Crosstalk between repair pathways elicits double-strand breaks in alkylated DNA and implications for the action of temozolomide
Figures
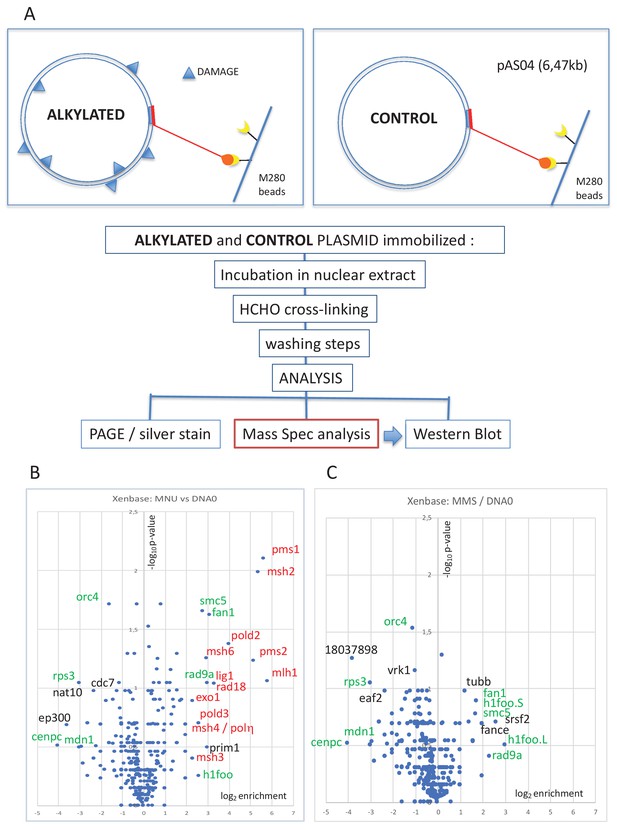
Pull-down of proteins that bind to alkylated versus untreated plasmid DNA.
(A) Experimental workflow. Plasmid DNA (pAS04, 6.5 kb) was treated with alkylating agents under conditions leading to a similar extent of N-alkylation (≈ one alkaline cleavage site every 500 nt) (Figure 1—figure supplement 1A). Immobilized plasmid DNA was incubated in Xenopus nucleoplasmic extracts (NPE) for 10 min at room temperature under mild agitation. The reaction was stopped by addition of formaldehyde (0.8% final) to cross-link the protein-DNA complexes. The beads were processed and analyzed by polyacrylamide gel electrophoresis (PAGE) or by mass spectrometry (MS) as described in 'Materials and methods'. (B) Relative abundance of proteins captured on N-methyl-N-nitrosourea (MNU)-treated versus -untreated DNA0. Proteins captured on equal amounts of MNU-treated or -untreated plasmid were analyzed by label-free MS in triplicate. For all proteins, average spectral count values in the MNU-treated plasmid sample were divided by the average spectral count values in the DNA0 sample. The resulting ratio is plotted as its log2 value along x-axis. The statistical significance of the data is estimated by the p-value in the Student’s t-test and plotted as -log10p along y-axis. Proteins enriched on MNU versus untreated plasmid DNA appear on the right-side top corner and essentially turn out to be mismatch repair (MMR) proteins labeled in red (B). Data shown are analyzed using Xenbase database. (C) Relative abundance of proteins captured on methyl-methane sulfonate (MMS)-treated versus -untreated DNA0. Proteins captured on equal amounts of MMS-treated or -untreated plasmid were analyzed by label-free MS in triplicate. The data are analyzed and plotted as in panel (B) for MNU using Xenbase database. Proteins (labeled in green in B and C) are found enriched or excluded in both MMS versus DNA0 and MNU versus DNA0 plasmids. We suggest these proteins are recruited or excluded from binding to DNA by the abundant class of N-alkylation adducts that both MMS- and MNU-treated plasmids share in common (~27 N-alkyl adducts per plasmid).
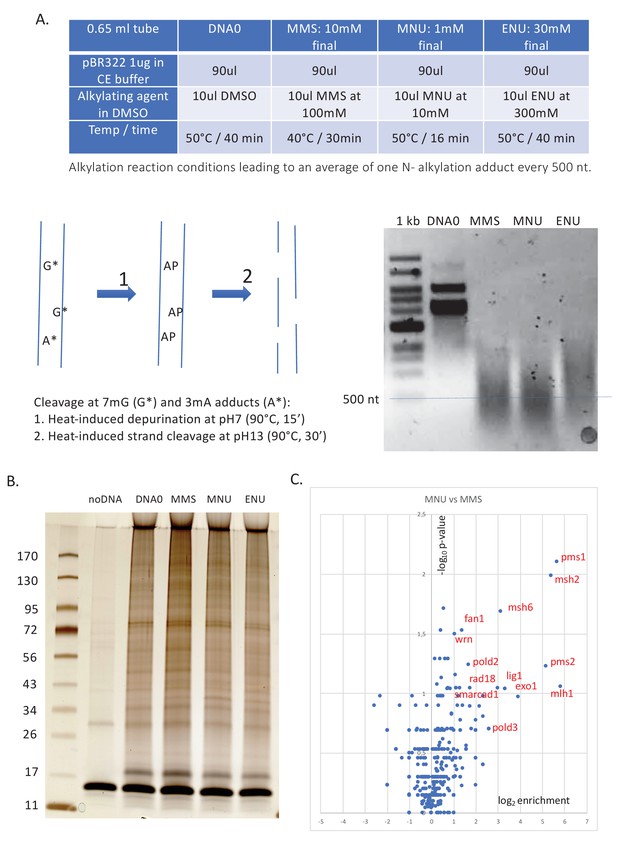
Alkylation reaction conditions and differential protein capture data.
( A) Alkylation reaction conditions and alkaline cleavage. The alkylation reaction conditions (concentration, reaction temperature, and time) were adjusted by successive trials until an average of one N-alkyl adduct per 500 nt was reached. The desired level of alkylation (≈1 N-alkyl adduct/500nt) was attested using the alkaline cleavage procedure, followed by neutral agarose gel electrophoresis (for details, see 'Materials and methods'). (B) Polyacrylamide gel electrophoresis (PAGE) analysis and silver staining of the captured proteins. Proteins captured on equal amounts of immobilized plasmid DNA incubated in nucleoplasmic extracts (NPE) (as described in Figure 1A) were resolved by denaturing gel electrophoresis and silver stained. Samples, corresponding to ≈30 ng of immobilized input plasmid DNA, were loaded on a 4–15% PAGE (Biorad pre-cast) gel, run at 200 volts for 32 min, and silver stained. Lanes DNA0, MMS, MNU, and ENU display a complex pattern of proteins (total amount of proteins per lane estimated at 100–200 ng). To monitor non-specific protein binding to beads, we included a negative control containing the same amount of M280 beads in the absence of plasmid DNA (noDNA). Importantly, in the noDNA lane, only a low amount of residual protein is visible, indicating efficient removal of non-specifically bound proteins during the washing procedure. (C) Relative abundance of proteins captured on N-methyl-N-nitrosourea (MNU)- versus methyl-methane sulfonate (MMS)-treated plasmids. Proteins captured on equal amounts of MNU- or MMS-treated plasmids were analyzed by label-free mass spectrometry (MS) in triplicate. The data are analyzed and plotted as described in Figure 1B using Xenbase database. Proteins enriched on MNU- versus MMS-treated DNA appear on the right-side top corner and essentially turn out to be mismatch repair (MMR) proteins labeled in red (Figure 1B).
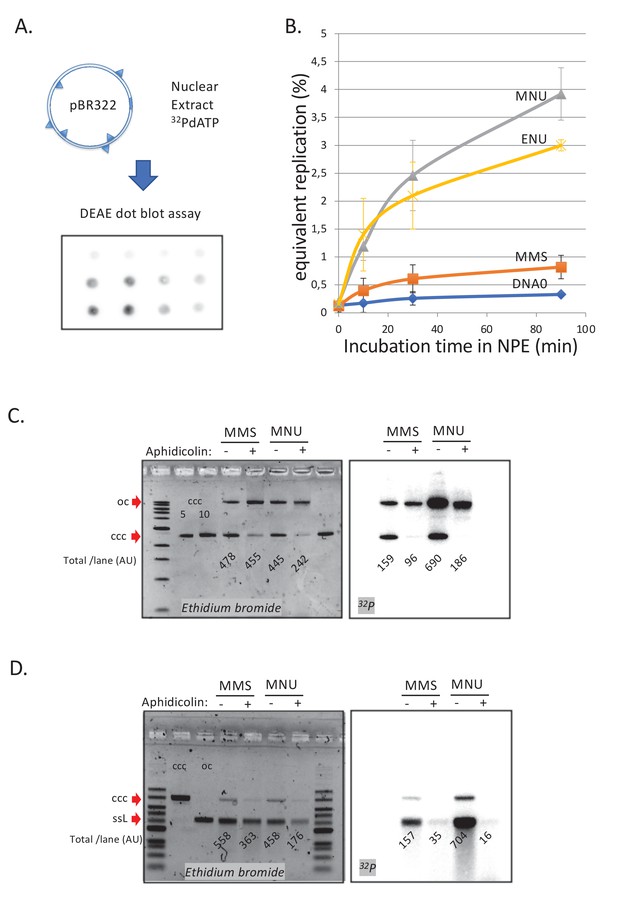
DNA repair synthesis in alkylated and undamaged control plasmid DNA in NPE.
(A) Outline of the spot assay. Plasmids were incubated in nuclear extracts supplemented with α32P-dATP; at various time points, an aliquot of the reaction mixture was spotted on DEAE paper (see 'Materials and methods'). The dot blot is shown for the sake of illustration only. (B) Plasmid DNA pBR322 (4.3 kb) samples, modified to a similar extent with -MMS, -MNU and -ENU, were incubated in nucleoplasmic extracts (NPE) supplemented with α32P-dATP at room temperature; incorporation of radioactivity was monitored as a function of time using the spot assay described above (A). Undamaged plasmid DNA0 was used as a control. At each time point, the average values and standard deviation from three independent experiments were plotted. The y-axis represents DNA repair synthesis expressed as a fraction of input plasmid replication (i.e., 10% means that the observed extent of repair synthesis is equivalent to 10% of input plasmid replication). This value was determined knowing the average concentration of dATP in the extract (50 μM) and the amount of added α32P-dATP. (C) N-methyl-N-nitrosourea (MMS)- and N-methyl-N-nitrosourea (MNU)-treated plasmids were incubated in NPE, supplemented or not, by aphidicolin (150 μM final). After 1 hr of incubation, plasmids were purified and analyzed by agarose gel electrophoresis under neutral loading conditions. The gel was imaged by fluorescence (left: ethidium bromide image) and by autoradiography (right: 32P image). The number below each lane indicates the total amount of signals per lane (expressed in arbitrary units [AU]). Aphidicolin treatment decreases incorporation into MNU-treated plasmid close to fourfold, while it affected incorporation into MMS-treated plasmid only 1.6-fold. (D) Samples as in (C). Gel loading is performed under alkaline conditions to denature DNA before entering the neutral agarose gel, allowing single-stranded nicks present in DNA to be revealed. The number below each lane indicates the amount of signals per lane (AU).
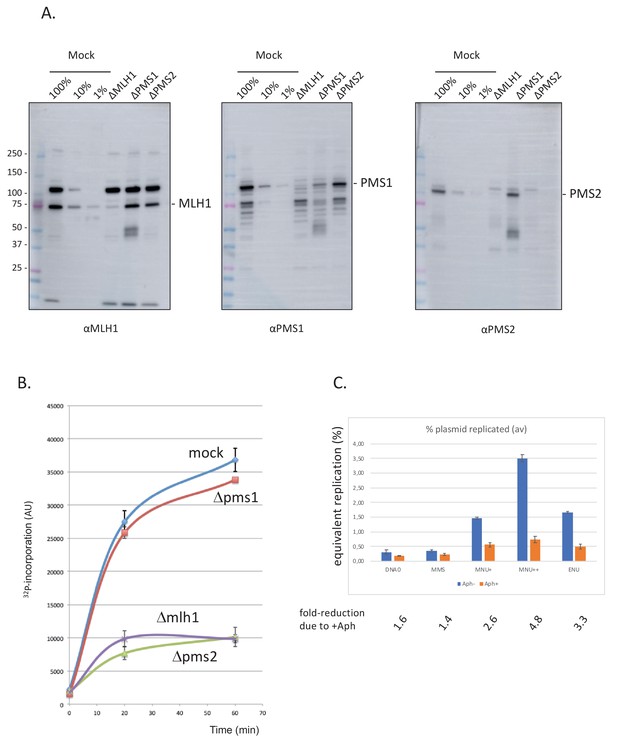
Involvement of mismatch repair in repair synthesis and effect of aphidicolin.
(A) Analysis by western blotting (WB) of the nucleoplasmic extracts (NPE) depleted for mismatch repair (MMR) proteins. Antibodies against Mlh1, Pms2, and Pms1 were used as previously described (Kato et al., 2017; Kawasoe et al., 2016). Extracts were depleted, with the respective antibodies, for three rounds at 4°C at a ratio bead: antibody: extract = 1:3:5. (B) Depletion experiments show that α32P-dATP incorporation into N-methyl-N-nitrosourea (MNU) plasmid is mediated by mismatch repair. NPE was depleted by antibodies against Pms1, Pms2, Mlh1, or mock depleted. Upon incubation at room temperature in different NPE, incorporation of α32P-dATP in MNU plasmid was monitored by the spot assay as a function of time. At each time point, the average values and standard deviations from two independent experiments were plotted. As expected from previous data (Figure 2B), robust incorporation was observed for MNU plasmid incubated in mock-depleted extracts. In contrast, radioactive dATP incorporation was severely reduced when the plasmid was incubated in extracts depleted with antibodies against Mlh1 or Pms2. These data strongly suggest that the incorporation seen in mock-depleted NPE is mediated by MMR, as both Mlh1 and Pms2 assemble into the functional MMR complex MutLα. In contrast, depletion with antibodies against Pms1 did not affect incorporation kinetics as Pms1 is reported not to function in MMR (Jiricny, 2006). (C) Effect of aphidicolin: alkylated plasmids (pEL97: 11.3 kb) were incubated in NPE supplemented or not by aphidicolin (150 μM final) in the presence of 32P-dATP for 1 hr at room temperature (RT). Analysis was performed by spot assay as described in 'Materials and methods'. The y-axis represents the percentage of radioactive DNA synthesis expressed in % of full plasmid replication. Addition of aphidicolin to NPE (150 μM final) more severely reduces incorporation in MNU+, MNU++, and ENU plasmids, compared to control and methyl-methane sulfonate (MMS) plasmids. Fold reduction in incorporation under aphidicolin conditions is shown.
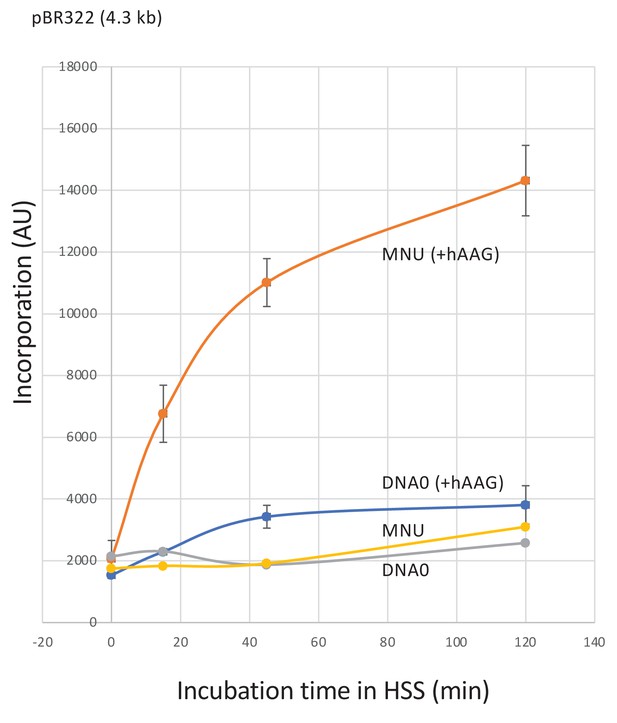
Repair synthesis in HSS extracts.
Plasmid treated with N-methyl-N-nitrosourea (MNU) (≈1N-alkyl adduct/500nt) was incubated in high-speed supernatant (HSS) extracts in the presence of α32P-dATP; repair synthesis was monitored at room temperature as a function of time using the spot assay. Both DNA0 and MNU plasmids exhibited background incorporation levels. Robust unscheduled DNA synthesis (UDS) was observed in MNU plasmid upon addition of hAAG glycosylase (150 nM). Average values and standard deviations from two independent experiments were plotted.
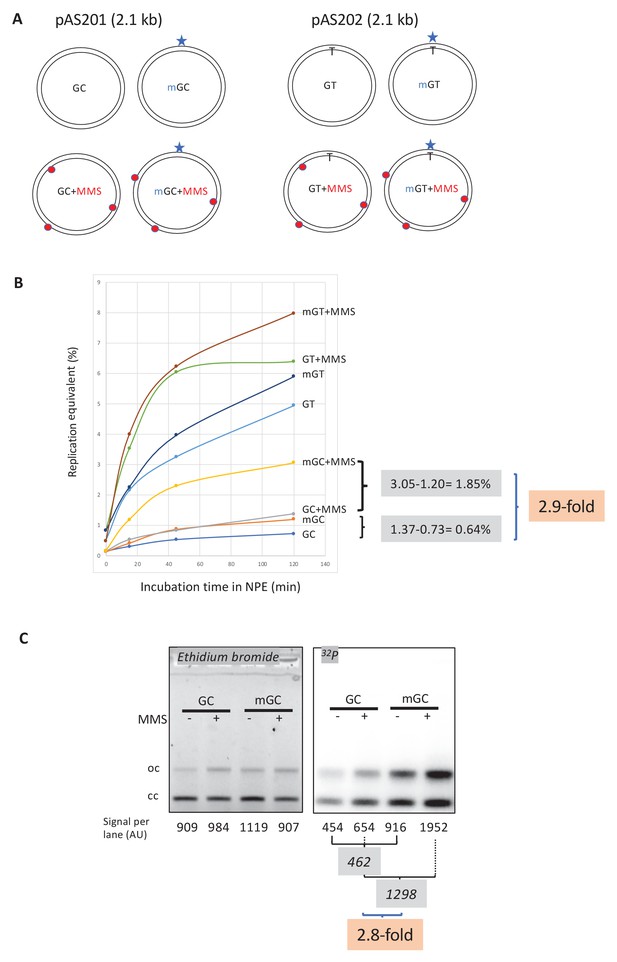
Stimulation of MMR at a single O6mG site by N-alkyl adducts in cis.
(A) Covalently closed circular (ccc) plasmids (pAS200.2, 2.1 kb) containing a site-specific O6mG:C base pair (plasmid mGC) and the corresponding lesion-free control (plasmid GC) were constructed (Isogawa et al., 2020). Similarly, plasmids with a site-specific GT or a O6mG:T mismatch were constructed. All the four constructs were treated with methyl-methane sulfonate (MMS) in order to introduce random N-alkyl (7mG and 3mA) adducts, generating plasmids GC+MMS, mGC+MMS, GT+MMS, and mGT+MMS. We adjusted the MMS reaction conditions as to introduce ≈ nine adducts per plasmid (i.e., one N-alkylation adduct every ≈500 nt). The resulting proportion of O-alk and N-alkyl adducts mimics the proportion in N-methyl-N-nitrosourea (MNU)-treated plasmids. The single O6mG adduct and the randomly located N-alkyl adducts are represented by a star and red dots, respectively. (B) Plasmids described above were incubated in nucleoplasmic extracts (NPE) supplemented with α32P-dATP at room temperature; incorporation of radioactivity was monitored as a function of time using the spot assay. The y-axis represents the percentage of DNA repair synthesis with respect to input DNA (i.e., 10% means that the observed extent of repair synthesis is equivalent to 10% of input plasmid replication). Overall, incorporation into GT and mGT plasmids is higher than incorporation into their GC and mGC counterparts. Incorporation attributable to repair at the O6mG:C lesion is increased close to threefold due to the presence of random N-alkyl lesions introduced by MMS treatment. The stimulatory effect of random N-alkyl lesions on GT and mGT repair is observed but is slightly less pronounced than for mGC. (C) The same plasmids were incubated for 2 hr in NPE, purified, resolved by agarose gel electrophoresis, and revealed by ethidium bromide fluorescence and 32P autoradiography. The total amount of signals per lane is indicated (arbitrary units [AU]). As expected, the amount of plasmid extracted from each incubation mix is relatively constant, as quantified below the ethidium bromide image. Increase in repair at the O6mG:C lesion due to MMS treatment (2.8-fold) is in good agreement with data in (B).
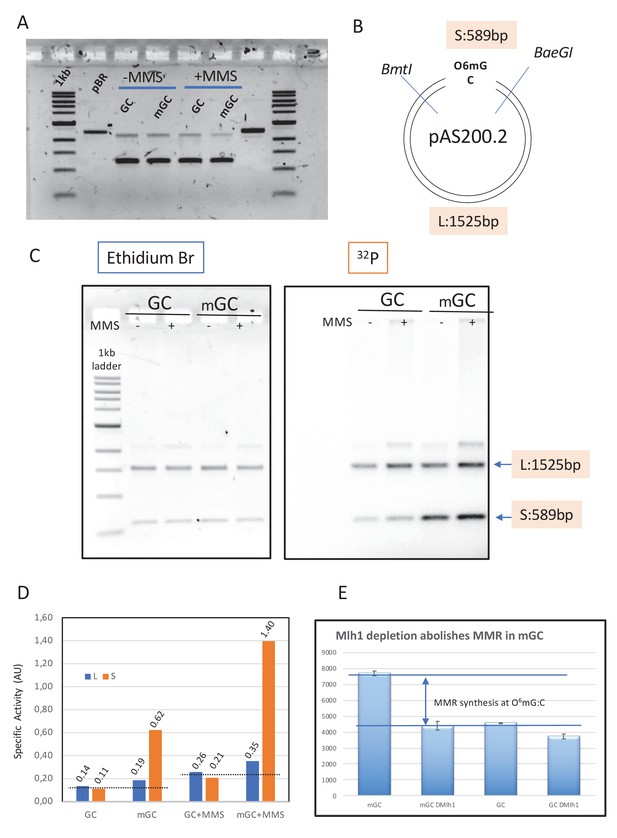
Mapping repair synthesis in the vicinity of a single O6mG adduct.
(A) Agarose gel electrophoresis of the native form of GC, mGC, GC+MMS, and mGC+MMS. (B) Restriction map of pAS201; the two single cutter enzymes BmtI and BaeGI yield a short S (589 bp) and a long L (1525 bp) fragment, respectively. Fragment S contains the single O6mG adduct. (C) Plasmids GC, mGC, GC+MMS, and mGC+MMS were incubated in nucleoplasmic extracts (NPE) in the presence of α32P-dATP and extracted after 2 hr. The purified plasmids were digested with restriction enzymes BmtI and BaeGI. Digested plasmids were analyzed by agarose gel electrophoresis and visualized by ethidium bromide staining and by 32P imaging. (D) The specific activity (SA) of a given fragment (S or L) was quantified as the amount of 32P incorporated divided by the amount of DNA deduced from the ethidium bromide fluorescence image (expressed in arbitrary units [AU]). In control plasmid GC, a similar SA value was observed for both fragments as expected for residual background incorporation; similarly, in methyl-methane sulfonate (MMS)-treated plasmids (GC+MMS), the SA value was similar for S and L fragments. The slightly increased SA value in GC+MMS compared to GC is compatible with repair synthesis by base excision repair (BER) at randomly located MMS-induced lesions. For plasmids containing a single O6mG:C lesion (mGC and mGC+MMS), there is a robust increase in SA of the short compared to the long fragments. Incorporation, above background, due to O6mG, in the absence of MMS, amounts to 0.065 and 0.495 AU for long and short fragments, respectively (signal above the dotted line in S3D). Similarly, in the context of MMS lesions, incorporation, above background, due to O6mG, amounts to 0.115 and 1.17 AU for long and short fragments, respectively (signal above the dotted line in S3D). Taken together, these results clearly show that O6mGC-mediated repair specifically takes place in the S fragment, with only modest spill-over into the L fragment (10–15%). (E) Plasmids with a single O6mG:C lesion site (mGC) and control plasmids (GC) were incubated in NPE depleted with anti-Mlh1 antibodies and mock-treated NPE. Incorporation was measured into whole plasmids. The increase in incorporation into mGC compared to GC observed upon incubation in NPE fully disappeared when Mlh1 was depleted. This experiment shows that the increase in incorporation observed in mGC compared to incorporation in control GC plasmid can specifically be attributed to mismatch repair activity at the single O6mG:C lesion.
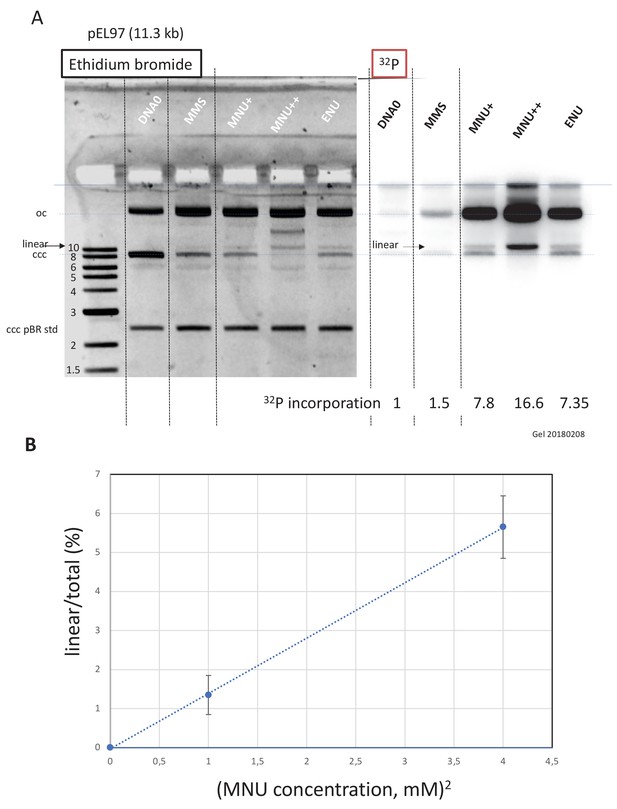
Double-strand breaks occur in MNU-treated plasmids during incubation in extracts.
(A) Analysis by agarose gel electrophoresis (AGE) of alkylated plasmids (pEL97: 11.3 kb) incubated in nucleoplasmic extracts (NPE) in the presence of α32P-dATP. Plasmid pEL97 was treated with methyl-methane sulfonate (MMS), N-methyl-N-nitrosourea (MNU)+, and ENU as to introduce ≈ one alkylation event, on average, every 500 nt. For MNU, a plasmid with twice the level of alkylation (MNU++, one lesion every 250 nt) was also produced (Figure 4—figure supplement 1). Alkylation of these plasmids essentially not affected their migration on agarose gels (Figure 4—figure supplement 2A). After 2 hr of incubation, the reaction was stopped and a known amount of pBR322 (10 ng) plasmid was added as an internal standard. Ethidium bromide image: in different lanes, the internal standard band, pBR (covalently closed circular [ccc]), appears to be of similar intensity (1158 +/- 95 arbitrary units [AU]), assessing reproducible DNA extraction. For the alkylated plasmids, incubation in NPE led to massive conversion from ccc to relaxed plasmids. 32P image: little incorporation of 32P-dATP is seen in DNA0 and in MMS-treated plasmids compared to MNU- and ENU-treated plasmids as shown by the relative incorporation levels normalized to one for untreated plasmid (DNA0). As expected, the MNU++ sample exhibits about twice the amount of incorporated radioactivity compared to MNU+. In both ethidium bromide and 32P images, a small amount of linear plasmid is seen mostly in the MNU++ sample. This band is also visible in the MNU+ and ENU lanes although at a weaker intensity. (B) Quadratic dose-response for double-strand break (DSB) formation. When the % of linear form (linear/(linear + oc)) is plotted as a function of the square dose of MNU (mM2) for untreated, MNU+, and MNU++ plasmids, we observed a straight line (y = 1.4173x - 0.0288; R² = 0.9999).
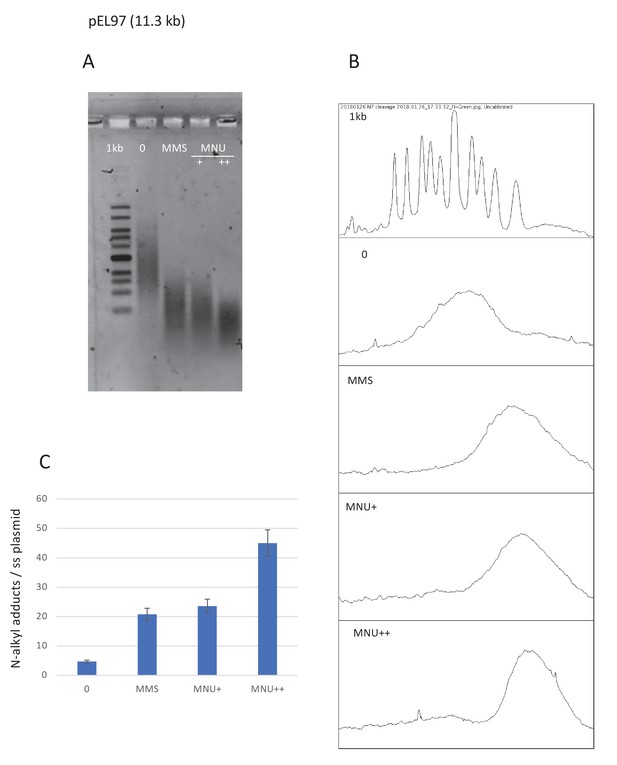
Estimation of N-alkylation levels of modification by MMS and MNU.
(A) Alkaline fragmentation of alkylated plasmid (pEL97, 11.3 kb) DNA is analyzed by agarose gel electrophoresis. (B) Densitometry of the ethidium bromide-stained agarose gel. (C) Estimation of the average number of alkali cleavage sites per plasmid strand. The data reveal that the average number of cleavage sites per plasmid strand is 21–23 for both methyl-methane sulfonate (MMS) and N-methyl-N-nitrosourea (MNU)+ reaction conditions (the average distance between two sites is ≈510 nt). For MNU++, the average distance between two cleavage sites is ≈254 nt.
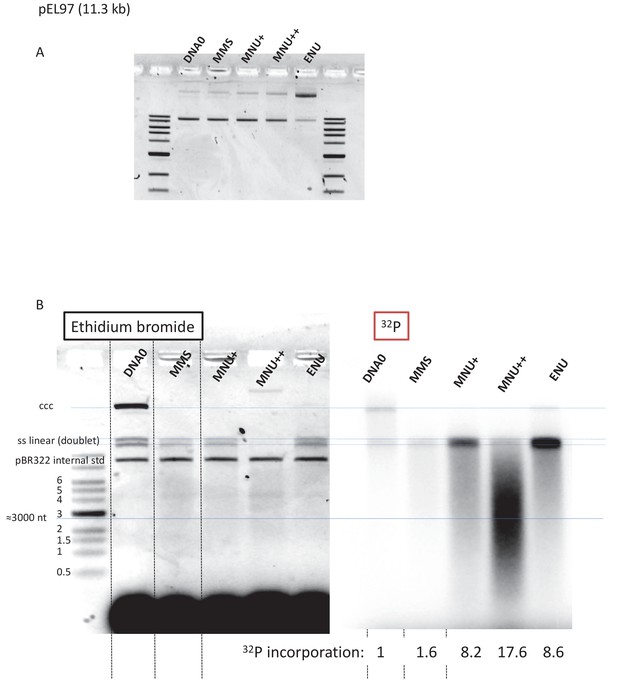
Fragmentation of alkylated plasmid as analyzed on AGE loaded under alkaline conditions.
(A) Alkylation did not affect plasmid topology except for ENU treatment that increases relaxation as seen in the agarose gel electrophoresis (AGE) image. (B) Analysis by AGE, under alkaline loading conditions, of alkylated plasmids (pEL97: 11.3 kb) incubated in nucleoplasmic extracts (NPE) in the presence of α32P-dATP (same samples as in Figure 4A). Loading under alkaline conditions allowed single-stranded nicks to be revealed. In the different lanes, an internal standard band, pBR (covalently closed circular [ccc]), was introduced before the extraction procedure in order to assess the consistent recovery of DNA during extraction (as seen by ethidium bromide staining). For the alkylated plasmids, incubation in NPE led to massive conversion of plasmid DNA into a smear with a low amount of DNA present as a single-stranded linear band (doublet). For the MNU++ sample, no linear single-stranded DNA (ssDNA) was seen; all DNA was converted into a smear. The smearing can best be seen in the 32P image. We suggest that the ssDNA fragments that form the smear arise between a gap caused by a mismatch repair (MMR) event at an O6mG site and an uncompleted base excision repair (BER) event at a neighboring lesion. It cannot be excluded that some fragments result from MMR attempts at two O6mG lesions. The intense 32P labeling of the fragments in the smear results from incorporation of up to several hundred nt during a single MMR repair synthesis. Relative 32P incorporation normalized to one for DNA0 is indicated below the 32P image.
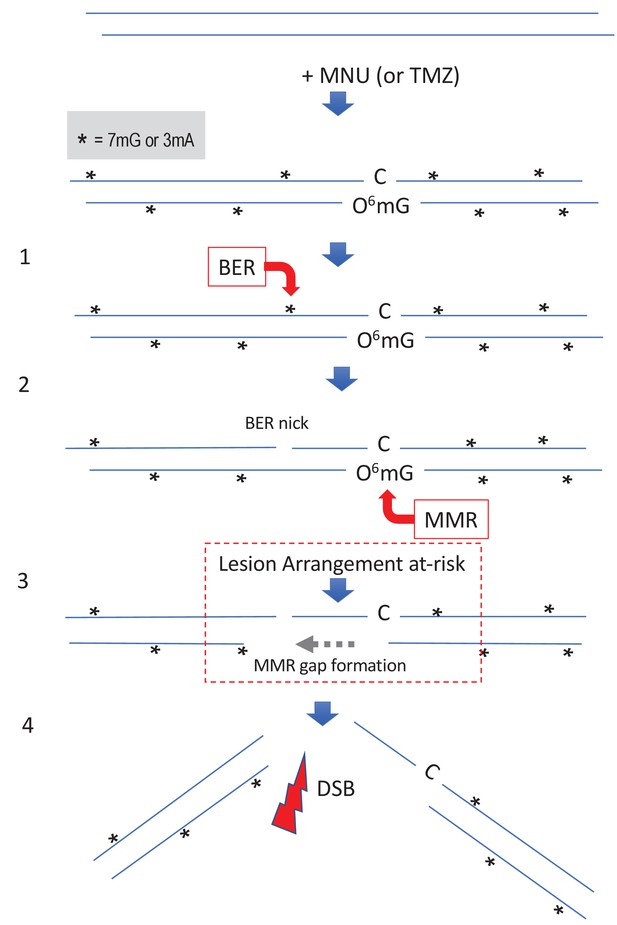
Simultaneous repair of two closely spaced MNU-induced lesions may lead to a DSB.
Such a situation occurs when an N-alkyl lesion located within ≈500 nt of an O6mG lesion is processed simultaneously (‘Lesion Arrangement at-risk’). Note that the mismatch repair (MMR) excision track can occur on either strand as described for noncanonical MMR (Peña-Diaz et al., 2012). Reaction of N-methyl-N-nitrosourea (MNU) with double-stranded DNA induces N-alkylation adducts, mostly 7mG and 3mA shown as * and O-alkylation adducts (O6mG), at a ratio of 10:1 approximately. Step 1: a base excision repair (BER) event is initiated at an N-alkyl adduct, creating a nick. Step 2: concomitantly, an MMR event takes place, in the opposite strand, at a nearby O6mG:C site. Step 3: the MMR machinery extends the nick into a several hundred nt-long gap by means of Exo1 action. Step 4: the two independently initiated repair events lead to a double-strand break (DSB), if the MMR gap reaches the BER-initiated nick before resealing.
Additional files
-
Source data 1
Original image data used in Figures and Figure supplements.
- https://cdn.elifesciences.org/articles/69544/elife-69544-data1-v2.zip
-
Source data 2
Image data before trimming used in Figures and Figure supplements.
- https://cdn.elifesciences.org/articles/69544/elife-69544-data2-v2.zip
-
Source data 3
Resource data of MS analysis.
- https://cdn.elifesciences.org/articles/69544/elife-69544-data3-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/69544/elife-69544-transrepform-v2.docx





