) | (1:200) |
| Commercial assay or kit | Tyramide signal amplification system | Thermo Fisher Scientific | Cat #B40912 |





