Protein allocation and utilization in the versatile chemolithoautotroph Cupriavidus necator
Peer review process
This article was accepted for publication as part of eLife's original publishing model.
History
- Version of Record updated
- Version of Record published
- Accepted Manuscript published
- Accepted
- Received
- Preprint posted
Decision letter
-
Gisela StorzSenior and Reviewing Editor; National Institute of Child Health and Human Development, United States
-
Jörg ToepelReviewer; Helmholtz Centre for Environmental Research - UFZ, Germany
-
Nico ClaassensReviewer; Wageningen University & Research, Netherlands
Our editorial process produces two outputs: (i) public reviews designed to be posted alongside the preprint for the benefit of readers; (ii) feedback on the manuscript for the authors, including requests for revisions, shown below. We also include an acceptance summary that explains what the editors found interesting or important about the work.
Decision letter after peer review:
Thank you for submitting your article "Protein allocation and utilization in the versatile chemolithoautotroph Cupriavidus necator" for consideration by eLife. Your article has been reviewed by 3 peer reviewers, and the evaluation has been overseen by Gisela Storz serving as Reviewing and Senior Editor. The following individuals involved in review of your submission have agreed to reveal their identity: Jörg Toepel (Reviewer #1); Nico Claassens (Reviewer #2).
The reviewers have discussed their reviews with one another, and the Reviewing Editor has drafted this to help you prepare a revised submission.
Essential revisions:
– Determine biomass per OD600 for different conditions.
– Examine PHB production, particularly if the authors want to include the ammonia condition. Otherwise maybe they should leave out claims regarding growth on that condition.
– As detailed in the individual reviews, the text would be improved by some rewriting, including some tempering of conclusions.
Reviewer #1 (Recommendations for the authors):
Introduction:
Try to replace wondering at least some times, it sounds very repetitive.
Results:
Figure 3: The significant tests are just reveal that the differences are just between two groups and not between all of the groups, please indicate that, or did I miss something?
Discussion:
The conclusion regarding the up-regulation of the cbb cycle under heterotrophic growth is weak and not supported by the data and controversy to the cited paper. The authors should state that in detail (as they did but it i am not sure if sufficient).
What are the consequences for biotechnological applications? Deletion of large parts of the genome? Please explain and give some ideas.
Delete the last sentence of the discussion it is somehow arbitrary. You also can explain it in a separate paragraph in more detail
Methods:
Biomass per OD highly depends on the growth condition and bears the risk or error
Reviewer #2 (Recommendations for the authors):
The paper is well-written , and could be interesting for the readership of eLife if several major issues are tackled by the authors first.
1. One of the only clear ' questions' the researchers are after is to determine if co-utilization of the CCB cycle for growth on fructose is useful. The RBA and fitness analysis shows this is not the case for fructose and any of the other (even more oxidized) substrates. This could also be already expected theoretically. If you consider fructose has a reduction degree per carbon of 4 and biomass about 4.2, it is clear that no 'electrons' are left over to reduce extra CO2. This would only be interesting on very reduced substrates like glycerol (degree of reduction > biomass). However, real biology may behave differently than what 'theoretical stoichiometries predict', C. necator may perform' overoxidation in the TCA cycle, and this could potentially explain a benefit of the CO2 refixation, even though on paper it's a wasteful detour. Other papers suggest this overactivity of TCA cycle, also in autotrophic growth of C. necator on hydrogen (10.1016/j.procbio.2017.07.007), where limiting the oxygen concentration increased biomass yields. A similar situation may be going on in heterotrophic growth and explain observations on a small impact. I suggest the authors look at that more closely and revisit some conclusions on this.
2. The authors state that the deletion of ccbR has no fitness cost for growth on fructose, however in Figure 5F I still observe a minor, but significant fitness cost for growth on fructose, hence I suggest to rephrase the text slightly to better reflect that. This may actually change the conclusion of the paper on the 'useless' role of the CCB on fructose and sucrose.
3. The authors should better explain the differences between their findings and Shimizu et al., 2015 paper in Scientific reports.
4. Page 16, line 19 and 32: please check if you are not referring to figure 4 instead.
5. P21 L17 " Another conditionally essential gene on formate was ppc,
encoding the PEP-carboxylase (PPC). The reaction has no other annotated (iso-) enzymes and was predicted by RBA to carry most flux on formate compared to the other carbon sources (Figure 6 B)." It would be interesting to explain the biochemical logic why this anaplerotic reaction would be used more than in other conditions, where anaplerosis is also needed. I guess it has to do with the fact that there is generally lower TCA flux during growth on formate, NADH is generated directly from NADH rather than the TCA cycle. However, anaplerosis is needed in all conditions to generate oxaloacetate etc. So interesting to explain this.
6. P25 L14 " P25 L14 " Of all central carbon metabolism, the TCA cycle enzymes
showed on average lowest abundance, variability and utilization. This is similar to E. coli, where the sole flux capacity demand suggested lower enzyme abundance than what was measured experimentally [Noor et al., 2016]. Only when reverse flux (reactions with low thermodynamic driving force) and l ow enzyme saturation, estimated from metabolite levels was taken in to account, was the calculated enzyme demand similar to the measured levels [Noor et al., 2016]."
This is an important point the authors raise in the discussion, and also makes wonder why the authors did not take into account reverse fluxes in their RBA analysis, is this because the lack of kinetic parameters known, thermodynamic parameters to perform this also for C. necator should be available generally, right? Good to specify this (or repeat that analysis including reverse fluxes, e.g. only based on thermodynamics).
7. Be clearer about what sugars C. necator can use. On P25 L25 you state " For C.
necator, hexose sugars are only one out of many possible substrate classes and the flux" However, it can only accept one hexose sugar (fructose) as is stated before twice:
– " The only sugar that supports growth 27 is fructose, which i s metabolized via the Entner-Doudoroff (ED) pathway [Alagesan et al., 28 2018]."
– " Fructose was chosen as it is the only known sugar (apart from sugar alcohols) that C. necator utilizes [Orita et al., 2012]."
Overall, this is not so clear, Orita et al. do not discuss sugar alcohols and I am not sure which sugar alcohols C. necator can consume (it can consume some sugar acids, but that's something different, at least glycolate). In addition, C. necator does consume N-acetylglucosamine, which is defined as sugar. Maybe just rephrase that C. necator is only known to grow on one hexose sugar, being fructose and not on glucose. And a better reference could be the 2008 review of Cramm, which states " Organic carbon and energy sources for heterotrophic growth include TCA cycle intermediates, sugar acids, fatty acids, amino acids, alcohols, and aromatic compounds, while utilization of sugars is restricted to fructose and N -acetylglucosamine [Johnson and Stanier, 1971; Kersters and De Ley, 1984]"
8. The authors speculate about the evolutionary nature of the two copies of the Calvin cycle operon, as well as their data show the key role of cbbR. Can the authors maybe also mention and discuss the presence of an inactive ccbR copy in the 2nd CCB operon on pHG1, which is now not mentioned, but also indicates the 'recent' and ' unoptimized' acquisition of these CBB operons. Would also be good to know what is the abundance of this inactive cbbR copy from pHG1 in the proteome?
Reviewer #3 (Recommendations for the authors):
This study dealt with comprehensive analyses regarding protein quantitation and enzyme utilization in the versatile chemolithoautotrophic bacterium C. necator. The experiments and analyses were well considered and designed, and the authors obtained quite interesting information such as presence of many under-utilized enzymes as well as excess amount of utilized enzymes, suggesting the highly robust properties of this bacterium against environmental perturbations.
I have no comments in the experiments, results, and discussion obtained by carbon-limited chemostat cultivation, although I here address a few concerns in the nitrogen-limited cultivation as below, which should be considered by the authors before publication.
1) It has been well known that C. necator H16 synthesizes and accumulates PHB with in the cells under nitrogen-limited conditions. In many cases of PHB-producing cells, the accumulation of intracellular PHB granule reflects on apparent OD owing to the changes in cell size and morphology. In the nitrogen-limited cultivation in this study, I think that the constant OD was a result of steady states of both cell growth and PHB synthesis. Did the authors determine PHB accumulation under the nitrogen-limited condition?
2) PHB is a water-insoluble polyester, thus it can be escaped from cytosolic equilibrium once after polymerized from water-soluble monomers. Did the RBA model used in this study include PHB synthesis? That is, in the nitrogen-limited condition, did the biomass yield calculated by RBA simulation contain PHB?
3) The authors concluded that reassimilation of CO2 by CBB pathway does not provide a fitness benefit for heterotrophic growth. I agreed with this conclusion, because it is feasible that energy- and reducing equivalents-consuming CBB cycle is not required when carbons other than CO2 are available. While, I read the previous report by Shimizu et al. They demonstrated advantage of CBB cycle under heterotrophic growth in synthesis of the storage polyester unassociated with cell growth, and never proposed advantage in the heterotrophic growth. It is not adequate to discuss about this matter by applying the results in the current study focusing on growth to the previous study focusing on growth-unassociated PHB synthesis. I recommend the authors to correct/modify the relevant descriptions in order to avoid readers' misunderstanding.
[Editors' note: further revisions were suggested prior to acceptance, as described below.]
Thank you for resubmitting your work entitled "Protein allocation and utilization in the versatile chemolithoautotroph Cupriavidus necator" for further consideration by eLife. Your revised article has been reviewed by 2 peer reviewers and the evaluation has been overseen by Gisela Storz serving as Senior and Reviewing Editor.
I very much appreciate the effort you have taken to carefully address the previous reviews. There are just a few more points outlined below that should be considered in the final version for eLife.
Reviewer #2 (Recommendations for the authors):
The authors did a very good job in revising the manuscript and adding additional experiments and modelling on dry weight and PHB production.
The only answer that puzzled us was the answer on PEP carboxylase, we were puzzled by the explanation that oxaloacetate flux goes to glycine via reserve GCV operation (does that mean that this goes via threonine).
Furthermore, they satisfied all our queries well, so I recommend publication.
Reviewer #3 (Recommendations for the authors):
I respect the authors' great effort in re-examination of bioreactor experiment and replacement of metabolic modeling regarding the N-limitation condition. I agree with the most revisions so think that the revised manuscript is valuable for publication by eLife, but I have still a few concerns as below, which are expected to be considered by the authors prior to the publication.
1) Were the g-DCW values used for calculation of m-protein (g/g-DCW) in Figure 4, Y (gDCW/g-S) in Figure 5C (and so on) under the N-limitation condition the cell mass excluding PHB (PHB-subtracted biomass)? The m-protein graph under the N-limitation in Figure 4 in the revised manuscript looks like to be the same as the previous version.
2) The rate of intracellular PHB synthesis usually becomes maximum when the cell growth has been stopped by nitrogen depletion, so the increase in PHB production by the function of CBB cycle during the heterotrophic condition on fructose, reported by Shimizu et al., may become significant during the growth-unassociated PHB synthesis. The authors carried re-simulation to estimate the advantage of the heterotrophic CBB cycle (lines 373-385 in the revised manuscript), but this re-simulation was done still under growth conditions. Is it possible to simulate the growth-unassociated PHB synthesis on fructose under N-depletion? Fructose-uptake by C. necator was supposed to be weakened after the growth phase due to marked down-regulation of expression of EM and ED pathway genes (reported in previous transcriptome analyses). Considering this, I'm not sure whether the constant fructose uptake rate of 4.0 mmol/gDCW/h was adequate or not for the growth-unassociated PHB synthesis.
https://doi.org/10.7554/eLife.69019.sa1Author response
Essential revisions:
– Determine biomass per OD600 for different conditions.
The biomass in g dry cell weight (DCW) was determined for all substrates. For fructose, succinate and formate, biomass yield was determined using shake flask experiments, as the biomass yield was constant in previous chemostat cultivations (no change in optical density at different growth rates). For nitrogen limitation, such a change in optical density was observed, indicating an increase in total biomass with increasing N-limitation rate due to the production of PHB. The ammonium-limited cultivations were therefore reproduced in chemostats and DCW as well as PHB concentration determined for each growth rate individually.
– Examine PHB production, particularly if the authors want to include the ammonia condition. Otherwise maybe they should leave out claims regarding growth on that condition.
The PHB and DCW measurements for all substrates were added to the manuscript as new Figure 1—figure supplement 2. The new results show that C. necator does not produce PHB during carbon limitation, while it does produce an increasing amount of PHB with decreasing growth rate in the nitrogen-limited chemostat (up to 80% of biomass was PHB), in line with previous findings in the literature. The metabolic modeling was updated with the corrected biomass and PHB concentrations. In summary, the following changes were made to the manuscript:
– yield and substrate uptake rates are now lower than the previous estimation
– some fitted model parameters (e.g. kapp) have slightly changed due to changed input
– PHB production was explicitly included in the RBA model
– some figures show slight changes due to updated modeling results
– new Figure 1—figure supplement 2 summarizes PHB and DCW quantification
– new Figure 3—figure supplement 2 shows enzyme utilization of PHB biosynthesis pathway
– Figure 3D was updated with detailed mass and utilization per substrate instead of average values
– As detailed in the individual reviews, the text would be improved by some rewriting, including some tempering of conclusions.
All criticized text sections were changed according to the reviewer's suggestions as outlined in our response to the reviewer's comments.
Reviewer #1 (Recommendations for the authors):
Introduction:
Try to replace wondering at least some times, it sounds very repetitive.
The wording was changed in the introduction.
Results:
Figure 3: The significant tests are just reveal that the differences are just between two groups and not between all of the groups, please indicate that, or did I miss something?
Yes, the significance test is a pairwise comparison of two groups (moderate and high utilization) to the reference (low utilization). We have added horizontal bars to Figures 3A and B to indicate which groups were compared, and added an explanation to the figure legend.
Discussion:
The conclusion regarding the up-regulation of the cbb cycle under heterotrophic growth is weak and not supported by the data and controversy to the cited paper. The authors should state that in detail (as they did but it i am not sure if sufficient).
We assume that this criticism refers to our original conclusion that the expression of the cbb operon in heterotrophic conditions (fructose) does not provide a fitness benefit. This issue was also raised by the other reviewers. We have toned down our conclusions regarding the benefit of the cbb operon after considering the ambiguity of some of the fitness results, and revisiting the claims from Shimizu et al. (see more detailed response to comments from reviewer 2 and 3).
What are the consequences for biotechnological applications? Deletion of large parts of the genome? Please explain and give some ideas.
We have added the following paragraph about biotechnological implications of our work to the discussion:
“Our results imply that C. neactor is in its current state far from being an ideal host for biotech applications. […] Alternatively, laboratory evolution could be employed to select mutants with beneficial traits such as tolerance to formic acid.”
Delete the last sentence of the discussion it is somehow arbitrary. You also can explain it in a separate paragraph in more detail
The sentence was removed.
Methods:
Biomass per OD highly depends on the growth condition and bears the risk or error
We agree with the reviewer. The reason we used OD measurement as a proxy for biomass concentration was the limited volume of our chemostat cultures (65 mL); dry cell weight measurement makes it necessary to sacrifice a substantial amount of culture volume. We have now performed a new set of cultivations for all carbon and nitrogen limitations (fructose, succinate, formate in shake flasks, ammonium in chemostat), and determined g DCW and PHB from these cultures. The updated substrate uptake rates and the PHB production were included in the model.
Reviewer #2 (Recommendations for the authors):
The paper is well-written , and could be interesting for the readership of eLife if several major issues are tackled by the authors first.
1. One of the only clear ' questions' the researchers are after is to determine if co-utilization of the CCB cycle for growth on fructose is useful. The RBA and fitness analysis shows this is not the case for fructose and any of the other (even more oxidized) substrates. This could also be already expected theoretically. If you consider fructose has a reduction degree per carbon of 4 and biomass about 4.2, it is clear that no 'electrons' are left over to reduce extra CO2. This would only be interesting on very reduced substrates like glycerol (degree of reduction > biomass). However, real biology may behave differently than what 'theoretical stoichiometries predict', C. necator may perform' overoxidation in the TCA cycle, and this could potentially explain a benefit of the CO2 refixation, even though on paper it's a wasteful detour. Other papers suggest this overactivity of TCA cycle, also in autotrophic growth of C. necator on hydrogen (10.1016/j.procbio.2017.07.007), where limiting the oxygen concentration increased biomass yields. A similar situation may be going on in heterotrophic growth and explain observations on a small impact. I suggest the authors look at that more closely and revisit some conclusions on this.
We agree with the reviewer that theoretical considerations like degree of reduction and reaction stoichiometry can already give hints on the usefulness of the CBB cycle on different substrates. We have included a comment on the degree of reduction for substrate (fructose) and product (biomass). Other factors such as utilization of more protein-(in-)efficient pathways can under some circumstances improve growth rate on the cost of biomass yield, or vice versa (Basan et al., Nature, 2015). In our study we did not find evidence for such a growth rate/biomass yield tradeoff.
Another factor influencing biomass yield is the activity of futile cycles or other energy-wasting reactions. The reference from Lu et al., 2017, falls in this category. Their results show that only O2 but no other limitation resulted in higher energy efficiency (H2 consumed per CO2) during lithoautotrophic growth (see also Yu et al., Int J Hyd Energy, 2013, https://doi.org/10.1016/j.ijhydene.2013.04.153). The final biomass yield was unchanged between different limitations (~1 g DCW / g H2). We also note that their reported yield and H2 energy efficiency was highest in non-limited growth, so that O2 limitation only improved yield in relative comparison to other limitations. The authors speculated about changes in energy efficiency being caused by a shift in membrane bound- versus soluble hydrogenase (affecting NADH/ATP ratio), or increased TCA cycle flux oxidizing biomass precursors for energy (p. 155, last paragraph). Unfortunately neither of these hypotheses were tested experimentally.
From our own simulations (Figure 5D), increased flux through the TCA cycle was in fact one of the predicted consequences of forcing CBB activity during growth on fructose. C. necator seems to have no yield or rate benefit from that alone. The idea of an overactive TCA cycle is intriguing, where fixed carbon is reoxidized and NADH used in respiration. However, when we tested the effect of O2 limitation on C. necator cultivated in fructose- and formate-fed turbidostats we did not see increased yield (OD600 nm) or growth rate during oxygen limitation, see Author response image 1. To us, this data would go against the occurrence of wasteful oxidation of fixed carbon in the TCA cycle during normal conditions.

O2-limitation does not increase biomass yield, but reduces growth rate.
C. necator was grown in fructose or formate-fed turbidostats (0.5 and 1.5 g/L substrate, respectively). The oxygen concentration was stepwise reduced by mixing air with molecular nitrogen (N2).
2. The authors state that the deletion of ccbR has no fitness cost for growth on fructose, however in Figure 5F I still observe a minor, but significant fitness cost for growth on fructose, hence I suggest to rephrase the text slightly to better reflect that. This may actually change the conclusion of the paper on the 'useless' role of the CCB on fructose and sucrose.
This comment was addressed in a previous response.
3. The authors should better explain the differences between their findings and Shimizu et al., 2015 paper in Scientific reports.
This comment was addressed in a previous response.
4. Page 16, line 19 and 32: please check if you are not referring to figure 4 instead.
The figure references were corrected. We thank the reviewer for the suggestion.
5. P21 L17 " Another conditionally essential gene on formate was ppc, encoding the PEP-carboxylase (PPC). The reaction has no other annotated (iso-) enzymes and was predicted by RBA to carry most flux on formate compared to the other carbon
sources (Figure 6 B)." It would be interesting to explain the biochemical logic why this anaplerotic reaction would be used more than in other conditions, where anaplerosis is also needed. I guess it has to do with the fact that there is generally lower TCA flux during growth on formate, NADH is generated directly from NADH rather than the TCA cycle. However, anaplerosis is needed in all conditions to generate oxaloacetate etc. So interesting to explain this.
This is an interesting question. In order to answer it, we have closely inspected the fluxes from PEP to oxaloacetate (OA) via PPC and further down to different amino acid biosynthesis routes. On formate, 75% of flux from PEP went to OA while only 12% proceeded to pyruvate. Flux through the TCA was generally low on formate. As the reviewer points out, OA is a hub for amino acid biosynthesis reactions and around 90% of it was transaminated to aspartate, which was used for protein biosynthesis (23%), but also further metabolized to other amino acids (thr, ala, lys, ile, met, asn) and peptidoglycan. However, we also noticed that a large portion of the flux from OA (40%) went to the serine hydroxymethyltransferase and the glycine cleavage system (reactions GHMT3 and GLYAMT), generating glycine and serine by running reverse of the canonical direction. While this is thermodynamically possible as recently demonstrated by engineering the reductive glycine cycle in C. necator (Claassens et al., Met Eng, 2020), it is unlikely to happen without a high concentration of methyl-THF "pushing" these reactions. This error was corrected by constraining the reaction directionality in the model.
With this change, the flux from PEP to OA on formate is lower (45% from PEP to OA, 55% to pyruvate, see updated Figure 6B). PPC nevertheless has a more important role on formate compared to fructose, which could explain its stronger effect on fitness. It carries 70% of the total flux towards the TCA cycle while it only carries 35% on fructose (50% of flux from ED pathway goes to pyruvate and enters the TCA downstream of PPC). In line with this, the ppc knock-out did not only affect growth on formate (fitness = -4.2, table in Supplementary file 2), but also on fructose (fitness = -2.7), while there was no effect on succinate (fitness = -0.1). The fitness associated with ppc is therefore directly correlated to the predicted flux through PPC. We have updated the respective text section and Figure 6B-D with the corrected results.
6. P25 L14 " P25 L14 " Of all central carbon metabolism, the TCA cycle enzymes showed on average lowest abundance, variability and utilization. This is similar to E. coli, where the sole flux capacity demand suggested lower enzyme abundance than what was measured experimentally [Noor et al., 2016]. Only when reverse flux (reactions with low thermodynamic driving force) and l ow enzyme saturation, estimated from metabolite levels was taken in to account, was the calculated enzyme demand similar to the measured levels [Noor et al., 2016]."
This is an important point the authors raise in the discussion, and also makes wonder why the authors did not take into account reverse fluxes in their RBA analysis, is this because the lack of kinetic parameters known, thermodynamic parameters to perform this also for C. necator should be available generally, right? Good to specify this (or repeat that analysis including reverse fluxes, e.g. only based on thermodynamics).
Protein constrained metabolic modeling is a relatively recent development, and only a handful of frameworks exist. Of the four ones we know (ME-model, RBA, EFTL, GECKO), only one -EFTL- currently supports thermodynamic constraints. However, the effort to set up such a model is considerable and the availability of input parameters is limited. In this work we chose RBA for its good documentation, numerical efficiency and semi-automated model generation, although it does not include thermodynamic constraints. Such constraints may be included in future updates.
7. Be clearer about what sugars C. necator can use. On P25 L25 you state " For C. necator, hexose sugars are only one out of many possible substrate classes and the flux" However, it can only accept one hexose sugar (fructose) as is stated before twice:
– " The only sugar that supports growth 27 is fructose, which i s metabolized via the Entner-Doudoroff (ED) pathway [Alagesan et al., 28 2018]."
– " Fructose was chosen as it is the only known sugar (apart from sugar alcohols) that C. necator utilizes [Orita et al., 2012]."
Overall, this is not so clear, Orita et al. do not discuss sugar alcohols and I am not sure which sugar alcohols C. necator can consume (it can consume some sugar acids, but that's something different, at least glycolate). In addition, C. necator does consume N-acetylglucosamine, which is defined as sugar. Maybe just rephrase that C. necator is only known to grow on one hexose sugar, being fructose and not on glucose. And a better reference could be the 2008 review of Cramm, which states " Organic carbon and energy sources for heterotrophic growth include TCA cycle intermediates, sugar acids, fatty acids, amino acids, alcohols, and aromatic compounds, while utilization of sugars is restricted to fructose and N -acetylglucosamine [Johnson and Stanier, 1971; Kersters and De Ley, 1984]"
We agree and have changed the respective paragraphs to make clear that fructose and N-acetylglucosamine are the only consumed sugars in the wild type. C. necator can also grow on glycerol, which is a sugar alcohol albeit a short chain one [Alagesan et al., 2018]. The reference to sugar alcohols was nevertheless removed because it is of no importance for this study.
8. The authors speculate about the evolutionary nature of the two copies of the Calvin cycle operon, as well as their data show the key role of cbbR. Can the authors maybe also mention and discuss the presence of an inactive ccbR copy in the 2nd CCB operon on pHG1, which is now not mentioned, but also indicates the 'recent' and ' unoptimized' acquisition of these CBB operons. Would also be good to know what is the abundance of this inactive cbbR copy from pHG1 in the proteome?
We have added a sentence mentioning the inactive, truncated cbbRP copy on pHG1 to the discussion. The gene of this potential protein is not included in the standard genome annotation of C. necator that we obtained from Uniprot. We did therefore not detect the gene product in our data. It is possible though that identical peptides of the cbbRP protein contribute to the measured abundance of cbbRC if it is expressed.
Reviewer #3 (Recommendations for the authors):
This study dealt with comprehensive analyses regarding protein quantitation and enzyme utilization in the versatile chemolithoautotrophic bacterium C. necator. The experiments and analyses were well considered and designed, and the authors obtained quite interesting information such as presence of many under-utilized enzymes as well as excess amount of utilized enzymes, suggesting the highly robust properties of this bacterium against environmental perturbations.
I have no comments in the experiments, results, and discussion obtained by carbon-limited chemostat cultivation, although I here address a few concerns in the nitrogen-limited cultivation as below, which should be considered by the authors before publication.
1) It has been well known that C. necator H16 synthesizes and accumulates PHB with in the cells under nitrogen-limited conditions. In many cases of PHB-producing cells, the accumulation of intracellular PHB granule reflects on apparent OD owing to the changes in cell size and morphology. In the nitrogen-limited cultivation in this study, I think that the constant OD was a result of steady states of both cell growth and PHB synthesis. Did the authors determine PHB accumulation under the nitrogen-limited condition?
There was indeed a subtle change in optical density over the course of the original cultivation, from higher OD (~0.4) to lower OD (~0.2) when limitation was relieved (Figure 1—figure supplement 1A). Revisiting this result, it suggests a different PHB content at different levels of limitation. As commented before, we redid the nitrogen limitation experiment and determined absolute biomass in g DCW/L and PHB content in g/g DCW (new Figure 1—figure supplement 2).
2) PHB is a water-insoluble polyester, thus it can be escaped from cytosolic equilibrium once after polymerized from water-soluble monomers. Did the RBA model used in this study include PHB synthesis? That is, in the nitrogen-limited condition, did the biomass yield calculated by RBA simulation contain PHB?
Yes, the RBA model contains the pathway to synthesize PHB. For simplification, the chain length of the PHB polymer is not taken into account, so that the molar PHB concentration is identical to the molar concentration of the 3-hydroxybutyrate-CoA monomer. PHB is then secreted via an exchange reaction and does not count as biomass. In our previous version of the RBA model, we had included a constant growth-rate dependent flux towards PHB in order to match the simulated biomass yield with experimentally determined biomass yield. In our revised manuscript, this inaccurate behavior was replaced with a more realistic simulation of PHB production based on the new measurements of biomass and PHB concentration. The flux towards PHB during nitrogen limitation is now explicitly included in the model.
3) The authors concluded that reassimilation of CO2 by CBB pathway does not provide a fitness benefit for heterotrophic growth. I agreed with this conclusion, because it is feasible that energy – and reducing equivalents-consuming CBB cycle is not required when carbons other than CO2 are available. While, I read the previous report by Shimizu et al. They demonstrated advantage of CBB cycle under heterotrophic growth in synthesis of the storage polyester unassociated with cell growth, and never proposed advantage in the heterotrophic growth. It is not adequate to discuss about this matter by applying the results in the current study focusing on growth to the previous study focusing on growth-unassociated PHB synthesis. I recommend the authors to correct/modify the relevant descriptions in order to avoid readers' misunderstanding.
We agree with the reviewer. Our previous discussion of the findings from Shimizu et al. could make the impression that the authors claimed a general yield or fitness benefit of the CBB cycle on fructose. The picture is more nuanced in that the study from Shimizu et al. found an increase in PHB yield (+20.6% compared to a Rubisco knockout strain), without commenting on total biomass yield. The authors showed that during growth on sugar, the emitted and reassimilated CO2 was preferentially incorporated into PHB, and likely caused an increase in PHB-synthesis. Here, we find that fixation of emitted CO2 does not improve biomass yield on sugar.
Our results suggest that any additional activity of the CBB cycle during metabolism of fructose is a wasteful process, regardless if the carbon is invested into biomass or PHB. It is possible that the increased PHB yield that the authors reported when Rubisco is present is achieved on the cost of the (PHB-subtracted) biomass yield. However, our results did not allow us to draw conclusions on the effect of heterotrophic CBB activity on PHB or biomass yield. We deem the resolution of the transposon library experiments too low to decide if loss of CBB activity (cbbR knock-out) confers a fitness advantage or not. We have therefore toned down our claims regarding the fitness benefit of CBB cycle and corrected our interpretation of the results from Shimizu et al. as suggested by the reviewers (see Results section 5, and discussion).
[Editors' note: further revisions were suggested prior to acceptance, as described below.]
I very much appreciate the effort you have taken to carefully address the previous reviews. There are just a few more points outlined below that should be considered in the final version for eLife.
Reviewer #2 (Recommendations for the authors):
The authors did a very good job in revising the manuscript and adding additional experiments and modelling on dry weight and PHB production.
The only answer that puzzled us was the answer on PEP carboxylase, we were puzzled by the explanation that oxaloacetate flux goes to glycine via reserve GCV operation (does that mean that this goes via threonine).
Furthermore, they satisfied all our queries well, so I recommend publication.
In the current manuscript the flux towards serine and glycine is taking the expected route, that means the canonical pathway starting from the glycolysis intermediate 3-phosphoglycerate (3PG) to 3-phosphohydroxypyruvate, O-phosphoserine, serine, and glycine + methyl-THF. The methyl-THF is ligated with CO2 by the glycine cleavage system to form one additional glycine.
Before we corrected this error (in our original submission), flux towards serine was taking the route from oxaloacetate to aspartate, aspartate semialdehyde, homoserine, phospho-homoserine, threonine, 2-amino-3-oxobutanoate, and finally glycine + acetyl-CoA. Serine would then be synthesized from 2 molecules glycine (one of it converted to methyl-THF via the glycine cleavage system). While probably possible, we deem this route unlikely to have flux in reality because it is much longer, and requires several enzymatic steps where the enzyme abundance and thermodynamic driving force might not be sufficient.
Reviewer #3 (Recommendations for the authors):
I respect the authors' great effort in re-examination of bioreactor experiment and replacement of metabolic modeling regarding the N-limitation condition. I agree with the most revisions so think that the revised manuscript is valuable for publication by eLife, but I have still a few concerns as below, which are expected to be considered by the authors prior to the publication.
1) Were the g-DCW values used for calculation of m-protein (g/g-DCW) in Figure 4, Y (gDCW/g-S) in Figure 5C (and so on) under the N-limitation condition the cell mass excluding PHB (PHB-subtracted biomass)? The m-protein graph under the N-limitation in Figure 4 in the revised manuscript looks like to be the same as the previous version.
The protein mass in g/gDCW was calculated by multiplying the relative protein mass fraction (%) with the average total protein content of 0.68 g / gDCW (Park et al., BMC, Sys Bio, 2011), with DCW being dry cell weight without PHB. The graph in Figure 4 looks similar to the previous version because the protein quantification is relative and only depends on the estimated protein content per DCW, which did not change. The experimentally measured gDCW was only used to determine the substrate uptake rate but does not influence the proteomics results.
2) The rate of intracellular PHB synthesis usually becomes maximum when the cell growth has been stopped by nitrogen depletion, so the increase in PHB production by the function of CBB cycle during the heterotrophic condition on fructose, reported by Shimizu et al., may become significant during the growth-unassociated PHB synthesis. The authors carried re-simulation to estimate the advantage of the heterotrophic CBB cycle (lines 373-385 in the revised manuscript), but this re-simulation was done still under growth conditions. Is it possible to simulate the growth-unassociated PHB synthesis on fructose under N-depletion? Fructose-uptake by C. necator was supposed to be weakened after the growth phase due to marked down-regulation of expression of EM and ED pathway genes (reported in previous transcriptome analyses). Considering this, I'm not sure whether the constant fructose uptake rate of 4.0 mmol/gDCW/h was adequate or not for the growth-unassociated PHB synthesis.
The reviewer wonders if PHB production can be modeled when growth has stopped completely. This is an interesting question, but it would require a different modeling framework. The RBA model that we used here assumes steady state growth (which we achieved with chemostat bioreactors) and currently only supports maximizing growth as objective. The flux towards PHB was forced by constraining the respective reactions. A model with the ability to predict flux towards PHB without forcing it would require a dynamic (time-dependent) setup. The objective would need to be maximizing final biomass, not growth rate, for simulation of a growth experiment. Such a model would then incentivize storing additional carbon as PHB during nitrogen limitation, because this strategy would generate more biomass as soon as nitrogen becomes available again.
Regarding the constant uptake rate of 4 mmol fructose h-1 gDCW-1 (Figure 4 A-D), we have also tested different nitrogen/fructose uptake rates in combination with different CO2 refixation rates in the RBA model but the results were similar, see Author response image 2. Here, we simulated a stepwise reduction of the nitrogen concentration which reduces growth and triggers PHB formation. The model simulations suggest that any additional CBB activity reduces growth rate and biomass yield even further, and has no advantage for PHB production. The reason is that – from a purely stoichiometric perspective – the ratio of CO2 re-fixation to fructose uptake rate determines the energetic efficiency, which decreases with increasing flux through the CBB cycle. In reality, an increase in PHB yield is nevertheless possible on the cost of the non-PHB biomass yield.
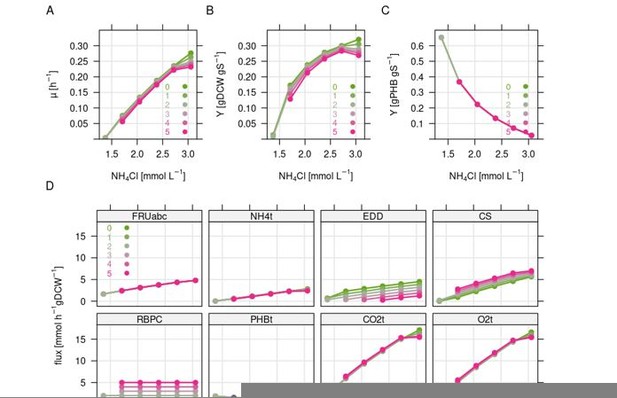
Simulation of the combined effect of nitrogen limitation and CO2 refixation via the CBB cycle.
Simulations were carried out using the resource balance analysis (RBA) model for Ralstonia eutropha as described previously. Nitrogen limitation was simulated by stepwise reduction of the nitrogen concentration in the medium, which triggers (forced) PHB formation. Colors indicate the amount of flux forced through the Calvin cycle from 0 to 5 mmol gDCW-1 h-1. Note that not all simulations were feasible due to non-positive growth rate. (A) Growth rate. (B) Biomass yield per g fructose. (C) PHB yield in g per g fructose. (D) Flux in mmol gDCW-1 h-1 for selected reactions. FRUabc, fructose transporter; NH4t, ammonium transporter; EDD, 6PG-hydratase representing ED flux; CS, citrate synthase representing TCA cycle flux; RBPC, Rubisco, representing CBB cycle flux; PHBt, PHB pseudo transport reaction; CO2t, CO2 transporter; O2t, O2 transporter.





