Reduced antibody cross-reactivity following infection with B.1.1.7 than with parental SARS-CoV-2 strains
Figures
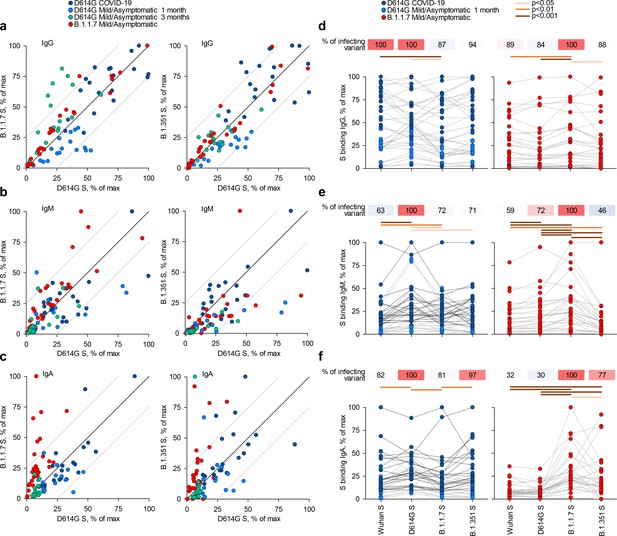
Recognition of distinct severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike glycoproteins by antibodies in D614G and B.1.1.7 sera.
(a-c) Correlation of IgG (a), IgM (b), and IgA (c) antibody levels to D614G and B.1.1.7 or B.1.351 spikes in the indicated groups of donors infected either with the D614G or B.1.1.7 strains. Each symbol represents an individual sample and levels are expressed as a percentage of the positive control. Black lines denote complete correlation and grey lines a 25% change in either direction. (d-f) Comparison of IgG (d), IgM (e), and IgA (f) antibody levels to the indicated spikes in groups of donors acutely infected either with the D614G or B.1.1.7 strains. Connected symbols represent individual donors. Numbers above the plots denote the average binding to each spike, expressed as a percentage of binding to the infecting spike.
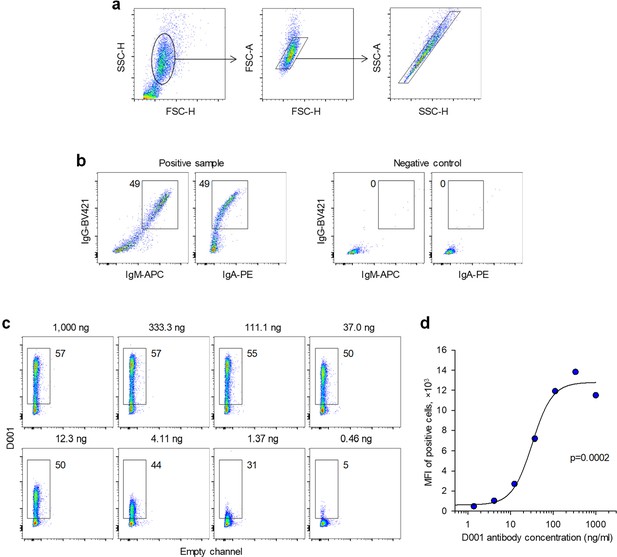
Flow cytometric detection of spike-binding antibodies.
HEK293T cells were transfected with expression plasmids encoding each severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variant spike and were used for flow cytometric analysis 2 days later. (a) Gating of HEK293T cells and of single cells in these mixed cell suspensions. (b) Example of IgG, IgM, and IgA staining in a positive sample and a negative control. Numbers within the plots denote the percentage of positive cells. (c) Staining of HEK293T cells transfected to express the Wuhan spike, with titrated amounts of the S2-specific D001 monoclonal antibody. Numbers above the plots denote the final D001 antibody concentration. (d) Median fluorescence intensity (MFI) of stained cells in c, according to the D001 antibody concentration.
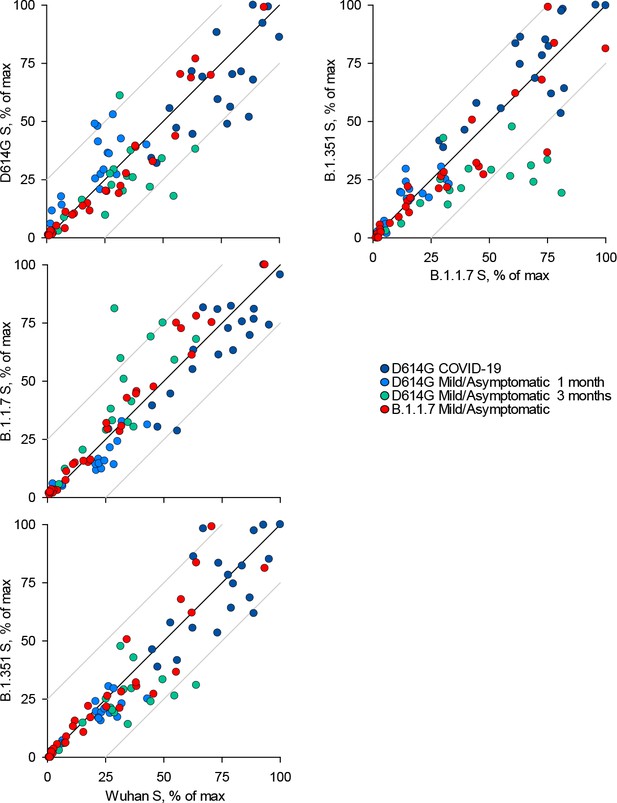
Recognition of distinct severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike glycoproteins by antibodies in D614G and B.1.1.7 sera.
Correlation of IgG antibody levels to Wuhan, D614G, B.1.1.7, and B.1.351 spikes in the indicated groups of donors infected either with the D614G or B.1.1.7 strains. Each symbol represents an individual sample and levels are expressed as a percentage of the positive control. Black lines denote complete correlation and grey lines a 25% change in either direction.
Recognition of distinct severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike glycoproteins by antibodies in D614G and B.1.1.7 sera.
Correlation of IgM antibody levels to Wuhan, D614G, B.1.1.7, and B.1.351 spikes in the indicated groups of donors infected either with the D614G or B.1.1.7 strains. Each symbol represents an individual sample and levels are expressed as a percentage of the positive control. Black lines denote complete correlation and grey lines a 25% change in either direction.
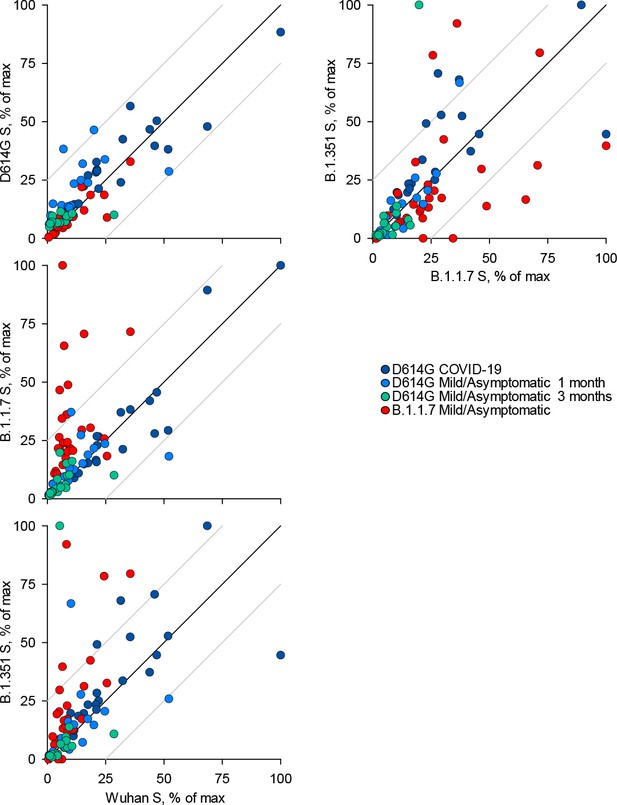
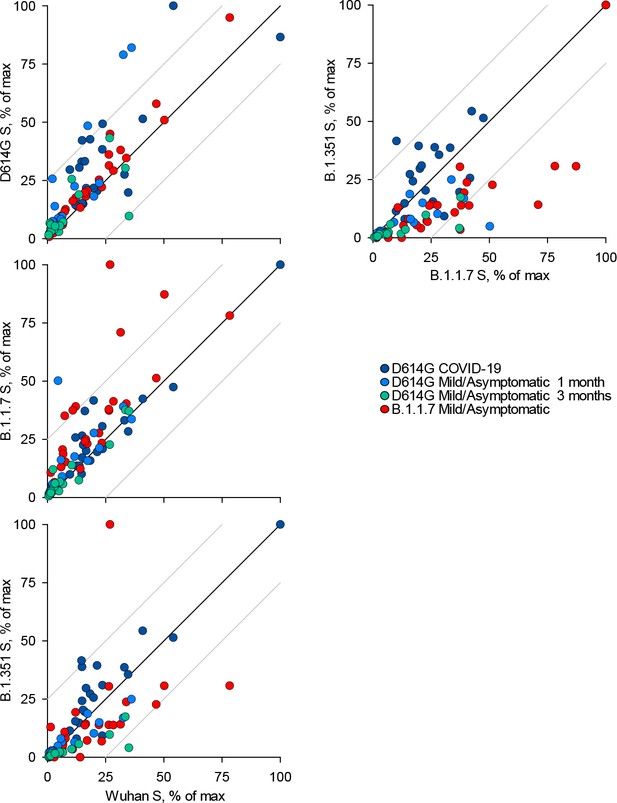
Recognition of distinct severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike glycoproteins by antibodies in D614G and B.1.1.7 sera.
Correlation of IgA antibody levels to Wuhan, D614G, B.1.1.7, and B.1.351 spikes in the indicated groups of donors infected either with the D614G or B.1.1.7 strains. Each symbol represents an individual sample and levels are expressed as a percentage of the positive control. Black lines denote complete correlation and grey lines a 25% change in either direction.
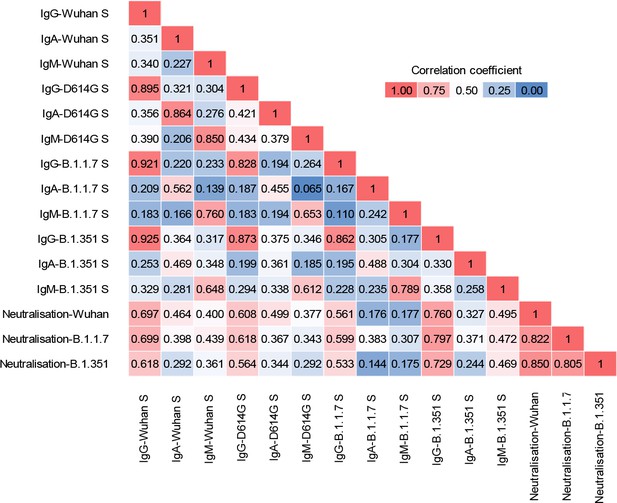
Matrix of correlation coefficients between binding and neutralising antibodies.
Levels of binding IgG, IgM, and IgA antibodies to the indicated spikes and levels of neutralising antibodies to the indicated strains were correlated using all the samples described in this work (n=83).
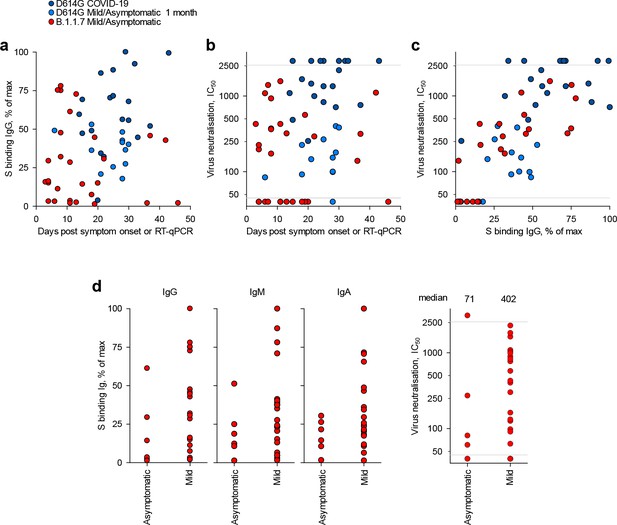
Kinetics and magnitude of the antibody response to D614G and B.1.1.7 infection.
(a) Levels of IgG antibodies to the spike of the infecting strain in sera from donors infected with the D614G or B.1.1.7 strains, over time since onset of symptoms (for symptomatic cases) or the first positive RT-qPCR diagnosis (for asymptomatic cases). Levels are expressed as a percentage of the positive control. (b) Neutralising antibody levels (IC50) against the closest infecting strain (Wuhan for D614G infection and B.1.1.7 for B.1.1.7 infection) in sera from donors infected with the D614G or B.1.1.7 strains, over time since onset of symptoms or since the first positive RT-qPCR diagnosis. (c) Correlation of binding IgG and neutralising antibody levels from a and b, respectively. (d) Comparison of binding IgG, IgM, and IgA antibody levels and of neutralising antibody levels (IC50) between B.1.1.7-infected asymptomatic donors and those with mild COVID-19 symptoms. Antibody binding and virus neutralisation were tested against the homologous B.1.1.7 spike and virus, respectively. Differences between the two groups were not statistically significant. Grey horizontal lines denote the lower and upper limit of detection. In (a-d), each symbol represents an individual sample.
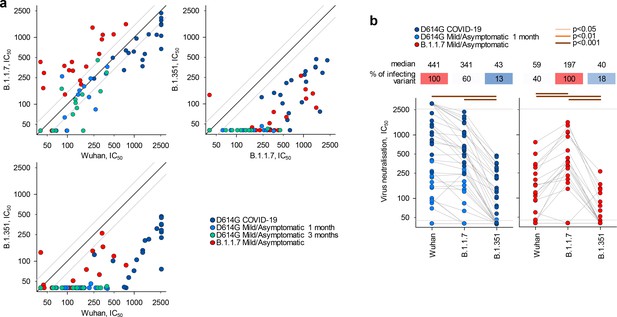
Neutralisation of distinct severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) strains by antibodies in D614G and B.1.1.7 sera.
(a) Correlation of neutralising antibody levels (IC50) against the Wuhan, B.1.1.7, or B.1.351 strains in the indicated groups of donors infected either with the D614G or B.1.1.7 strains. Each symbol represents an individual sample. Black lines denote complete correlation and grey lines a 50% (twofold) change in either direction. (b) Comparison of neutralising antibody levels (IC50) to the indicated SARS-CoV-2 strains in groups of donors acutely infected with either the D614G or B.1.1.7 strains. Connected symbols represent individual donors. Numbers above the plots denote the average IC50 against each strain, expressed as a percentage of IC50 against the infecting strain. Grey horizontal lines denote the lower and upper limit of detection.
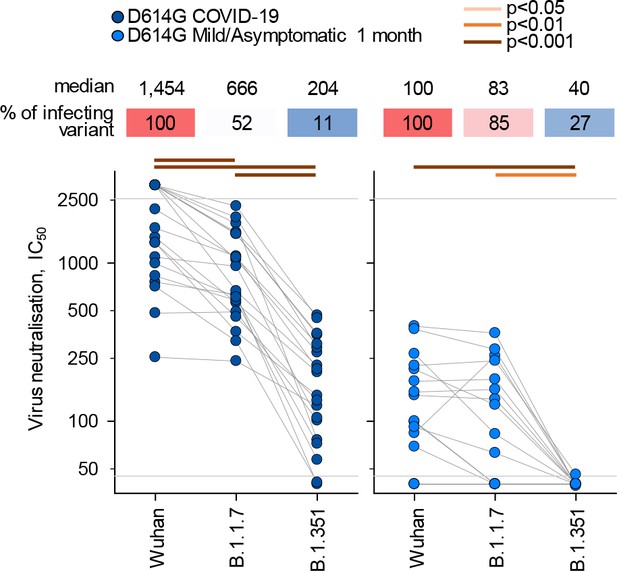
Neutralisation of distinct severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) strains by antibodies in D614G sera, according to severity of infection.
Comparison of neutralising antibody levels (IC50) to the indicated SARS-CoV-2 strains in donors acutely infected with the D614G strain, grouped according to the severity of the outcome. Connected symbols represent individual donors. Numbers above the plots denote the average IC50 against each strain, expressed as a percentage of IC50 against the infecting strain. Grey horizontal lines denote the lower and upper limit of detection.
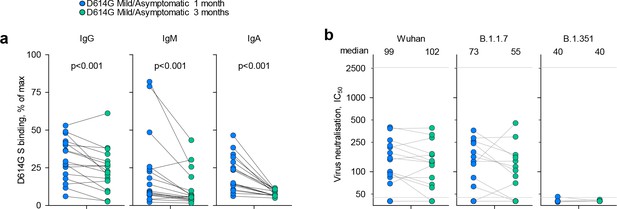
Binding and neutralising antibodies at a 3-month follow-up of mild/asymptomatic D614G infection.
(a) Levels of IgG, IgM, and IgA antibodies (expressed as a percentage of the positive control) to the D614G spike in sera from D614G-infected donors at 1 and 3 months post infection. (b) Neutralising antibody levels (IC50) against the Wuhan, B.1.1.7, or B.1.351 strains in same donors described in a. In a and b, connected symbols represent individual donors.
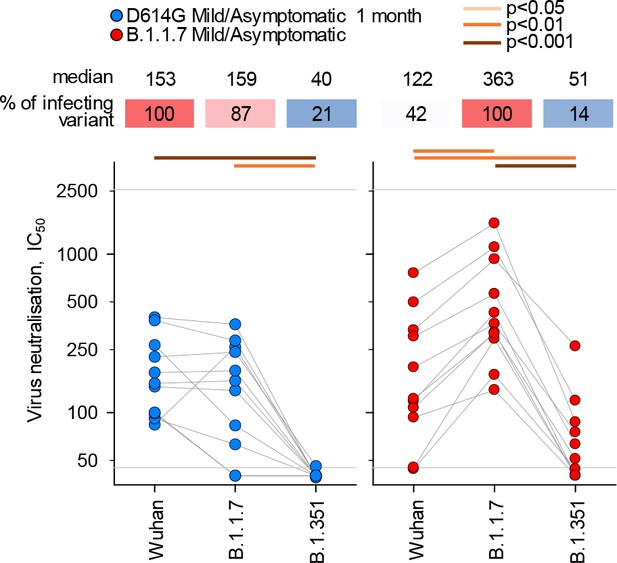
Neutralisation of distinct severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) strains by antibodies in D614G and B.1.1.7 sera from mild/asymptomatic infection.
Comparison of neutralising antibody levels (IC50) to the indicated SARS-CoV-2 strains in subgroups of donors acutely infected with either the D614G (n=11) or B.1.1.7 (n=11) strains, selected for comparable disease outcome and time since infection. Connected symbols represent individual donors. Numbers above the plots denote the average IC50 against each strain, expressed as a percentage of IC50 against the infecting strain. Grey horizontal lines denote the lower and upper limit of detection.
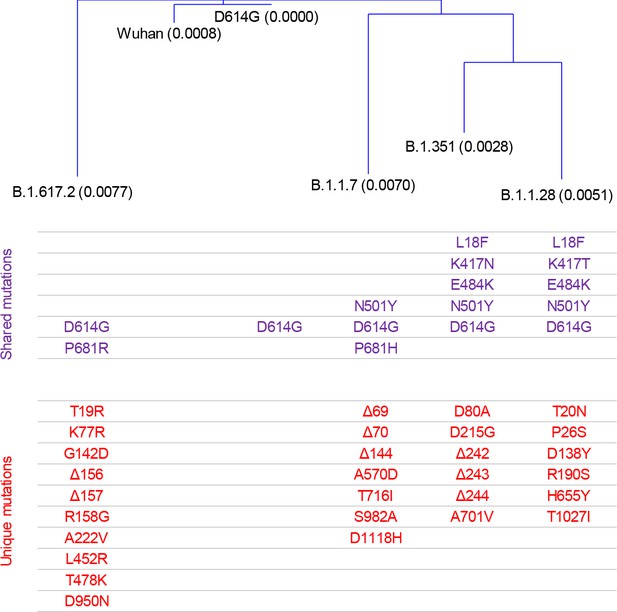
Spike sequence distance of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants.
Distance was calculated based on the sequence alignment of the full-length spike amino acid sequences of the indicated SARS-CoV-2 variants. Mutations of amino acid residues that are shared by at least two strains or are unique to specific strains are indicated in different colours. Mutations were considered shared if they affected the same amino acid position even if the change was not identical.
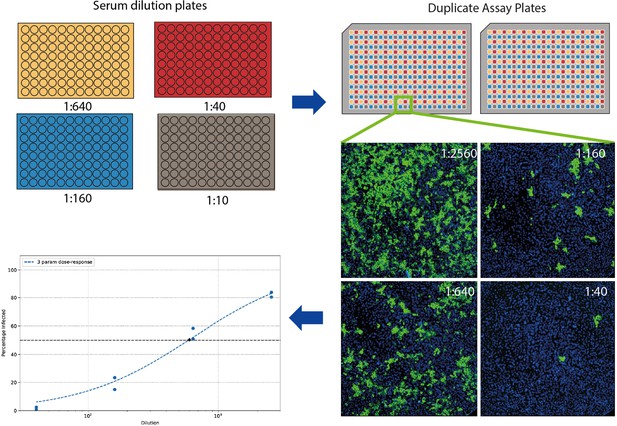
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) neutralisation assay setup.
The 96-well racks of serum samples including controls are serially diluted after an initial dilution of 1:10 to generate four total dilution plates. These are used to treat pre-seeded Vero E6 cells in 384-well assay plates in duplicate before infection with SARS-CoV-2 virus. After immunostaining with DAPI and a 488-conjugated monoclonal antibody against SARS-CoV-2 nucleoprotein, each well is imaged and infection area per area of cells calculated, followed by automated curve fitting and identification of serum dilution factor to achieve 50% neutralisation (IC50).
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | BV421 anti-human IgG (monoclonal) | Biolegend | RRID:AB_2562176; Cat# 409318 | FACS (1:200) |
| Antibody | APC anti-human IgM (monoclonal) | Biolegend | RRID:AB_493011; Cat# 314510 | FACS (1:200) |
| Antibody | PE anti-human IgA (monoclonal) | Miltenyi Biotech | RRID:AB_2733860; Cat# 130-114-002 | FACS (1:200) |
| Antibody | Anti-SARS-CoV-2 S2 clone D001 (monoclonal) | SinoBiological | RRID:AB_2857932; Cat# 40590-D001 | FACS |
| Antibody | Alexa488 anti-SARS-CoV-2 nucleoprotein (monoclonal) | Produced in-house | CR3009 | IF |
| Recombinant DNA reagent | pcDNA3-SARS-CoV-2_WT spike | Dr Massimo Pizzato, University of Trento, Italy | Wuhan spike sequence | Transfected construct |
| Recombinant DNA reagent | pcDNA3-SARS-CoV-2_D614G spike | Dr Massimo Pizzato, University of Trento, Italy | Wuhan spike sequence with D614G mutation and cytoplasmic tail deletion | Transfected construct |
| Recombinant DNA reagent | pcDNA3-SARS-CoV-2_B.1.1.7 spike | This paper | B.1.1.7 spike sequence | Transfected construct |
| Recombinant DNA reagent | pcDNA3-SARS-CoV-2_ B.1.351 spike | This paper | B.1.351 spike sequence | Transfected construct |
| Cell line (Homo sapiens) | HEK293T | Cell Services facility at the Francis Crick Institute | RRID:CVCL_0063; CVCL_0063 | |
| Cell line (Chlorocebus sp.) | Vero E6 | Dr Björn Meyer, Institut Pasteur, Paris, France | CRL-1586 | |
| Cell line (Chlorocebus sp.) | Vero V1 | Prof. Steve Goodbourn, St. George’s, University of London, London, UK | CCL-81 | |
| Other | SARS-CoV-2 | hCoV-19/England/02/2020 | Respiratory Virus Unit, Public Health England, UK | Wuhan strain |
| Other | SARS-CoV-2 | hCoV-19/England/204690005/2020 | Public Health England (PHE), UK, through Prof. Wendy Barclay, Imperial College London, London, UK | B.1.1.7 strain |
| Other | SARS-CoV-2 | 501Y.V2.HV001 Cele et al., 2021 | B.1.351 strain |
Additional files
-
Source data 1
Binding and neutralising titre values.
- https://cdn.elifesciences.org/articles/69317/elife-69317-data1-v2.xlsx
-
Supplementary file 1
Donor and patient characteristics.
This table lists the number, median age (and range), gender proportion, and the median time (and range) post infection for the donors and patients studied.
- https://cdn.elifesciences.org/articles/69317/elife-69317-supp1-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/69317/elife-69317-transrepform-v2.pdf





