Single-molecule imaging of chromatin remodelers reveals role of ATPase in promoting fast kinetics of target search and dissociation from chromatin
Figures
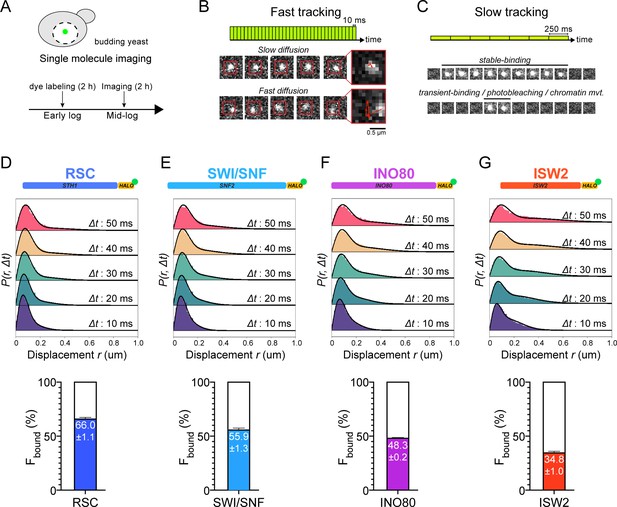
Chromatin-binding and chromatin-free fractions of RSC, SWI/SNF, INO80, and ISW2.
(A) Experimental scheme. (B) Fast-tracking imaging regime uses short exposures (10 ms) at high laser power to distinguish slow (chromatin-bound) and fast (chromatin-free) diffusing populations. (C) Slow-tracking regime directly observes the dwell times of chromatin-bound molecules using 250 ms exposures at low laser power. (D–G) (Top) Raw displacement histograms over the first 5 time frames (Δt: 10, 20, 30, 40, 50 ms). A two-state kinetic model was used for fitting the CDF [black lines] in Spot-On. (Bottom) Spot-On kinetic modeling results based on displacement distribution histograms for Sth1-Halo (D), Snf2-Halo (E), Ino80-Halo (F), and Isw2-Halo (G). Solid colored bar with indicated value represents % chromatin-bound molecules; open bar represents % chromatin-free. Error bars are standard deviations from 2 [or 3 for ISW2] biological replicates.
-
Figure 1—source data 1
MSD-based kinetic analysis.
- https://cdn.elifesciences.org/articles/69387/elife-69387-fig1-data1-v2.xlsx
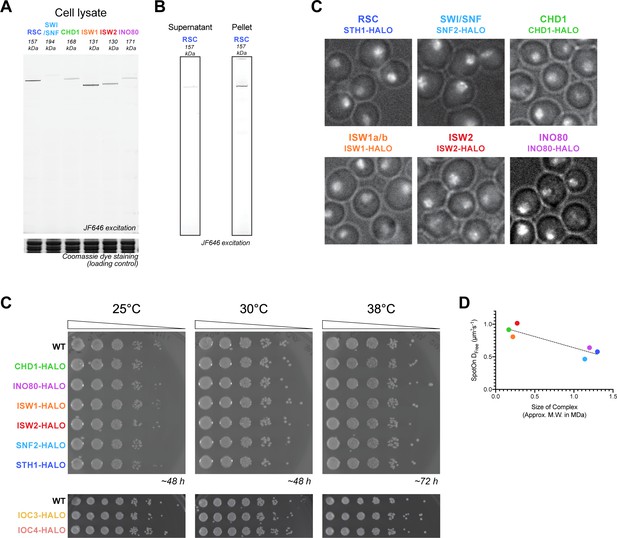
Cell growth, integrity, and localization of HaloTagged remodeler subunits.
(A) SDS-PAGE gel scanned for JF646 dye fluorescence (top) and imaged after Coomassie staining (bottom). Cell lysates are prepared by NaOH protein extraction method (Amberg et al., 2006) after treatment with JF646 at a saturating dye concentration (20 nM) for 2 hr at 30°C. (B) JF646 dye fluorescence in the soluble supernatant (as shown in A) and insoluble pellet for Sth1-Halo strain. Substantial Sth1-Halo protein is retained the insoluble pellet. (C) Overlay of Phase Contrast image and initial nuclear fluorescence captured by JF552 dye excitation of yeast stained with JF552. (D) Fivefold dilutions of HaloTag fusion and wildtype strains are plated on YPAD plates at the indicated temperatures for 2–3 days. (E) Relationship between Dfree values determined by Spot-On analysis and the calculated molecular weights of chromatin remodeling complexes.
-
Figure 1—figure supplement 1—source data 1
Original gel images for Figure 1—figure supplement 1A.
- https://cdn.elifesciences.org/articles/69387/elife-69387-fig1-figsupp1-data1-v2.zip
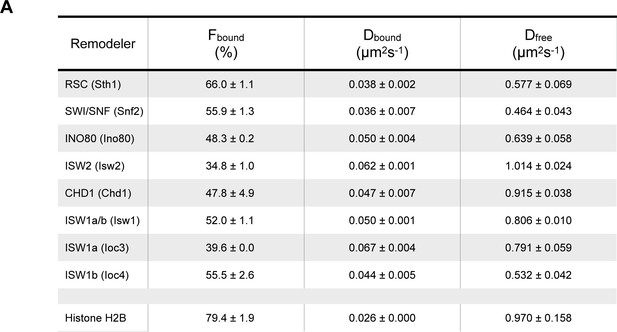
Spot-On kinetic modeling analysis.
(A) Kinetic parameters (Fbound, Dbound, Dfree) determined by Spot-On analysis. Errors represent standard deviation between 2 [or three for ISW2] biological replicates.
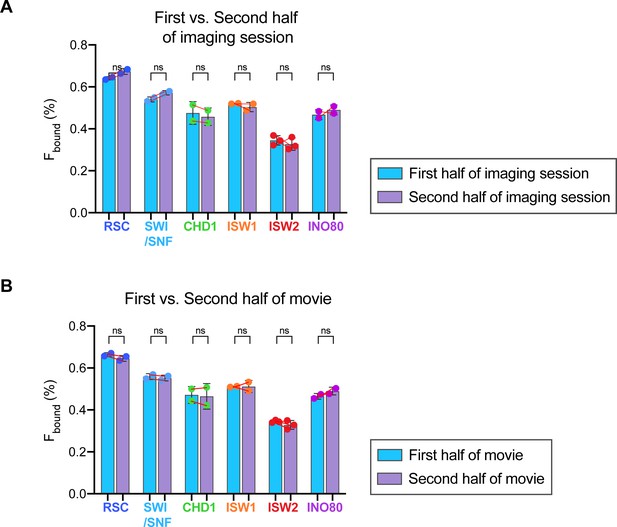
Yeast culture during imaging and laser illumination do not have obvious effects on remodeler diffusion.
(A) Spot-On analyses were performed on movies acquired either in the ‘first half’ or ‘second half’ of an imaging session to compare the Fbound values between the two groups. (B) Spot-On analyses were performed on the ‘first half’ [1–2500 frames] and ‘second half’ [2501–5000 frames] of each movie ‘substack’ to compare the Fbound values between the two groups. Circles represent individual biological replicates with red lines indicating before-after changes for each replicate. Errors represent standard deviation between biological replicates. Comparison between ‘first half’ and ‘second half’ groups for each remodeler by ordinary one-way ANOVA test (ns: not significant).
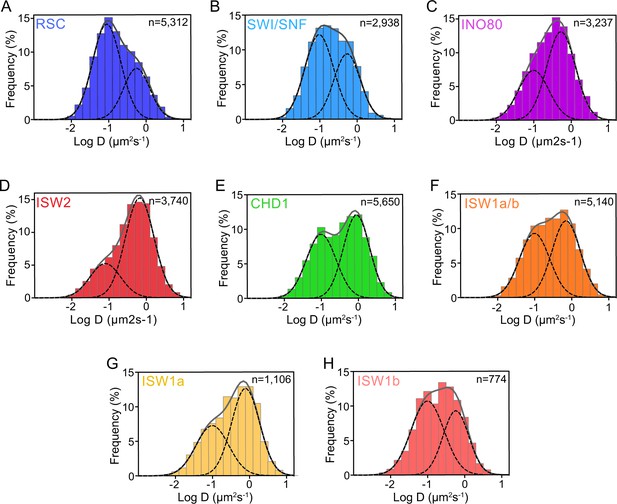
Log10D histograms for six remodelers and Gaussian fitting.
(A–H) Normalized histograms of log10 diffusion coefficients of single-molecule trajectories fitted to two Gaussian distribution functions (solid gray line: sum of two Gaussians; dashed lines: individual Gaussian curves representing chromatin-bound and chromatin-free populations) for Sth1-Halo (A), Snf2-Halo (B), Ino80-Halo (C), and Isw2-Halo (D), Chd1-Halo (E), and Isw1-Halo (F), Ioc3-Halo (G), and Ioc4-Halo (H). The two components are not well-resolved.
Fast-tracking movie (10 ms exposure/frame) for Sth1-Halo strain.
Related to Figure 1. Live-cell, single-molecule imaging of Sth1-Halo yeast cells stained with JF552. Movies with 10 ms exposure/frame were acquired with continuous 555 nm laser irradiation. Timestamp and scale bar (1 μm) are shown in the movie. White circles represent single nuclei. Last frame is an overlay of DIC image and initial nuclear fluorescence. Pixel: 107 nm.
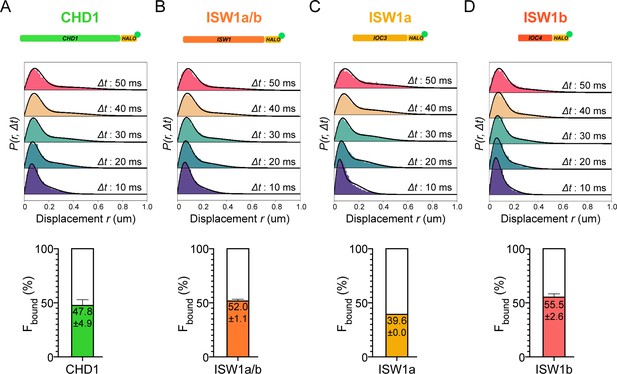
Chromatin-binding and chromatin-free populations of CHD1 and ISW1.
(A–B) Spot-On analysis as described in Figure 1 for the catalytic subunits Chd1-Halo (A) and Isw1-Halo (B). (C–D) Spot-On analysis of the accessory subunits of ISW1a and ISW1b complexes: Ioc3-Halo (C) and Ioc4-Halo (D).
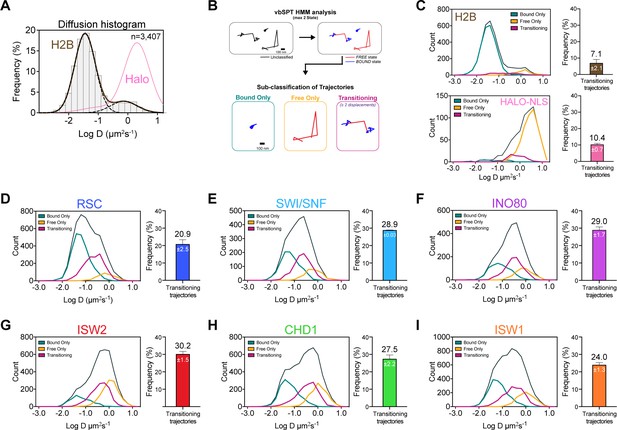
Remodelers undergo frequent transitions between bound and free states.
(A) Halo-H2B (brown) and Halo-NLS (pink) molecules display well-separated peaks in their diffusion coefficient histograms. (B) An overview of displacement-based HMM classification (vbSPT) to identify transitioning trajectories. After classifying each displacement as either in bound or free state, each trajectory is sub-classified as ‘bound only’, ‘free only’, or ‘transitioning’. (C–I) Left: Overlay of raw histograms of log10 diffusion coefficients for ‘Bsound only’ (turquoise), ‘Free only’ (yellow), ‘Transitioning’ (purple), and total trajectories (thin black). Right: Quantification (%) of transitioning trajectories in the diffusion coefficient histogram, where errors represent standard deviation between two [or three for ISW2] biological replicates. (C) Transitioning trajectories for Halo-H2B (top) and Halo-NLS (bottom). (D–I) Transitioning trajectories for remodelers: Sth1-Halo (D), Snf2-Halo (E), Ino80-Halo (F), and Isw2-Halo (G), Chd1-Halo (H), and Isw1-Halo (I).
-
Figure 3—source data 1
vbSPT analysis.
- https://cdn.elifesciences.org/articles/69387/elife-69387-fig3-data1-v2.xlsx
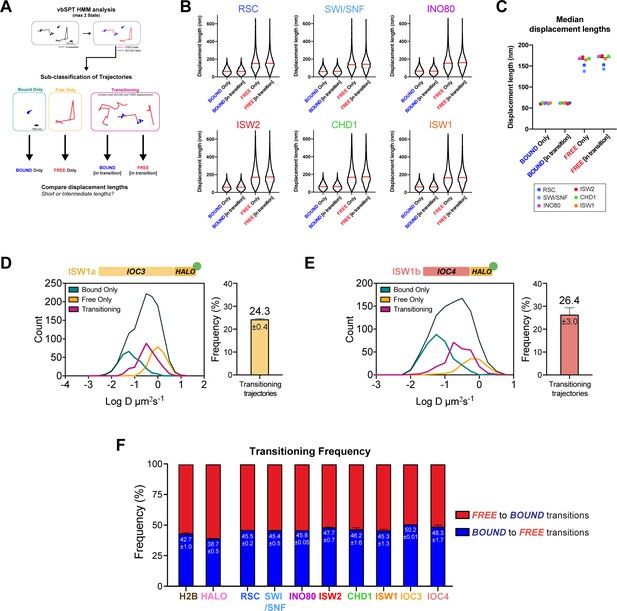
Validation of two diffusive states classified by vbSPT, and quantification of transitioning frequencies.
(A) Pipeline for classification and sub-classification of trajectories using vbSPT. Individual displacement lengths were determined for ‘bound only’, ‘free only’, and ‘transitioning’-subclassified trajectories to validate the two states and state transitions. (B–C) Violin plot (B) and median value (C) of individual displacement lengths for the Bound and Free states in non-transitioning and transitioning trajectories. For violin plots, thick red and dotted gray lines represent the median and two quartiles, respectively. (D–E) Transitioning trajectories for Ioc3-Halo (D) and Ioc4-Halo (E). Left: Overlay of raw histograms of log10 diffusion coefficients for ‘Bound only’ (turquoise), ‘Free only’ (yellow), ‘Transitioning’ (purple), and total trajectories (thin black). Right: Quantification (%) of transitioning trajectories in the diffusion coefficient histogram. (F) For all classified transitioning trajectories, ‘FREE’ to ‘BOUND’ and ‘BOUND’ to ‘FREE’ transition frequencies are indicated. For D–F, errors represent standard deviation between two [or three for ISW2] biological replicates.
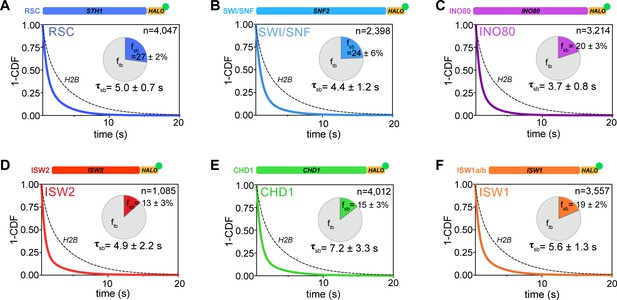
All remodelers have short-lived stable-binding residence times of 4–7 s.
(A–F) Fitted double exponential decay curves from 1-CDF plots of observed dwell times from individual binding events (n) imaged by slow-tracking, for Sth1-Halo (A) Snf2-Halo (B), Ino80-Halo (C), and Isw2-Halo (D), Chd1-Halo (E), and Isw1-Halo (F). Solid colored and dashed black fitted curves for indicated remodelers and H2B, respectively. Pie charts show the percentage (fsb) and average residence time (τsb) of the stable binding population after photobleaching correction. Errors represent bootstrap resampling errors after resampling 100 times (sb: stable-binding; tb: transient-binding).
-
Figure 4—source data 1
Kinetic parameters determined by slow-tracking.
- https://cdn.elifesciences.org/articles/69387/elife-69387-fig4-data1-v2.xlsx
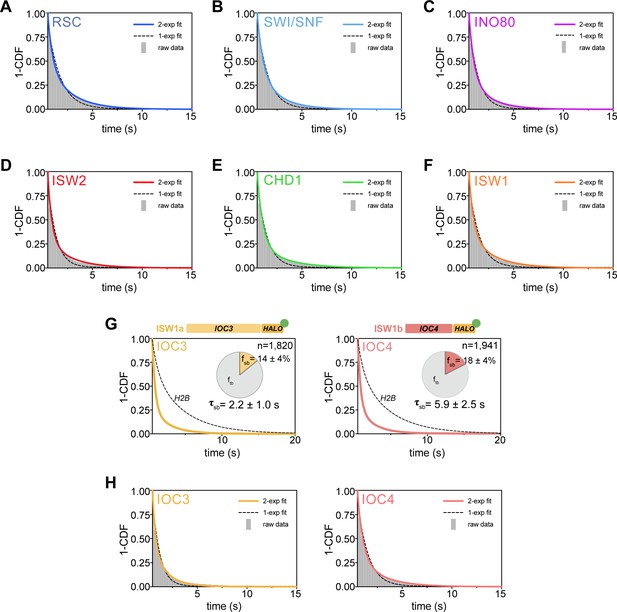
Survival plots [1-CDF] of dwell times showing 1- vs 2-component exponential decay fits.
(A–F) 1- and 2-component exponential decay fits to survival plots of dwell times for Sth1-Halo (A), Snf2-Halo (B), Ino80-Halo (C), and Isw2-Halo (D), Chd1-Halo (E), and Isw1-Halo (F). (G) 1-CDF plot, pie chart as in Figure 4, and residence times of Ioc3-Halo (Left) and Ioc4-Halo (Right). (H) One- and two-component exponential decay fits to survival plots of dwell times for Ioc3-Halo (Left) and Ioc4-Halo (Right).
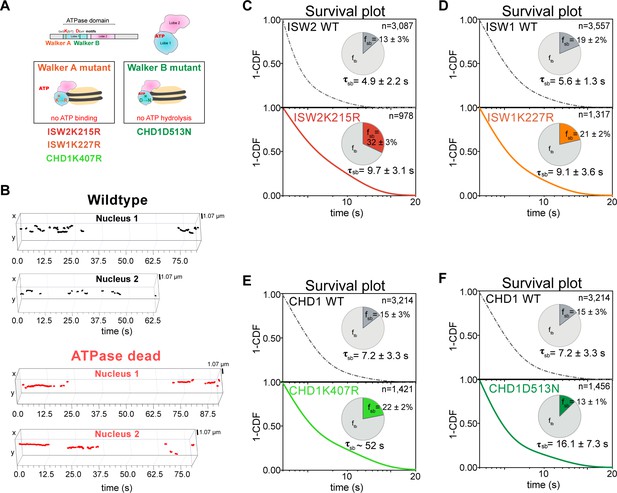
ATP hydrolysis is responsible for rapid chromatin dissociation.
(A) Bar diagram and cartoons for remodelers mutated in the ‘Walker A’ and ‘Walker B’ motifs, respectively. (B) Representative 3D plots of trajectories imaged by slow-tracking for wildtype (Chd1-Halo, black) and ATPase-dead mutant (Chd1K407R-Halo, red). Each plot shows all trajectories (≥ three frames) from single nucleus where lines represent apparent durations of chromatin-binding events. (C–F) 1-CDF plot, pie chart, and residence times of wild-type (top) and ATPase-dead mutants (bottom) for Isw2 (C), Isw1 (D), and Chd1 (E,F).
-
Figure 5—source data 1
Slow-tracking for ATPase-dead mutants.
- https://cdn.elifesciences.org/articles/69387/elife-69387-fig5-data1-v2.xlsx
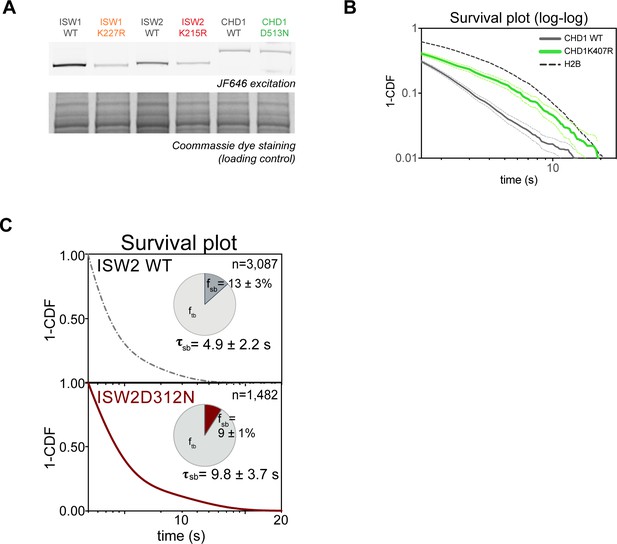
Expression levels and 1-CDF plots for wildtype and mutant ATPase-dead Isw2D312N.
(A) SDS-PAGE analysis; gel scanned for JF646 dye fluorescence (top) and imaged after Coomassie staining (bottom). Cell lysates of ATPase mutants (Isw1K227R-Halo, Isw2K215R-Halo and Chd1D513N-Halo) and their wild-type strains were prepared after treatment with JF646 at a saturating dye concentration (20 nM) for 2 hr at 30°C. (B–E) 1-CDF plot in log-log scale for Isw2K215R (B), Isw1K227R (C), Chd1K407R (D), and Chd1D513N (E) compared to wildtype. Colored dashed lines represent 95% confidence interval.
-
Figure 5—figure supplement 1—source data 1
Original gel images for Figure 5—figure supplement 1A.
- https://cdn.elifesciences.org/articles/69387/elife-69387-fig5-figsupp1-data1-v2.zip
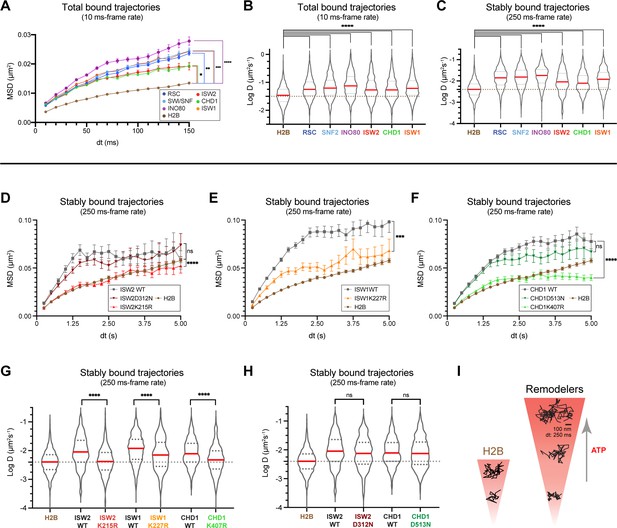
ATP utilization is responsible for enhanced mobility of chromatin-bound remodeler.
(A–B) Average MSD plot (A) and violin plot (B), of individual D values for ‘bound only’ trajectories imaged by fast-tracking, shown for six remodelers and H2B histone. (C) Violin plot showing distribution of individual D values imaged by slow-tracking for six remodelers and H2B histone. For (A–C) each wildtype remodeler is compared to H2B by the ordinary one-way ANOVA test (****p<0.0001, ***p<0.001, **p<0.01, *p<0.05). (D–H) MSD plot (D–F) and violin plot (G,H) of individual D values for ‘trajectories imaged by slow-tracking for wildtype, ATPase-dead mutant, and H2B. For violin plots, thick red and dotted gray lines represent the median and two quartiles, respectively. For D–H, mutants are compared to wildtype by the unpaired t test (****p<0.0001, ***p<0.001, ns: not significant). (I) Representative trajectories imaged by slow-tracking for H2B and remodelers. H2B displays low mobility, whereas remodelers display higher chromatin-associated diffusivity that is enhanced by ATP utilization.
-
Figure 6—source data 1
Number of molecules (N), statistical tests, and source data for Figure 6.
- https://cdn.elifesciences.org/articles/69387/elife-69387-fig6-data1-v2.xlsx
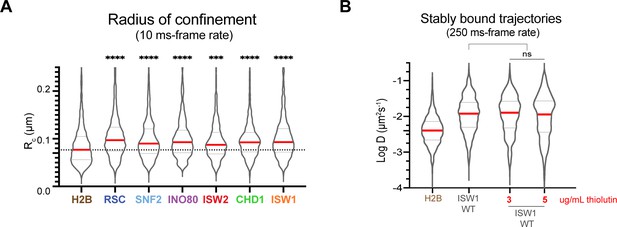
Chromatin-bound remodelers display higher radius of confinement (Rc) values than H2B.
(A) Radius of confinement values of bound trajectories in 10 ms exposure movies for histone H2B and chromatin remodelers. Violin plot showing distribution of Rc values, and comparison between histone H2B and each of wildtype remodelers by ordinary one-way ANOVA test. (B) Violin plot of individual D values by slow-tracking for Isw1-Halo after 30 min pre-treatment with 3 or 5 µg/mL thiolutin, and comparison between wildtype and thiolutin-treated samples by unpaired t test (ns: not significant). Thick red and dotted gray lines represent the median and two quartiles, respectively.
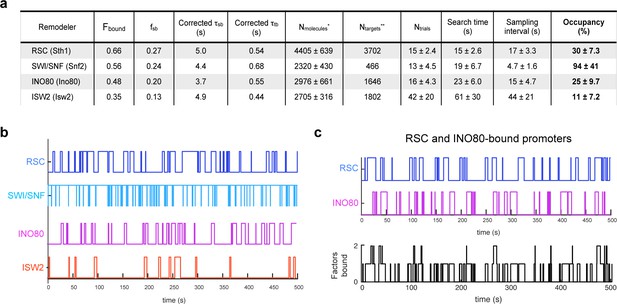
Remodelers show substantial temporal occupancies at chromatin targets.
(A) Key parameters measured in this study and acquired from the literature (Ho et al., 2018; Kubik et al., 2019) are used to calculate occupancy levels for gene promoter-acting remodelers. (B) Time trace simulations of temporal occupancy for individual remodelers at a target promoter region based on average τsb and sampling interval. Top and bottom bars represent occupied (on) and vacant (off) states, respectively, and vertical lines depict transitions between the two states. (C) Time trace simulations of occupancy at a RSC- and INO80-bound promoter region based on average τsb and sampling interval. Individual time trace simulations are shown above, and the cumulative simulated occupancy plot (black) shows either one or both remodelers bound in the time course of 500 s.
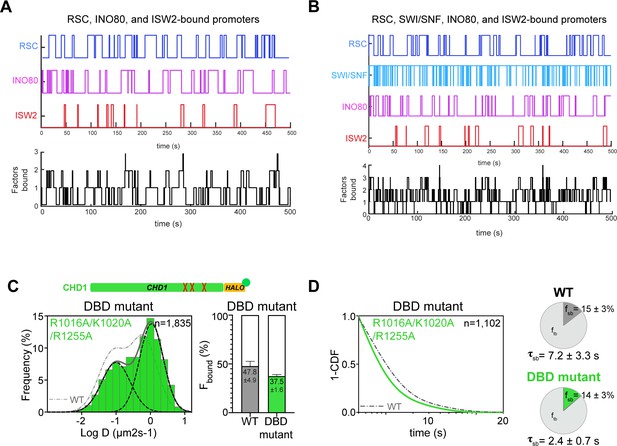
Time trace simulations of temporal occupancies at promoters bound by multiple remodelers, and analysis of CHD1 DNA-binding mutant.
(A–B) Time trace simulations of occupancy as in Figure 6, at a promoter region bound by RSC, INO80, and ISW2 remodelers (A) or by RSC, SWI/SNF, INO80, and ISW2 remodelers (B). Individual time trace simulations are shown above, and the cumulative simulated occupancy time trace (black) shows any one or multiple remodelers bound in the time course of 500 s. (C–D) Fast-tracking and slow-tracking results for CHD1 DNA-binding domain mutant (Chd1R1016A/K1020A/R1255A-Halo). (wildtype: dashed gray lines; DBD mutant: solid green lines). (C) Normalized histogram log10 diffusion coefficients (Left) and Spot-On kinetic modeling results (Right). (D) 1-CDF plot, pie chart, and residence times.
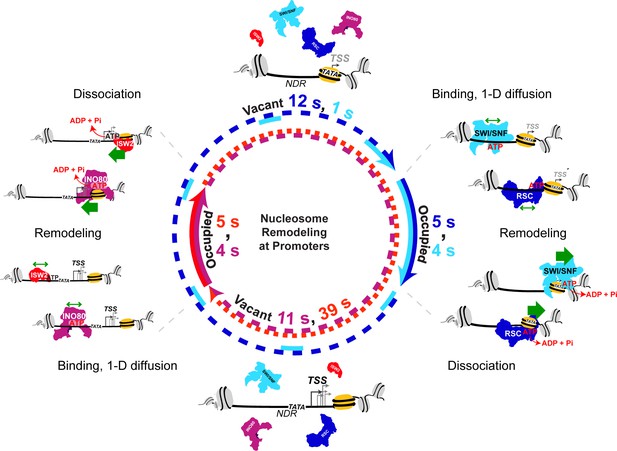
Nucleosome remodeling at promoters.
Model for nucleosome remodeling cycle at a gene promoter region targeted by RSC, SWI/SNF, INO80, and ISW2. The promoter region transitions between remodeler-occupied [solid arrow] and remodeler-vacant [dashed arrow] states, and their mean durations are indicated. After association with the NDR, remodelers undergo 1-D diffusion on chromatin facilitated by ATP binding, resulting in higher chromatin-associated mobility. Upon engaging a nucleosome substrate [e.g. the +1 nucleosome], RSC or SWI/SNF uses the energy of ATP hydrolysis to ‘push’ the nucleosome away from the NDR and INO80 or ISW2 to ‘pull’ the nucleosome into the NDR. ATP hydrolysis facilitates remodeler dissociation, and the promoter region becomes vacant for other factor interactions. The order of remodeler visitation is arbitrary, and simultaneous co-occupancy within the NDR can occur (see text for details).
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (S. cerevisiae) | Full list of yeast strains is presented in Supplementary file 1. | |||
| Recombinant DNA reagent | pBS-SK-Halo-NatMX (plasmid) | Ranjan et al., 2020 | ||
| Recombinant DNA reagent | pUG72 (plasmid) | Euroscarf, Germany | pUG72 (P30117)-Euroscarf | |
| Sequence-based reagent | Full list of oligonucleotides is presented in Supplementary file 2. | |||
| Chemical compound, drug | JF552-HaloTag ligand | Zheng et al., 2019 | N/A | |
| Chemical compound, drug | JF646-HaloTag ligand | Grimm et al., 2015 | N/A | |
| Chemical compound, drug | #1.5 Micro Coverglass – 25 mm Diameter | Electron Microscopy Sciences | 72225–01 | |
| Software, algorithm | ImageJ (1.52 p) | ImageJ | RRID:SCR_003070 https://imagej.net/ | |
| Software, algorithm | Diatrack 3.05 | Vallotton and Olivier, 2013 | http://www.diatrack.org/index.html | |
| Software, algorithm | GraphPad Prism version 8.4.2 | GraphPad Software, Inc. | RRID:SCR_002798 http://www.graphpad.com | |
| Software, algorithm | Sojourner package | Carl Wu lab | https://rdrr.io/github/sheng-liu/sojourner/ | |
| Software, algorithm | Spot-On | Hansen et al., 2018 | https://spoton.berkeley.edu/ | |
| Software, algorithm | vbSPT | Persson et al., 2013 | http://vbspt.sourceforge.net/ | |
| Software, algorithm | Radius of confinement calculation | Lerner et al., 2020 | https://data.mendeley.com/datasets/wctzwpp9h2/2 | |
| Software, algorithm | Custom Matlab script | This paper, Mendeley Data | http://dx.doi.org/10.17632/ydwcx9yhpp.2 |
Additional files
-
Supplementary file 1
List of yeast strains used in this study.
- https://cdn.elifesciences.org/articles/69387/elife-69387-supp1-v2.xlsx
-
Supplementary file 2
List of oligonucleotides used in this study.
- https://cdn.elifesciences.org/articles/69387/elife-69387-supp2-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/69387/elife-69387-transrepform-v2.docx







