Multi-syndrome, multi-gene risk modeling for individuals with a family history of cancer with the novel R package PanelPRO
Figures
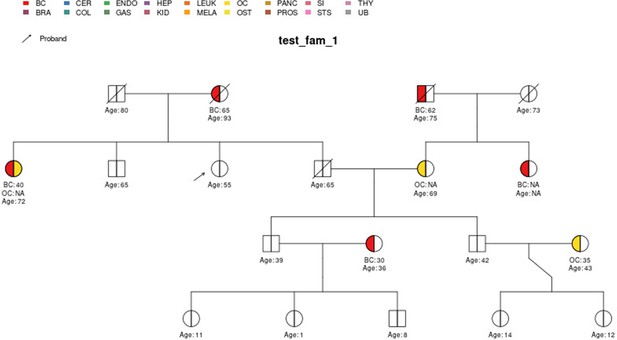
test_fam_1 sample pedigree as included in the PanelPRO package, plotted using the external visPed package.
The colors refer to cancer diagnoses in the legend. The age of diagnosis is shown below the individual if it is known.
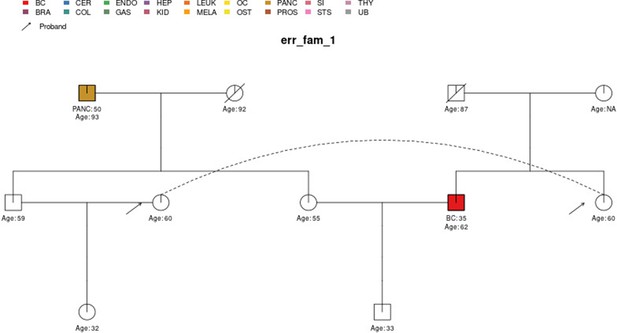
The sample pedigree err_fam_1 which contains a pedigree loop, due to the mating pattern of the siblings aged 59 and 55 with the siblings aged 60 and 62, respectively.
The two circles linked by a dotted line represent the same individual.
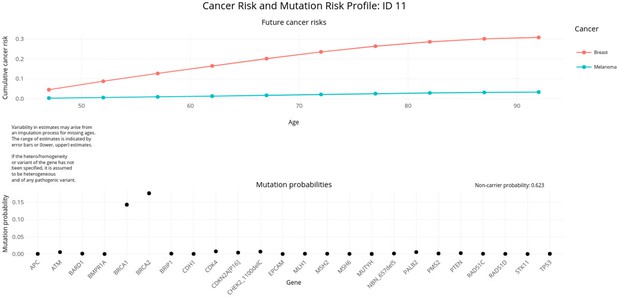
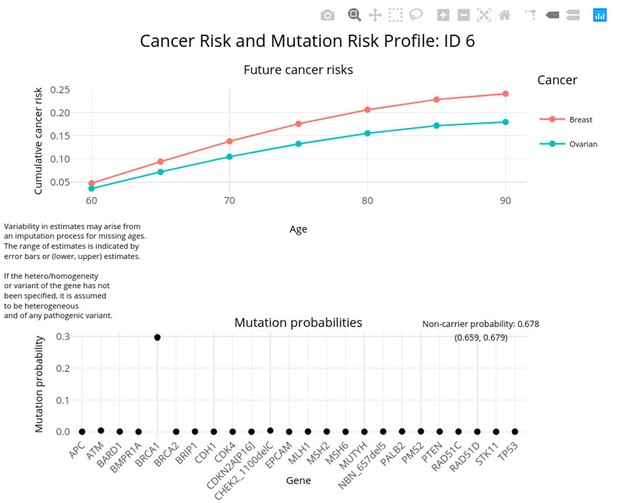
PanelPRO output with test_fam_5 as pedigree input.
Graphical output from the visRisk function in PanelPRO for test_fam_5.
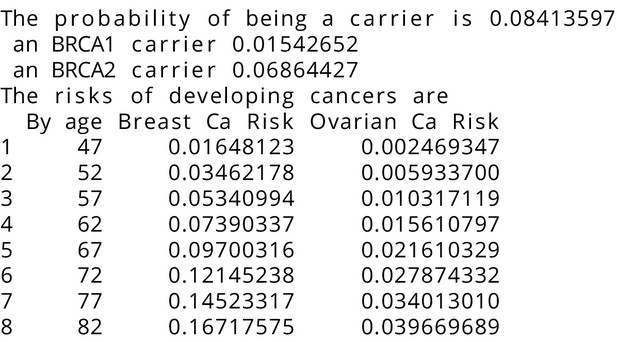
BRCAPRO output with test_fam_5 as pedigree input.
Text output from the BRCAPRO model for test_fam_5.

IBIS output with test_fam_5 as pedigree input.
Screenshot of output from IBIS for test_fam_5.
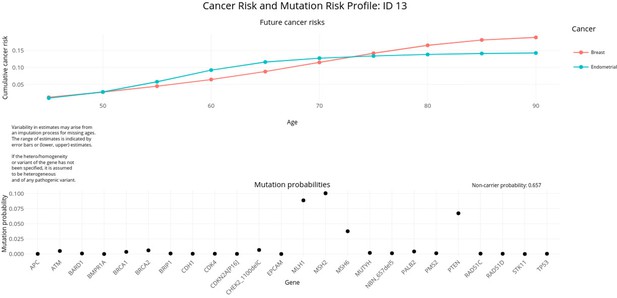
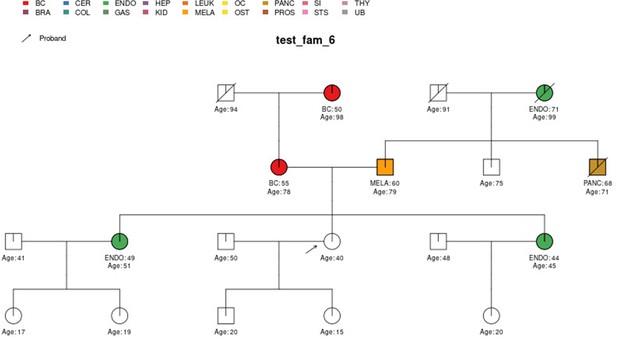
PanelPRO output with test_fam_6 as pedigree input.
Graphical output from the visRisk function in PanelPRO for test_fam_6.
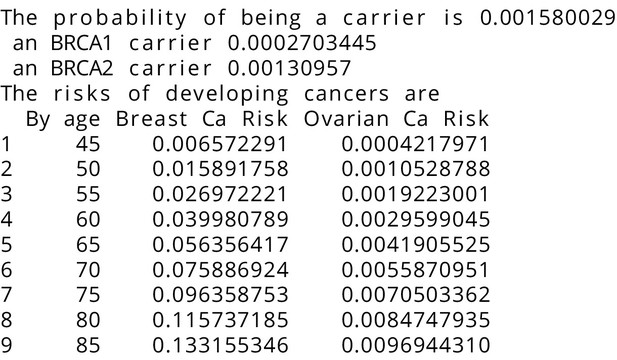
BRCAPRO output with test_fam_6 as pedigree input.
Text output from the BRCAPRO model for test_fam_6.

MMRPRO output with test_fam_6 as pedigree input.
Text output from the MMRPRO model for test_fam_6.
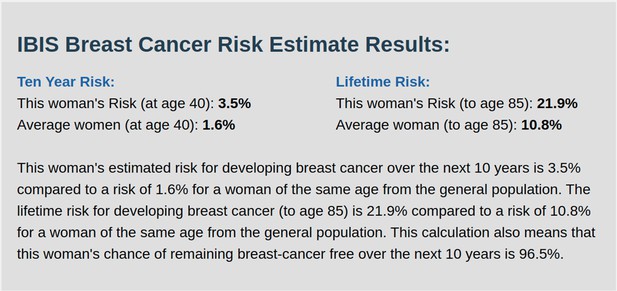
IBIS output with test_fam_6 as pedigree input.
Screenshot of output from IBIS for test_fam_6.

PREMM-5 output with test_fam_6 as pedigree input.
Screenshot of output from PREMM-5 for test_fam_6.
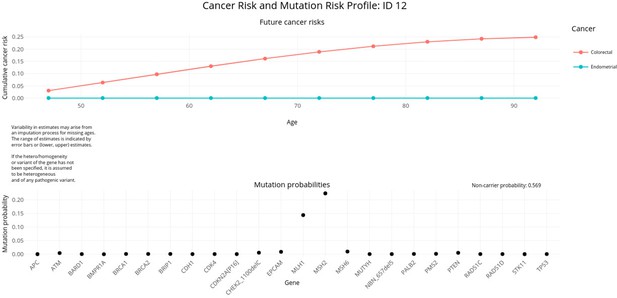
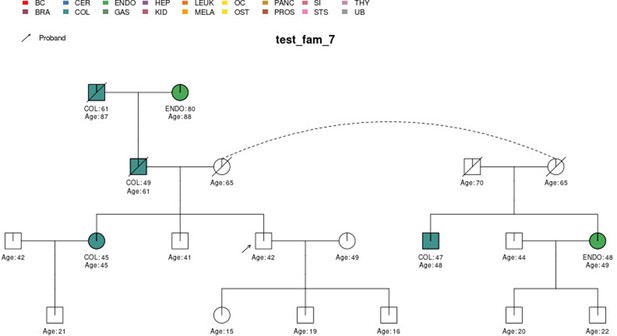
PanelPRO output with test_fam_7 as pedigree input.
Graphical output from the visRisk function in PanelPRO for test_fam_7.

MMRPRO output with test_fam_7 as pedigree input.
Text output from the MMRPRO model for test_fam_7.

PREMM-5 output with test_fam_7 as pedigree input.
Screenshot of output from PREMM-5 for test_fam_7.

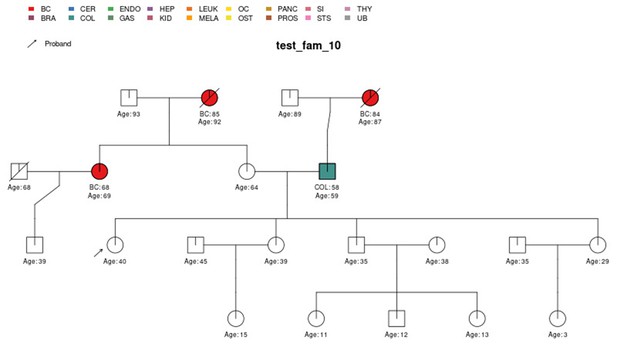
PanelPRO output with test_fam_10 as pedigree input.
Graphical output from the visRisk function in PanelPRO for test_fam_10.
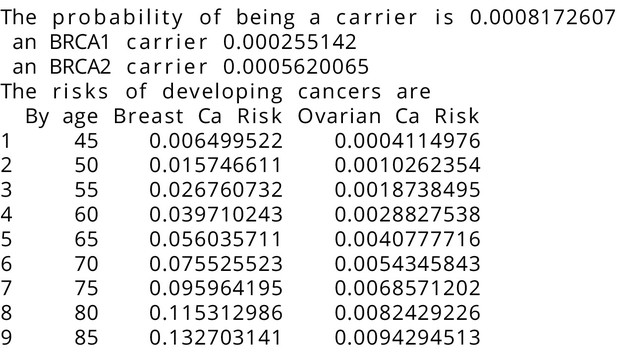
BRCAPRO output with test_fam_10 as pedigree input.
Text output from the BRCAPRO model for test_fam_10.
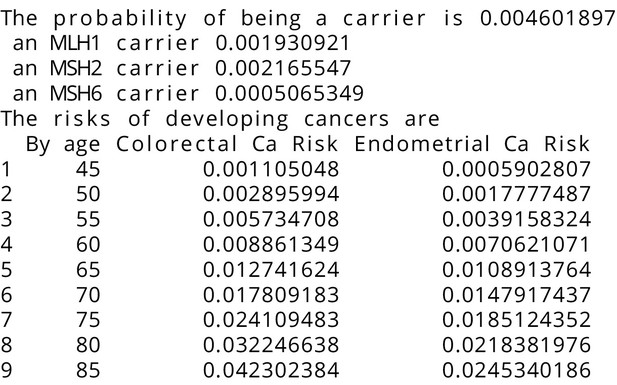
MMRPRO output with test_fam_10 as pedigree input.
Text output from the MMRPRO model for test_fam_10.
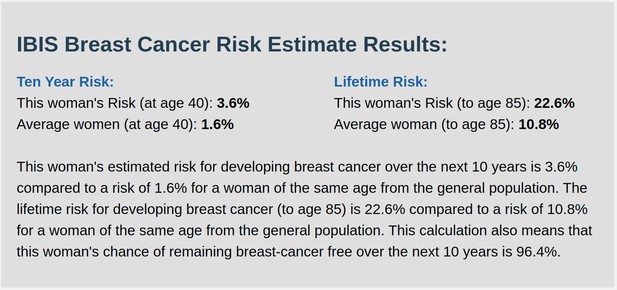
IBIS output with test_fam_1 as pedigree input.
Screenshot of output from IBIS for test_fam_10.

PREMM-5 output with test_fam_10 as pedigree input.
Screenshot of output from PREMM-5 for test_fam_10.
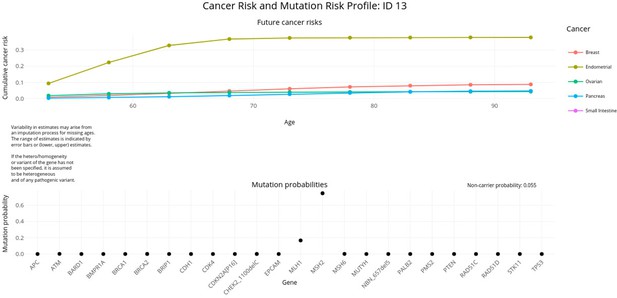
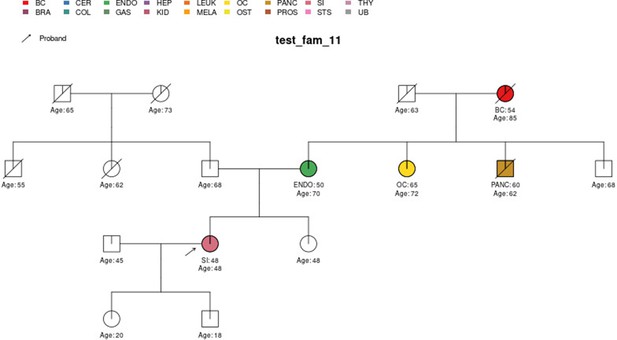
PanelPRO output with test_fam_11 as pedigree input.
Graphical output from the visRisk function in PanelPRO for test_fam_11.
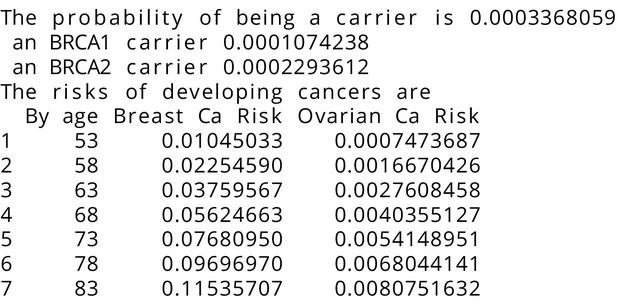
BRCAPRO output with test_fam_11 as pedigree input.
Text output from the MMRPRO model for test_fam_11.
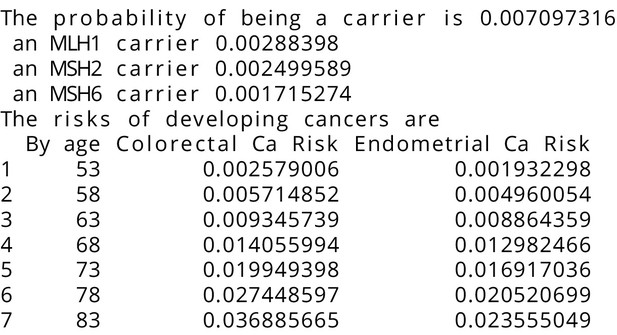
MMRPRO output with test_fam_11 as pedigree input.
Text output from the MMRPRO model for test_fam_11.
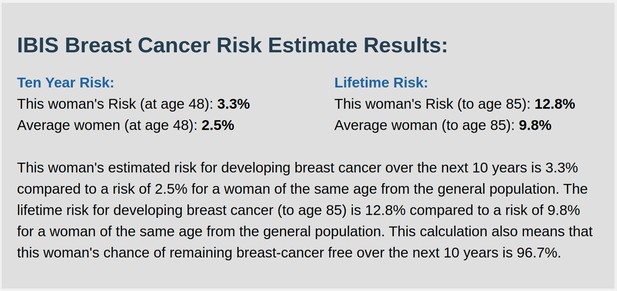
IBIS output with test_fam_11 as pedigree input.
Screenshot of output from IBIS for test_fam_11.

PREMM-5 output with test_fam_11 as pedigree input.
Screenshot of output from PREMM-5 for test_fam_11
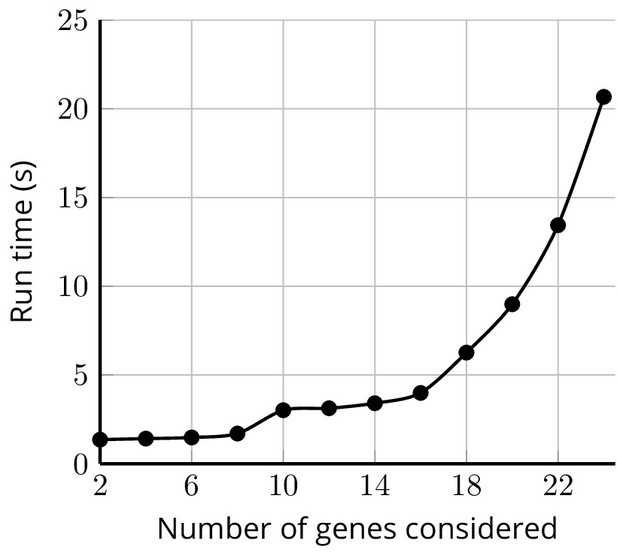
Sample run-times for test_fam_1 evaluated by PanelPRO on the default settings, as a function of the number of genes considered.
The paring parameter is set to 2. These run time experiments were performed on a 2020 Linux machine with an 11th Gen Intel(R) i7-1165G7 chip at 2.80 GHz.
Tables
Pedigree structure in PanelPRO.
| Column | Definition | Value |
|---|---|---|
| ID | Unique numeric identifier of each individual | Non-repeated strictly positive integer |
| MotherID | ID of one’s mother | Strictly positive integer or NA (missing) |
| FatherID | ID of one’s father | Strictly positive integer or NA (missing) |
| Sex | Sex of the individual: 1 for male, 0 for female | One of {0, 1} |
| isProband | Indicates the proband or counselee by 1 and 0 otherwise – multiple probands can be specified | One of {0, 1) |
| CurAge | Age of censoring: either the current age or death age, depending on isDead status | Positive integer or NA (missing) |
| isAff* | Affection status of cancer * | One of {0, 1} |
| Age* | Affection age of cancer * | Positive integer or NA (missing) |
| isDead | Whether someone has died | One of {0, 1, NA} |
| race | Race of individual (used to modify penetrance) | One of All_Races, AIAN, Asian, Black, White, Hispanic, WH, WNH, NA |
| Ancestry | Ancestry of individual (used to modify allele frequencies) | One of AJ, nonAJ, Italian, NA |
| Twins | Identifies siblings who are identical twins or multiple births | Each set is identified by a unique integer, and 0 otherwise |
| riskmod | Preventative interventions which modify penetrance | List, combination of "mastectomy", "hysterectomy", and "oophorectomy" |
| InterAge | Age of each preventative interventions | List, combination of integers |
| Gene name from GENE_TYPES | Germline testing result | One of {0, 1, NA} |
| Marker name from CK14, CK5.6, ER, PR, HER2, MSI | Marker testing result | One of {0, 1, NA} |
List of model options that the user can pass to PanelPRO, along with their defaults.
| Option | Default value | Possible values | Description |
|---|---|---|---|
| max.mut | NULL | Integers up to the number of genes | Number of maximum simultaneous mutations, also known as the paring parameter. If no integer has been input, it re-defaults to 2. |
| iterations | 20 | Integers from 1 upwards | In case of missing current or cancer ages in the pedigree, this is the number of times those ages will be imputed. |
| parallel | TRUE | TRUE or FALSE | If age imputations are needed, this parameter can be set to utilize multiple cores on one’s machine. |
| net | FALSE | TRUE or FALSE | Determines whether net or crude penetrances are used to compute future risk of cancer. Net penetrances exclude all other causes of death, apart from the affected cancer. |
| age.by | 5 | Integers from one upwards | The intervals of age used to report the future risk of cancer. |
List of main functions in PanelPRO.
| Category | Name | Description |
|---|---|---|
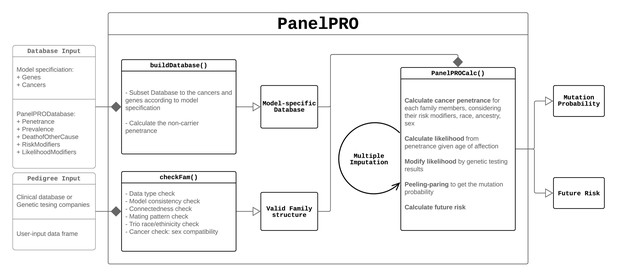
| Pre-processing | checkFam | Checks family structure as defined by the user. The inputs are a data.frame specifying the pedigree and a built database returned by buildDatabase. The output is a modified data.frame pedigree and list of imputed ages, if missing ages were imputed (see the Missing Data section). |
| Pre-processing | buildDatabase | Subsets the internal database PanelPRODatabase depending on the cancers and genes selected. The input is the list PanelPRODatabase. The output is another list which is a subset of PanelPRODatabase. |
| Algorithm | PanelPROCalc | Estimates the posterior carrier probabilities and future risks of the proband. The inputs are the outputs of checkFam. The outputs are lists of posterior probabilities and future risks for the proband. |
| Main function | PanelPRO | Runs main function. The inputs are the user-specified pedigree, a vector of cancers in the model, a vector of genes in the model, and other optional parameters. The output is a list of estimates of posterior carrier probabilities for each genotype, along with future cancer risks and ranges for each of these. |
Comparison between supported cancers and genes in PanelPRO and other platforms.
| Model or platform name | Version | Supported cancer input types | Supported gene carrier probability outputs | Supported future cancer risk outputs |
|---|---|---|---|---|
| PanelPRO | 0.2.0 | Brain, breast, cervical, colorectal, endometrial, gastric, kidney, leukemia, melanoma, ovarian, osteosarcoma, pancreatic, small intestine, soft tissue sarcoma, thyroid, urinary bladder, hepatobiliary | APC, ATM, BARD1, BMPR1A, BRCA1, BRCA2, BRIP1, CDH1, CDK4, CDKN2A, CHEK2, EPCAM, MLH1, MSH2, MSH6, MUTYH, NBN, PALB2, PMS2, PTEN, RAD51C, RAD51D, STK11, TP53 | same as cancer inputs |
| BRCAPRO | 2.1–7 | Breast, ovarian | BRCA1, BRCA2 | same as cancer inputs |
| MMRPRO | 2.1–7 | Colorectal, endometrial | MLH1, MSH2, MSH6 | same as cancer inputs |
| IBIS | 0.8b | Breast | NA | Breast |
| CanRisk | 1.2.3 | Breast, contralateral breast, ovarian, prostate, pancreatic | BRCA1, BRCA2, PALB2, CHEK2, ATM, RAD51D, RAD51C, BRIP1 | Breast, ovarian |
| PREMM-5 | NA | Colorectal, endometrial, other (group of ovarian, stomach, small intestine, urinary tract/bladder/kidney, bile ducts, brain, pancreas, sebaceous gland skin) | MLH1, MSH2, MSH6, PMS2, EPCAM | NA |
Notation for Mendelian Modeling for a model with genes and cancers and a family of members.
The subscript denotes the th family member.
| Variable and notation | Description | R object from user input, if applicable |
|---|---|---|
| Genotypes | ||
| Genotype of individual , where is the binary indicator for carrying a deleterious mutation in the th gene | ||
| Genotypes of all family members | ||
| Sex | ||
| Binary indicator that individual is male | Sex | |
| Binary male indicators for all family members | ||
| Cancer history | ||
| Age of diagnosis of the th cancer for individual | AgeXX | |
| Individual i’ s censoring age (current age or age of death) | CurAge | |
| Binary indicator that cancer occurs before the censoring age for individual | ||
| Observed history of the th cancer for individual , not including risk modifiers and interventions | ||
| All observed history for individual | ||
| Observed histories for all family members | ||
| Individual ’s age of death from causes other than cancer | ||
| Individual ’s age of first outcome, either cancer or death from causes other than cancer | ||
| Binary indicator that individual develops the th cancer | isAffXX |
Abbreviations of cancers in PanelPRO.
| Short name | Long name | Short name | Long name |
|---|---|---|---|
| BRA | Brain | OC | Ovarian |
| BC | Breast | OST | Osteosarcoma |
| CER | Cervical | PANC | Pancreas |
| COL | Colorectal | PROS | Prostate |
| ENDO | Endometrial | SI | Small Intestine |
| GAS | Gastric | STS | Soft Tissue Sarcoma |
| KID | Kidney | THY | Thyroid |
| LEUK | Leukemia | UB | Urinary Bladder |
| MELA | Melanoma | HEP | Heptobiliary |
Summary of sample pedigrees provided within the package.
| Pedigree name | Number of family members | Cancers present |
|---|---|---|
| test_fam_1 | 19 | BC, OC |
| test_fam_2 | 25 | ENDO, PANC, SI |
| test_fam_3 | 50 | BRA, BC, COL, ENDO, GAS, KID, MELA, OC, PANC, PROS, SI |
| test_fam_4 | 9 | BC, OC, BRA, COL, PROS, ENDO, SI, |
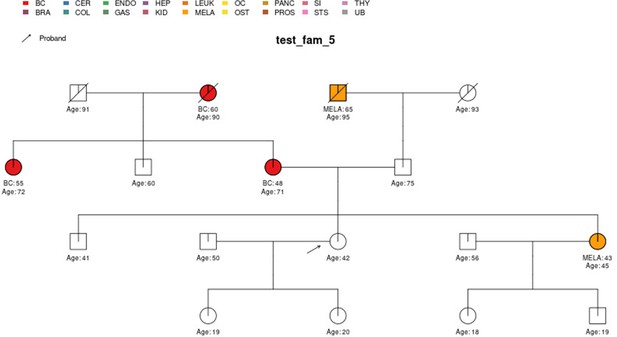
| test_fam_5 | 17 | BC, MELA |
| test_fam_6 | 19 | BC, ENDO, MELA, PANC |
| test_fam_7 | 19 | COL, ENDO |
| test_fam_8 | 20 | COL, PROS |
| test_fam_9 | 19 | BC, PROS |
| test_fam_10 | 21 | BC, COL |
| test_fam_11 | 16 | BC, ENDO, OC, PANC, SI |
| test_fam_12 | 21 | all cancers in Appendix 1—table 1 |
| err_fam_1 | 10 | BC, PANC |