The sleep-wake distribution contributes to the peripheral rhythms in PERIOD-2
Figures
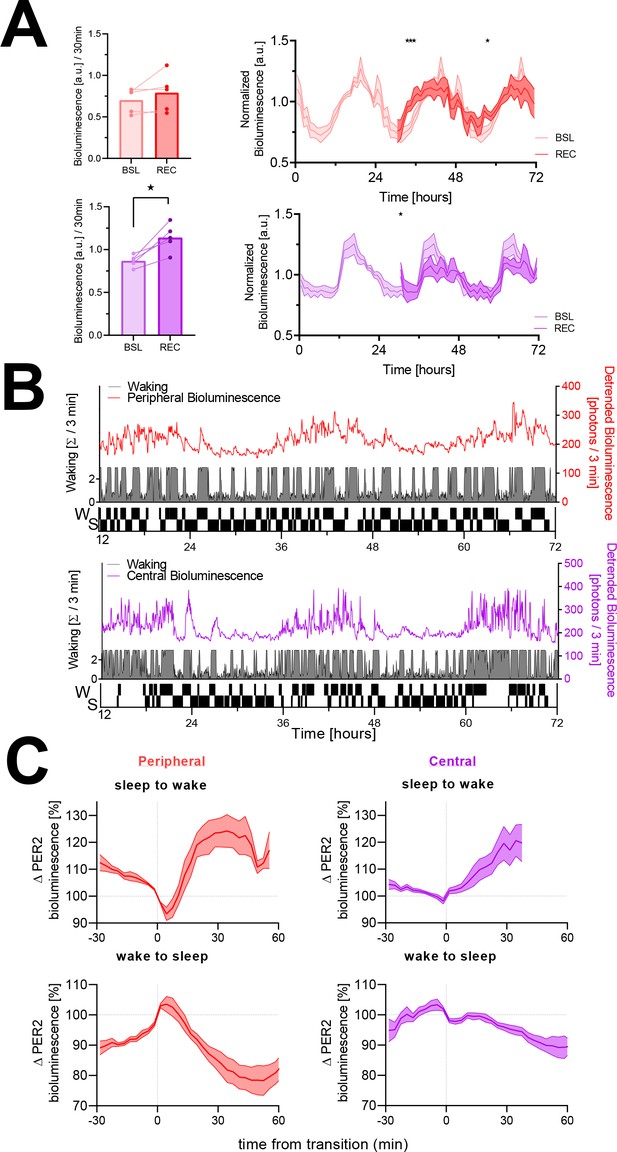
The sleep-wake distribution contributes to changes in PER2 bioluminescence.
Red, peripheral (kidney) bioluminescence; purple, central (cortical) bioluminescence. (A) Left panels: PER2 bioluminescence measured in the first 30 min of recovery (REC) after sleep deprivation compared to levels reached at this circadian time during baseline (BSL). Sleep deprivation elicited an acute response in central PER2 (lower bar graph; t(4) = 4.4, p=0.012), but not in the periphery (upper bar graph; t(4) = 1.5, p=0.21). Right panels: under baseline conditions (lighter graphs, time 0–24 hr repeated three times, average of 48 hr baseline), PER2 bioluminescence showed a circadian rhythm both in the periphery (left) and central (right). Sleep deprivation from ZT0 to –6 (times under preceding LD conditions) affected PER2 bioluminescence during recovery (two-way rANOVA w/ Condition × Time, periphery: F(41, 164) = 2.1, p=0.0007; central: F(41, 164) = 1.8, p=0.004). Asterisks indicate significant differences assessed by post-hoc paired t-tests. Bioluminescence is expressed as a fraction of the individual average bioluminescence during the experiment and depicted in 1 hr intervals as mean ± SEM for five mice (peripheral and central). (B) A sleep-wake state recording (gray area plot represents wakefulness in consecutive 3 min intervals) combined with peripheral (red line, upper graph) and central PER2 bioluminescence (purple line, lower graph) in two mice during baseline. Note that besides the circadian oscillation in PER2 bioluminescence, there are marked increases and decreases in PER2 bioluminescence. The ‘hypnogram’ (lower part of the graph) illustrates that the rapidly evoked changes in PER2 bioluminescence are related to periods of sleeping (S) and waking (W). This hypnogram is discontinuous as it depicts only the SW transitions selected in this mouse for the analysis in (C). (C) Changes in PER2 bioluminescence associated with transitions from sleep to wake (top) and wake to sleep (bottom) of peripheral (left, n = 5) and cortical (right, n = 6) origin. Data underlying figures can be found in Figure 1—source data 1.
-
Figure 1—source data 1
This file contains the numerical values on which the graphs in Figure 1A–C are based.
- https://cdn.elifesciences.org/articles/69773/elife-69773-fig1-data1-v2.xlsx
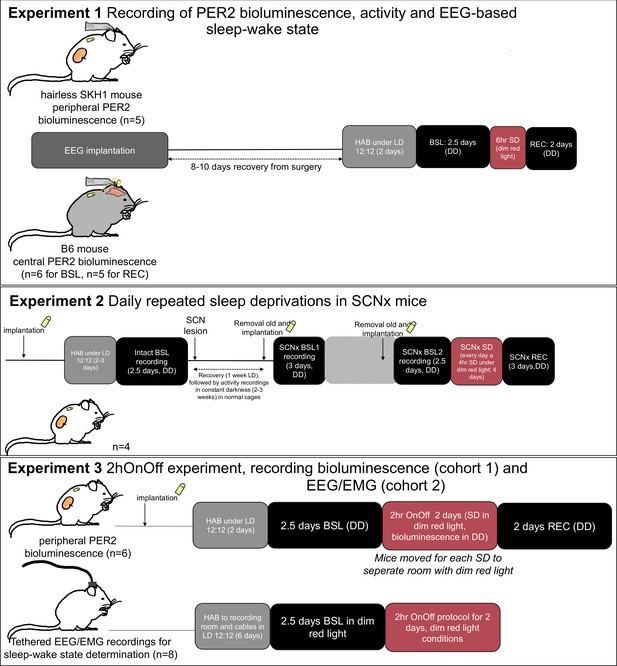
Experimental protocol for the three experiments.
HAB, habituation; BSL, baseline; SD, sleep deprivation; REC, recovery; SCNx, SCN lesion; DD, constant darkness conditions.
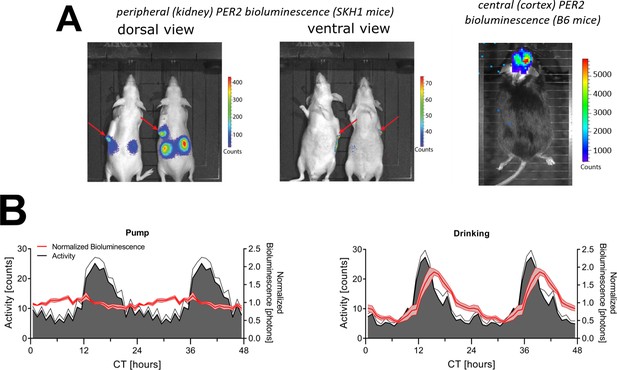
Sources of bioluminescence and different luciferin administration routes.
(A) Dorsal (left) and ventral (middle panel) in vivo imaging of peripheral PER2 bioluminescence in SKH1 mice using the IVIS Xenogen system. Note that the scale bar of the dorsal view has higher values than the scale bar of the ventral view. Images were taken between ZT6-8. The red arrows indicate the location of the osmotic mini-pump flow moderator. The right panel illustrates the central PER2-bioluminescence signal obtained in B6 mice carrying the Per2Luc KI construct using the same imaging system. B6 mice were not shaved, received luciferin centrally, and were mounted with a glass cone over a thinned skull assuring that the emitted bioluminescence reflects that of the brain and not of the periphery. (B) Mice constitutively expressing luciferase under the control of the synthetic CAG promoter (CAG-Luc mice) received luciferin via an osmotic mini-pump (left) or in their drinking water (right) (n = 4). Graphs are double plotted to aid visualization. CT, circadian time. Data underlying (B) can be found in Figure 1—figure supplement 2—source data 1.
-
Figure 1—figure supplement 2—source data 1
This file contains the numerical values on which the graphs in Figure 1—figure supplement 2B are based.
- https://cdn.elifesciences.org/articles/69773/elife-69773-fig1-figsupp2-data1-v2.xlsx

Sleep phenotyping of SKH1 mice.
Four-day time course of time spent asleep and electroencephalogram (EEG) delta power (1–4 Hz) during baseline, 6 hr sleep deprivation (SD; ZT0-6, t = 48–54 hr; pink area), and recovery in SKH1 mice (n = 8) under LD12:12 (gray areas denote the 12 hr D-periods) conditions. (A) Non-rapid eye movement (NREM) sleep (top) and REM sleep (middle), as mean minutes over hourly intervals (±1 SEM). Also depicted is REM sleep as % of total sleep (NREM + REM sleep; bottom panel). Dark gray line during SD and recovery represent baseline time course (mean over the two baseline days) for comparison. (B) Accumulated differences (min/hr) from baseline separately for recovery day 1 (t = 54–72 hr) and day 2 (t = 72–96 hr). During recovery day 1, mice accrue ca. + 38.8 ± 14.6 min extra NREM sleep, which is again lost during recovery day 2 (–31.5 ± 9.6 min). Mice lose –5.3 ± 1.7 min of REM sleep during first five recovery hours after SD. This additional deficit is offset during the remaining of recovery day 1, resulting in a nonsignificant increase of 5.5 ± 2.9 min. (C) Mean EEG delta power during NREM sleep (± 1 SEM) calculated for 12 and 6 percentiles during the L- and D-periods, respectively (except for L-period recovery day 1; eight percentiles) to which an equal number of 4 s epochs scored as NREM sleep contributed (per LD period and mouse). Delta power is considered an EEG correlate of sleep need decreasing when sleep prevails (L-period) and increasing when animals are mostly awake (D-periods). SD results in high immediate values that quickly revert to baseline. Red squares underneath curves represent hourly intervals in which values differed from baseline, post-hoc paired t-tests p<0.05. Data underlying this figure can be found in Figure 1—figure supplement 3—source data 1.
-
Figure 1—figure supplement 3—source data 1
This file contains the numerical values on which the graphs in Figure 1—figure supplement 3A–C are based.
- https://cdn.elifesciences.org/articles/69773/elife-69773-fig1-figsupp3-data1-v2.xlsx
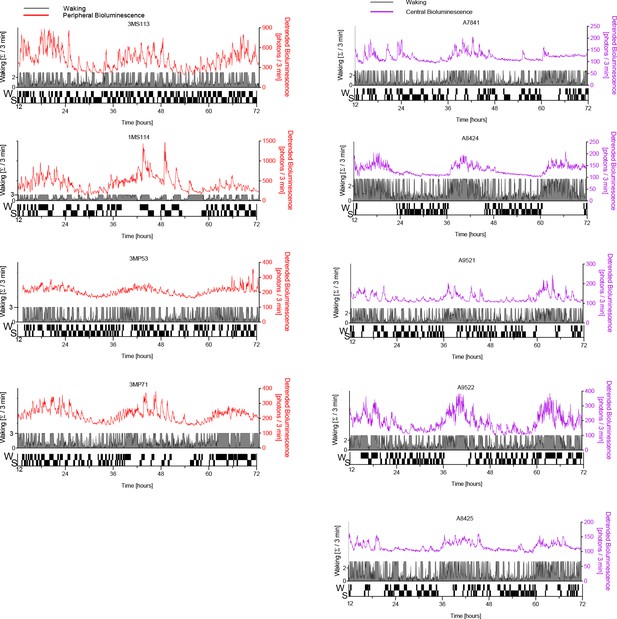
PER2 bioluminescence co-detected with EEG-based sleep-wake state in the other mice.
Left: peripheral (kidney) bioluminescence (red traces); right: central (cortical) bioluminescence (magenta traces). Legend as in Figure 1B. Data underlying this figure can be found in Figure 1—figure supplement 4—source data 1.
-
Figure 1—figure supplement 4—source data 1
This file contains the numerical values on which the nine graphs in Figure 1—figure supplement 4 are based.
- https://cdn.elifesciences.org/articles/69773/elife-69773-fig1-figsupp4-data1-v2.xlsx
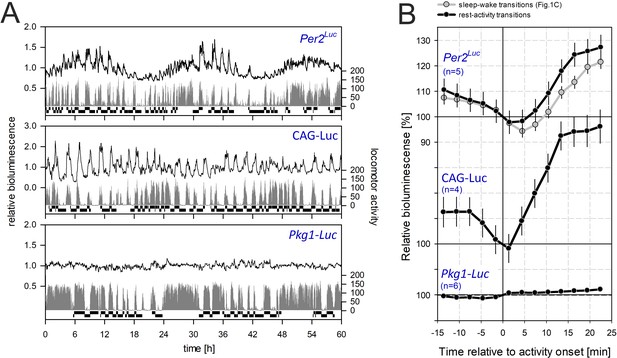
The effect of activity on bioluminescence in three different bioluminescence reporters.
(A) Three representative recordings of activity co-detected with bioluminescence in Per2Luc mice (top), reflecting both circadian and sleep-wake/activity-evoked changes, CAG-Luc mice (middle), representing activity-evoked changes, and Pkg1-Luc mice (bottom), not affected by changes in activity. The selected transitions are annotated on the bottom of each individual recording. Bioluminescence data are plotted relative to the mean bioluminescence of the individual’s recording. (B) Quantification of changes in bioluminescence around rest-activity transitions. For comparison, gray symbols and lines depict sleep-to-wake transition dynamics for PER2::LUC mice (same data as in Figure 1C). Note that the same scaling is used for all three constructs and that only the 100% level is indicated for the CAG-Luc and Pkg1-Luc mice. Data underlying this figure can be found in Figure 1—figure supplement 5—source data 1.
-
Figure 1—figure supplement 5—source data 1
This file contains the numerical values on which the graphs in Figure 1—figure supplement 5A and B are based.
- https://cdn.elifesciences.org/articles/69773/elife-69773-fig1-figsupp5-data1-v2.xlsx

Subcutaneous temperature is phase advanced relative to PER2 kidney bioluminescence.
(A) Two individual recordings of PER2 bioluminescence (red) and subcutaneous temperature (yellow) in SKH1 Per2Luc mice under constant darkness. In both mice, temperature is advanced relative to PER2 bioluminescence. Sinewave fitting revealed that the phase difference is similar in the two mice (3.7 hr and 3.5 hr). (B) Circular plot of the phase distribution of PER2 bioluminescence and temperature, where the peak of bioluminescence has been set at t = 18, reflecting the approximate peak time of PER2 bioluminescence relative to CT12 (activity onset), as observed in this article and Curie et al., 2015. Data underlying this figure can be found in Figure 1—figure supplement 6—source data 1.
-
Figure 1—figure supplement 6—source data 1
This file contains the numerical values on which the graphs in Figure 1—figure supplement 6A and B are based.
- https://cdn.elifesciences.org/articles/69773/elife-69773-fig1-figsupp6-data1-v2.xlsx
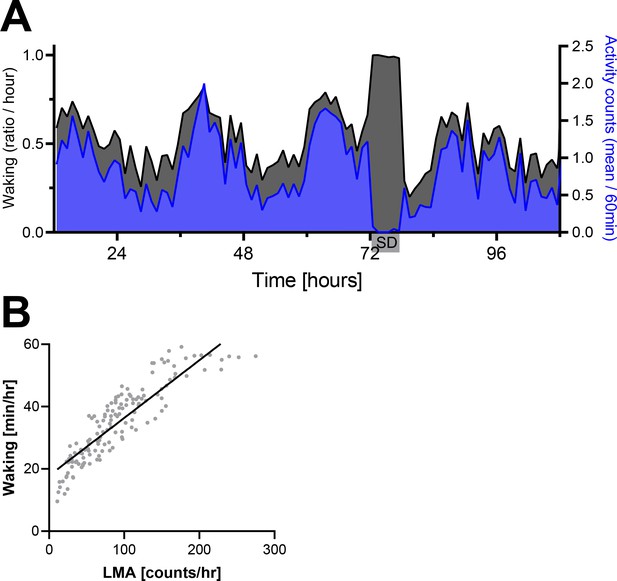
Locomotor activity (LMA) as proxy for wakefulness.
(A) Electroencephalogram (EEG)-based waking (black) and LMA (blue) from five mice used to determine central PER2 bioluminescence during a two-and-a-half-day baseline recording (12–72 hr), a sleep deprivation (72–78 hr), and the recovery days. Note that no activity was measured during the SD as mice were not in the cage with the activity sensor at that time. (B) LMA and waking measured in the 2hOnOff protocol EEG experiment, linear regression, p<0.0001, R2 = 0.79. Data underlying this figure can be found in Figure 1—figure supplement 7—source data 1.
-
Figure 1—figure supplement 7—source data 1
This file contains the numerical values on which the graphs in Figure 1—figure supplement 7A and B are based.
- https://cdn.elifesciences.org/articles/69773/elife-69773-fig1-figsupp7-data1-v2.xlsx
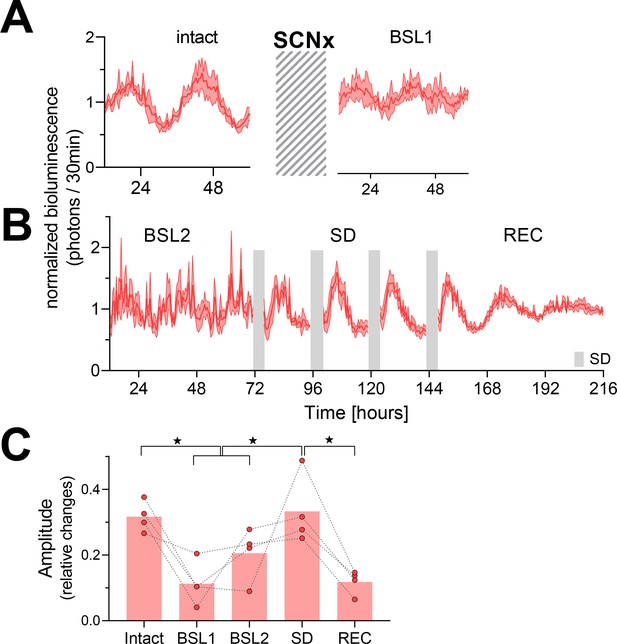
Temporary reinstatement of circadian PER2 oscillations in SCN lesion (SCNx) mice by repeated sleep deprivations (SDs).
(A) Time course of PER2 bioluminescence across 2.5 days under baseline conditions (left) and after the SCNx (right). Cross-hatched area represents an approximately 5-week interval separating the two recordings during which the success of SCNx on locomotor activity was verified under DD (Figure 2—figure supplement 1). (B) Four 4 h SDs (gray bars) repeated daily at the beginning of the light phase under the preceding LD conditions reinstate rhythmic PER2 bioluminescence (n = 4). Abbreviations of experimental conditions: intact, baseline prior to SCNx; BSL1, baseline after DD locomotor activity recordings; BSL2, baseline immediately preceding the SDs; SD, 20 hr recordings between the four 4 hr SDs; REC, recovery after the four SDs. Data are depicted as mean ± SEM (n = 4). (C) Effect of SCNx and SD on PER2-bioluminescence amplitude estimated by sinewave fitting (linear mixed model with fixed conditional effects [‘Intact,’ ‘BSL,’ ‘SD,’ and ‘REC’] and random intercept effect [‘Mouse’] followed by Tukey’s post-hoc tests; intact vs. BSL [BSL1 and BSL2], p=0.0045; intact vs. REC, p=0.0015; SD vs. BSL: p=0.0013; SD vs. REC: p<0.001; ★p<0.05). See Figure 2—figure supplement 2 for individual time courses and amplitude estimates of the separate recovery days. Data underlying figures can be found in Figure 2—source data 1.
-
Figure 2—source data 1
This file contains the numerical values on which the graphs in Figure 2A–C are based.
- https://cdn.elifesciences.org/articles/69773/elife-69773-fig2-data1-v2.xlsx
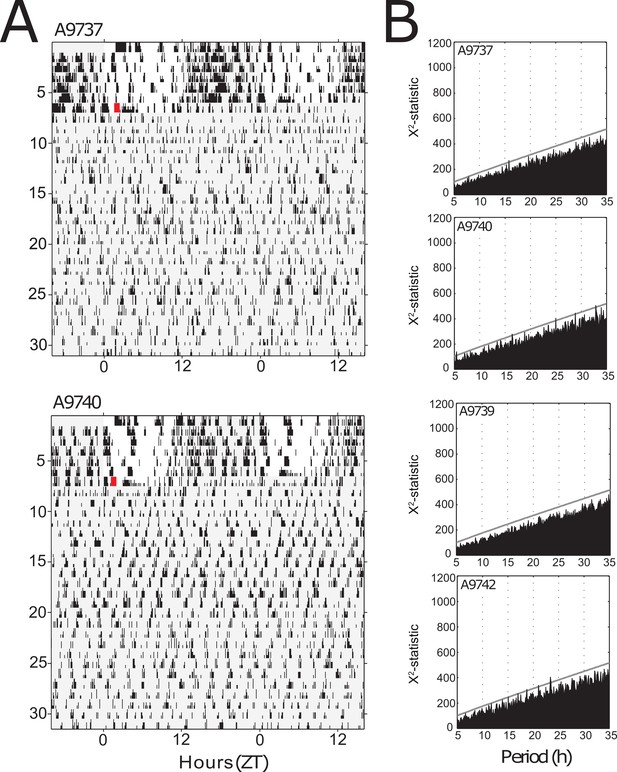
Suprachiasmatic nuclei (SCN) lesion eliminates circadian organization of locomotor activity.
(A) Examples of double-plotted actograms for two mice kept under LD12:12 condition (days 1–7 vertical axis) before bilateral lesion of the SCN (red square) and subsequently released under DD conditions (days 7–31). Light gray areas denote darkness. (B) Chi-square periodograms of locomotor activity under DD following SCN lesion. Analysis in the upper two graphs concerns the mice illustrated in (A). Gray lines in each panel indicate p=0.001 significance level. Data underlying this figure can be found in Figure 2—figure supplement 1—source data 1.
-
Figure 2—figure supplement 1—source data 1
This file contains the numerical values on which the graphs in Figure 2—figure supplement 1A and B are based.
- https://cdn.elifesciences.org/articles/69773/elife-69773-fig2-figsupp1-data1-v2.xlsx

Individual traces of PER2 kidney bioluminescence.
(A) PER2 bioluminescence in intact mice (left) and suprachiasmatic nuclei (SCN)-lesioned mice right. (B) SCN-lesioned mice prior to the sleep deprivation protocol (BSL2), during the SD protocol, and afterward (REC). (C) Amplitude estimates of 3 days (REC1-3) during the recovery following sleep deprivation (amplitude estimate with fixed 24 hr sinewave in Prism; pne-way ANOVA, F(2, 6) = 6.3, p=0.03, post-hoc paired t-test). Data underlying this figure can be found in Figure 2—figure supplement 2—source data 1.
-
Figure 2—figure supplement 2—source data 1
This file contains the numerical values on which the graphs in Figure 2—figure supplement 2A–C are based.
- https://cdn.elifesciences.org/articles/69773/elife-69773-fig2-figsupp2-data1-v2.xlsx
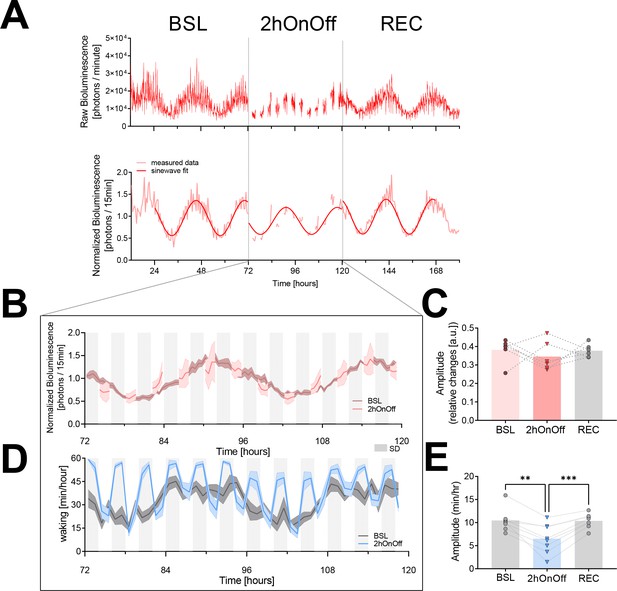
The 2hOnOff protocol reduced the circadian sleep-wake amplitude but did not consistently affect PER2 bioluminescence.
(A) An example of a PER2-bioluminescence recording under baseline, 2hOnOff, and recovery conditions. Upper graph shows bioluminescence data as collected, whereas in the lower graph, the same data are linearly detrended, normalized (relative to the individual overall mean), and averaged over 30 min intervals. A sinewave was fit to the data of the two baseline days, the two 2hOnOff days, and the two recovery days separately (see Materials and methods). (B) The time course of PER2 bioluminescence under baseline conditions and during the 2hOnOff protocol (n = 6, data depicted as mean ± SEM). Light gray squares below the graph mark the 2 hr sleep deprivations (SDs). (C) The amplitude of the PER2-bioluminescence rhythm decreased in four but increased in two mice, resulting in an overall lack of an effect of the 2hOnOff protocol (paired t-test, t(5) = 0.74, p=0.50; mean ± SEM; BSL: 0.38 ± 0.03; 2hOnOff: 0.35 ± 0.03; REC: 0.38 ± 0.01). For all six animals, the amplitude reverted to baseline during recovery. (D) The distribution of waking across the two baseline days (dark gray) and during the 2hOnOff protocol (blue) in EEG-implanted SKH1 mice (n = 8, hourly values depicted as mean ± SEM) and a sinewave fit through the hourly average for visual comparison of BSL to SD. (E) Individual estimates of the amplitude of circadian changes in wakefulness during the 2hOnOff protocol were obtained by fitting a sinewave function to the wakefulness present in consecutive 4 hr intervals (i.e., SD + SOW). This amplitude was smaller compared to the amplitude obtained in baseline using the same approach (one-way ANOVA, F(1.8, 12.3) = 21.9, p=0.0001; post-hoc paired t-test, BSL vs. 2hOnOff: t(7) = 4.9, p=0.002; 2hOnOff vs. REC: t(7) = 6.3, p=0.0004; mean ± SEM; BSL: 10.4 ± 0.9; 2hOnOff: 6.5 ± 1.1; REC: 10.4 ± 0.6). However, a circadian modulation was still present under the 2hOnOff protocol (amplitude > 0, one-sample t-test, t(7) = 5.8, p=0.0007). Note the overall higher levels of wakefulness during 2hOnOff compared to baseline. Data underlying figures can be found in Figure 3—source data 1. BSL, baseline; REC, recovery; SOW, sleep opportunity window.
-
Figure 3—source data 1
This file contains the numerical values on which the graphs in Figure 3A–E are based.
- https://cdn.elifesciences.org/articles/69773/elife-69773-fig3-data1-v2.xlsx
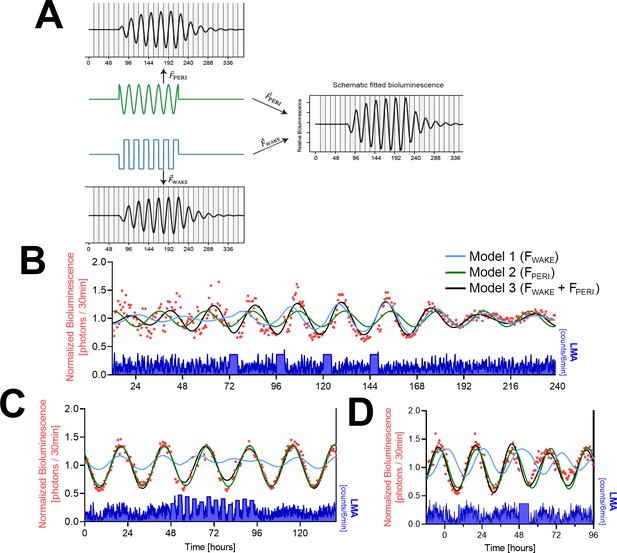
Mathematical modeling of PER2-bioluminescence dynamics.
(A) Schematic view of a driven damped harmonic oscillator. The oscillation (black) is assumed to be driven by two forces: (blue) and (green). In our model, is based on locomotor activity (LMA) (here simplified as a square wave) and on a sinewave. Left panels show the individual effect of each force on the oscillator. Both forces start at t = 72 hr and end at t = 216 hr, illustrating the waxing and waning of the resulting rhythm amplitude. Right panel shows the resulting changes in PER2 bioluminescence when combining both forces. Note that combining the two forces increased amplitude and changed the phase of the oscillation. In this example, the amplitude of the peripheral circadian force is flat at beginning and end to illustrate that the oscillation is not self-sustained. (B) Modeling of the SCNx experiment with the full model (model 3; black line) driving oscillations in PER2 bioluminescence or with either (model 1; blue line) or (model 2; green line) as the only driving force. Red symbols, 30 min PER2-biolumininescence averages. Lower graph in blue: 6 min LMA values. (C) Simulation of the PER2-biolumininescence in the 2hOnOff experiment using parameter estimates listed in Table 1. Details and legend as in (B). (D) Simulation of peripheral PER2-biolumininescence in the 6 hr SD experiment using parameter estimates obtained in (C) (see Table 1). Legend as in (B). Data underlying figures can be found in Figure 4—source data 1.
-
Figure 4—source data 1
This file contains the numerical values on which the graphs in Figure 4B–D are based.
- https://cdn.elifesciences.org/articles/69773/elife-69773-fig4-data1-v2.xlsx

Locomotor activity (LMA) as proxy for wakefulness.
(A) Similar residual sum of squares (RSS) for the prediction of PER2 dynamics were obtained when using waking (striped) or LMA (filled) as an input for in central (left) and peripheral (right) recordings. See also Figure 1—figure supplement 7 for the correlation between LMA and waking. Data underlying this figure can be found in Figure 4—figure supplement 1—source data 1.
-
Figure 4—figure supplement 1—source data 1
This file contains the numerical values on which the graph in Figure 4—figure supplement 1 is based.
- https://cdn.elifesciences.org/articles/69773/elife-69773-fig4-figsupp1-data1-v2.xlsx
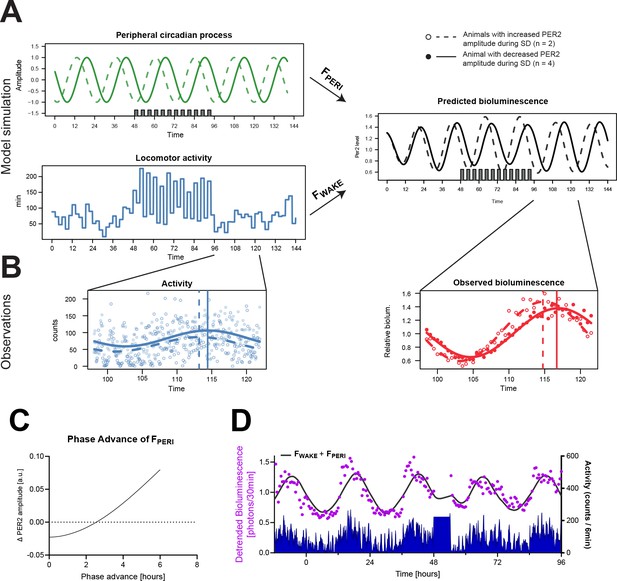
Predicting PER2 bioluminescence considering ’s phase (A–C) and bioluminescence emitted by cortical regions (D).
(A) Model simulation using fitted parameters as in Figure 4C (solid lines) and assuming a 6 hr phase advance of (dashed lines). Amplitude of predicted PER2-bioluminescence level (black lines) increased during 2hOnOff when phase was advanced (dashed), while it decreased using the fitted parameters (solid line). in green; in blue. (B) Phase advance observed for PER2 bioluminescence (red) and wake (blue) in mice at the end of sleep deprivation for mice that increased PER2-bioluminescence amplitude during 2hOnOff (empty circles, n = 2) vs. those in which amplitude decreased (filled circles, n = 4). Phase (vertical line) was calculated using sinewave fitting (solid and dashed lines). (C) Predicting the effect of a phase advance of on PER2 amplitude reveals that when the phase advance >2.5 hr, PER2 amplitude will increase. (D) PER2 bioluminescence predicted based on the parameters obtained for peripheral bioluminescence are not able to predict the earlier, sharp increase of PER2 bioluminescence at activity onset, nor the acute increase following sleep deprivation. Data underlying panels (C) and (D) can be found in Figure 4—figure supplement 2—source data 1.
-
Figure 4—figure supplement 2—source data 1
This file contains the numerical values on which the graphs in Figure 4—figure supplement 2C and D are based.
- https://cdn.elifesciences.org/articles/69773/elife-69773-fig4-figsupp2-data1-v2.xlsx
Tables
Parameter estimates obtained in the model optimization for the suprachiasmatic nuclei lesion (SCNx) and 2hOnOff experiments.
Damping constant, natural angular frequency, and the coefficient were optimized in the SCNx experiment and then used to predict the results of the 2hOnOff experiment. Amplitude and phase of were optimized for both experiments separately. The natural angular frequency defines the intrinsic period of the harmonic oscillation with 0.288 rad * hr–1 corresponding to a period of 21.8 hr and its square, is referred to as the string constant. Phase of the sine function describing is expressed as radians and corresponds to maximum values reached at time 11.8 and 19.4 hr for the SCNx and 2hOnOff experiment, respectively. Values in parenthesis represent the 95% CI.
| Parameters | SCNx | 2hOnOff |
|---|---|---|
| (damping constant) | 0.0155 (0.0103–0.0223) (hr–1) | |
| (natural angular frequency) | 0.288 (0.285–0.291) (rad * hr–1) | |
| (coefficient ) | 6.81e-5 (4.48e-5–9.01e-5) | |
| Model intercept | 0.92 (0.90–0.95) | 0.91 (0.89–0.92) |
| (amplitude ) | 2.92e-3 (2.58e-3–3.32e-3) | 3.87e-3 (3.80e-3–4.00e-3) |
| (phase ) | 4.73 (4.6–4.85) | 2.78 (2.78–2.82) |
Fit statistics for the suprachiasmatic nuclei lesion (SCNx), 2hOnOff, and 6 hr sleep deprivation (SD) experiments using a single force ( or ) or two forces combined ( and ).
Bayesian information criterion (BIC) for each model (lower is better; a BIC difference between two competing models larger than 10 is considered strong support for the model with the lower value). Residual sum of squares (RSS) minimized by the model (lower values reflect a better fit). Support of the driven harmonic model compared to a flat model using Bayes factor (>100 is considered a ‘decisive’ support for the driven harmonic model). Values should be compared only within the same experiment because variance and sample size differed among the experiments.
| BIC | RSS | Bayes factor(model vs. flat) | ||
|---|---|---|---|---|
| SCNx | = | –287.0 | 11.94 | 2.47e32 |
| = | –224.5 | 13.60 | 6.48e18 | |
| = | –427.5 | 8.36 | 7.87e62 | |
| 2hOnOff | = | –28.5 | 11.96 | 1.82e24 |
| = | –536.8 | 1.29 | 4.41e86 | |
| = | –540.1 | 1.27 | 2.27e87 | |
| 6 hr SD | = | 18.2 | 92.50 | 9.15e-17 |
| = | –205.9 | 4.16 | 5.80e48 | |
| = | –311.9 | 2.46 | 6.23e71 |
Additional files
-
Supplementary file 1
Sequence of the pCV100 viral vector construct containing Pkg1-Luc.
- https://cdn.elifesciences.org/articles/69773/elife-69773-supp1-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/69773/elife-69773-transrepform1-v2.pdf





