Unleashing a novel function of Endonuclease G in mitochondrial genome instability
Figures
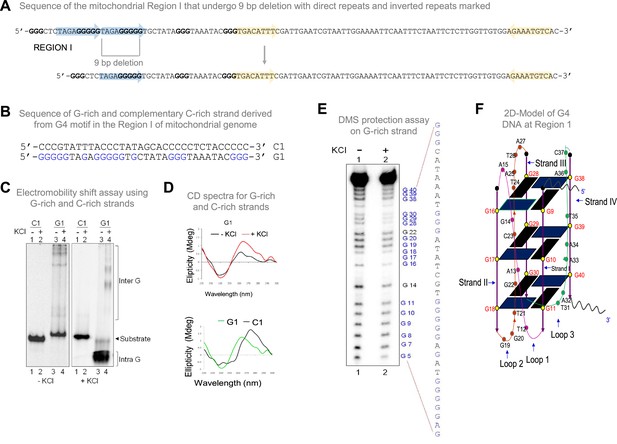
Biochemical studies to investigate formation of G-quadruplex structure in Region I of the mitochondrial genome.
(A) Sequence of the mitochondrial Region I with direct repeats and inverted repeats. Blue-shaded arrows represent the direct repeats, and yellow arrows represent the inverted repeats. The sequence in blue is G rich sequence predicted to form a G-quadruplex structure. The sequence after 9 bp deletion is also shown. (B) The oligomeric sequence of the G-rich strand (indicated by ‘G1’) and the complementary C-rich strand (indicated by ‘C1’). These oligomers are derived from the predicted mitochondrial G-quadruplex forming Region I. The stretches of guanines are marked in blue. (C) The G and C-rich strands were incubated in the presence of 100 mM KCl and resolved on 15% native polyacrylamide gels in the absence (-KCl) or presence (+KCl) of KCl (100 mM) in the gel and running buffer. The substrate, intramolecular (Intra G), and intermolecular (Inter G) quadruplex structures are indicated. (D) CD spectra for G1 in the presence (red) or absence of 100 mM KCl (black). CD spectra for G1 (green) and C1 (black) in the presence of 100 mM KCl. In each case where KCl was added, the respective oligomer resuspended in Tris-EDTA buffer was incubated for 1 h at 37 °C and spectra were recorded using JASCO J-810 spectropolarimeter (scan range of 220–300 nm). (E) DMS protection assay for the Region I of mitochondria. The oligomer was allowed to fold into G4 DNA and then treated with DMS, followed by cleavage with piperidine. The products were resolved on a 15% denaturing PAGE. Substrate indicates the gel-purified DNA from the reaction incubated with or without KCl. All the positions of guanines are indicated. (F) A representative 2D model for intramolecular G-quadruplex structure formed at mitochondrial Region I based on the reactivity of guanine to DMS and CD studies for determining the orientation of DNA fold. Refer also Figure 1—figure supplement 1.
-
Figure 1—source data 1
EMSA and CD studies to show formation of G-quadruplex structure in Region I.
- https://cdn.elifesciences.org/articles/69916/elife-69916-fig1-data1-v3.zip
-
Figure 1—source data 2
Gel shift assay to show abrogation of G-quadruplex structure in mutants of Region I.
- https://cdn.elifesciences.org/articles/69916/elife-69916-fig1-data2-v3.zip
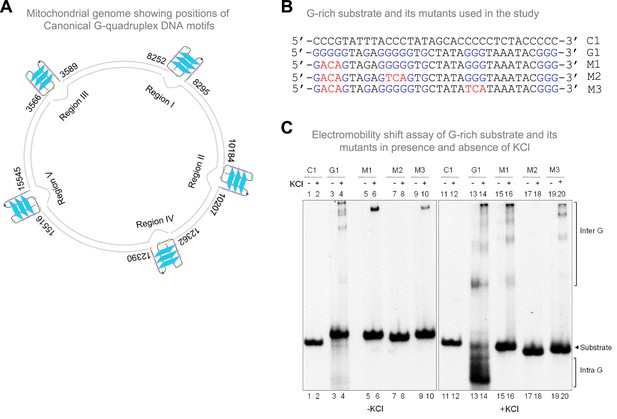
Evaluation of G-quadruplex formation at mitochondrial Region I.
Related to Figure 1. (A) Schematic representation of mitochondrial genome showing positions of five G-quadruplex DNA motifs. (B) List of oligomers used for studying effect of mutation of G stretches on G-quadruplex formation. Guanine stretches are indicated in blue, while sequences in red are mutations introduced to G stretches. (C) Gel shift assay in the absence (left panel) and presence (right panel) of KCl showing the impact of mutation of G stretches on intramolecular G4 DNA formation.
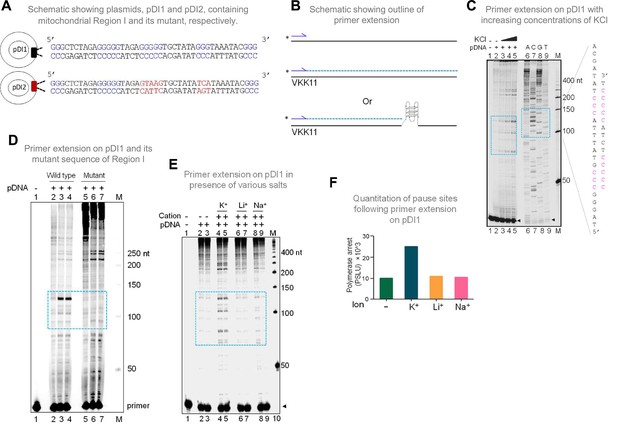
Evaluation of formation of G-quadruplex DNA in Region I of the mitochondrial DNA when cloned into a plasmid.
(A) Schematic showing cloning of the mitochondrial Region I and its mutant to generate plasmids pDI1 and pDI2, respectively. The duplex region containing the G stretches is depicted in blue, while the mutated nucleotides are marked in red. (B) Schematic showing the primer extension across plasmid DNA containing mitochondrial Region I. Positions of primer, VKK11 is indicated. The radiolabeled primer (indicated with an asterisk) binds to one of the strands of the plasmid DNA upon heat denaturation and extends till it encounters a non-B DNA, as it blocks the progression of the polymerase. The products (dotted lines) are then resolved on a denaturing PAGE. (C) The plasmid, pDI1, containing the mitochondrial Region I, was used for primer extension studies using radiolabeled primers for the G-rich (VKK11) in a KCl (25, 50, 100 mM) dependent manner. A sequencing ladder was prepared using VKK11 by the chain termination method of sequencing. Pause sites are shown with dotted rectangles in turquoise. The sequence corresponding to the pause site is boxed, and sequence complementary to G-quadruplex forming motifs are indicated in pink. Sequencing ladder was used as marker. (D) Plasmids, pDI1 and its mutant pDI2, containing mutation in G4 motif, were used for primer extension studies using radiolabelled primer (VKK11). Pause sites are shown with dotted rectangles (turquoise). (E) Evaluation of the effect of different cations (Na+, K+, Li+) on non-B DNA formation at mitochondrial Region I when present on a plasmid (pDI1) by using primer VKK11 in a primer extension assay. The pause sites are indicated in dotted rectangle (turquoise). In panels C-F, ‘M’ denotes the 50 nt ladder. (F) Bar diagram showing the effect of ions in primer extension assay. The values are expressed in PSLU (photo-stimulated luminescence units) representing the extent of pause. Refer also Figure 2—figure supplement 1.
-
Figure 2—source data 1
Primer extension studies to show formation of G-quadruplex structure in Region I containing plasmid.
- https://cdn.elifesciences.org/articles/69916/elife-69916-fig2-data1-v3.zip

Schematic showing cloning of the mitochondrial Region I and its mutant to generate plasmids, pDI1 and pDI2.
Related to Figure 2. The duplex region containing the G stretches are depicted in blue, while the mutated nucleotides are marked in red. A 68 nucleotide sequence is also shown with inverted repeats marked in orange.
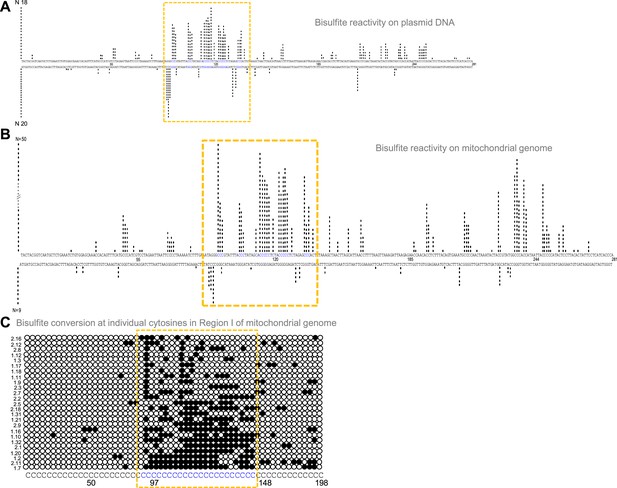
Evaluation of G-quadruplex formation at mitochondrial Region I.
(A) Bisulphite modification assay on plasmid (pDI1) containing mitochondrial Region I. Vertical bar represents the number of times the respective cytosine in the top and bottom strands of mitochondrial Region I is converted to thymine after deamination when treated with sodium bisulphite, followed by PCR. (B, C) Bisulphite sequencing on the mitochondrial genome for determining the formation of G4 DNA at Region I. Each vertical bar represents the conversion of a cytosine to uracil in the top strand or bottom strand. A total of 59 clones were sequenced from both the top and bottom strands (B). Single strandedness was observed in a DNA fragment of 198 nt containing Region I of the mitochondrial genome following bisulphite modification assay (C). Each row represents cytosines present in a DNA molecule. Each dark circle represents the conversion of cytosine to thymine on the top strand after deamination upon treatment with sodium bisulphite, followed by PCR and DNA sequencing. Of the 59 clones sequenced, the most reactive 25 molecules are shown (C). In all the panels, sequences corresponding to the G-quadruplex forming motif are indicated in a dotted rectangular box (mustard yellow). Refer also Figure 3—figure supplement 1.
-
Figure 3—source data 1
Sequence of clones after bisulphite treatment in a plasmid containing Region I of mitochondria.
- https://cdn.elifesciences.org/articles/69916/elife-69916-fig3-data1-v3.zip
-
Figure 3—source data 2
Sequence of clones after bisulphite treatment in a region I of mitochondria.
- https://cdn.elifesciences.org/articles/69916/elife-69916-fig3-data2-v3.zip
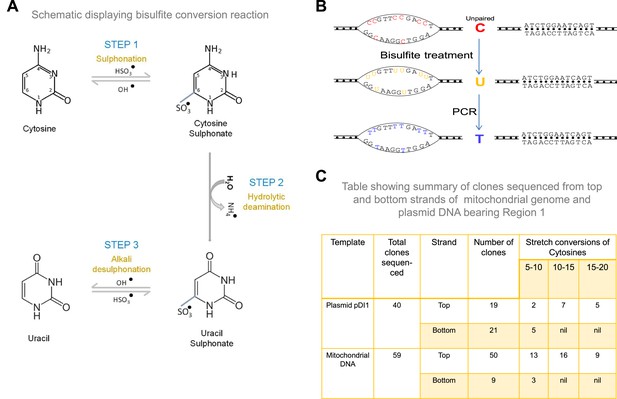
Schematic showing conversion of cytosine to thymine following bisulfite modification assay.
Related to Figure 3. (A, B) Treatment with sodium bisulfite can result in deamination of cytosine, leading to uracil when present on a single-stranded DNA (A). This C to U conversion can lead to C to T change following PCR and sequencing (B). (C) Table showing the summary of clone sequenced from mitochondrial region containing plasmid and mitochondrial genome for bisulfite sequencing for both top and bottom strand.
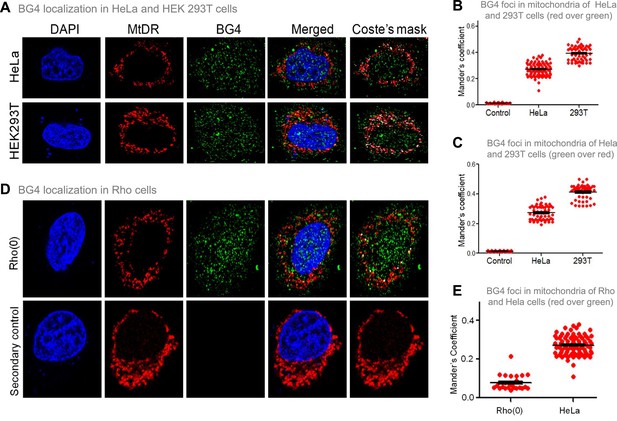
Evaluation of G-quadruplex structure in mitochondria of cells.
(A) Representative images of HeLa and HEK293T cells showing colocalisation of BG4, the G4 binding antibody to mitochondria following immunofluorescence assay. The nucleus is stained with DAPI (blue colour), mitochondria with MitoTracker DR (red) and BG4 with Alexa-Fluor 488 (green). A merged image is shown with a merge of red and green, as depicted by Coste’s mask (colocalisation is represented as a white dot). (B, C) Quantitation showing colocalisation of BG4 with MitoTracker indicated as dot plots. The colocalisation was quantified using Mander’s colocalisation coefficient (ImageJ software) analysing a minimum of 100 cells as red over green (B) and green over red (C). (D) Representative image of Rho(0) cells showing localization of BG4 to mitochondria following immunofluorescence as investigated in panel (A). (E) Quantitation showing comparison of colocalization of BG4 between HeLa cells and Rho(0) cells shown as dot plots. The colocalization was quantified using Mander’s colocalization coefficient analyzing a minimum of 50 cells as red over green. Refer also Figure 4—figure supplement 1.
-
Figure 4—source data 1
Localization of BG4 to mitochondria.
- https://cdn.elifesciences.org/articles/69916/elife-69916-fig4-data1-v3.zip
-
Figure 4—source data 2
Immunofluorescence showing the localization of BG4 to mitochondria.
- https://cdn.elifesciences.org/articles/69916/elife-69916-fig4-data2-v3.zip
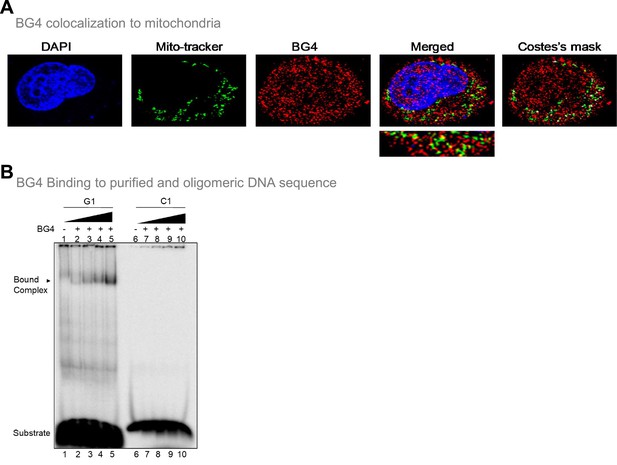
Evaluation of existence of G-quadruplex in mitochondria of cells.
Related to Figure 4. (A) Immunofluorescence study showing colocalization of BG4 (antibody that binds to G4 DNA) and mitochondrial genome. Nucleus from HeLa cells is stained with DAPI (blue color). Mitochondria are stained with MitoTracker green FM and G4 DNA with BG4 (Alexa-Fluor 568), a merged image is shown. A merge of red and green is depicted by Coste’s mask (colocalization is represented as a white dot). The experiments were done independently in HeLa and HEK 293T cells and the quantitation of colocalization of BG4 with MitoTracker is shown in Figure 2. (B) Increasing concentrations (0.2, 0.5, 1, and 2 µg) of purified BG4 was incubated with G-rich (G1) and C-rich (C1) oligomers and products were resolved on 5% native polyacrylamide gel. In each case a lane without protein (lane 1 and 6) served as control. The substrate and the bound complex are marked.
Evaluation of BG4 binding to G-quadruplex structure in mitochondrial genome.
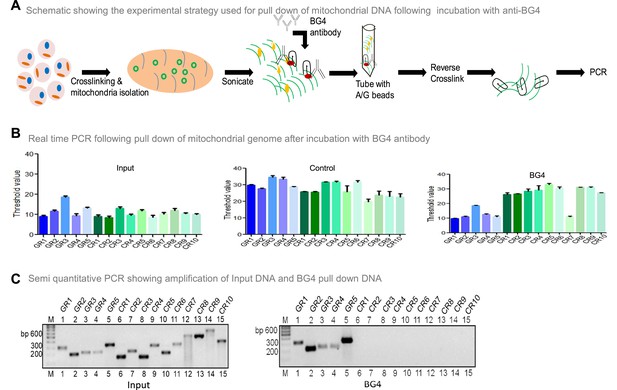
(A) Schematic showing the experimental strategy used for mito-IP using anti-BG4. Briefly, cells were crosslinked and then mitochondria were isolated and sonicated to obtain the small fragments of mitochondrial DNA. Purified BG4 antibody was used along with protein A/G agarose beads to pull down the BG4 bound regions. (B) BG4 bound mtDNA was purified after reverse crosslinking and used for real-time PCR using primers derived from different regions of the mitochondrial genome, which include 5 G-quadruplex forming regions and 10 random regions. Input DNA served as template control. No antibody control was also used. Bars in blue (first 5) are for G-quadruplex forming regions, while green (last 10) are for random regions. Y-axis depicts threshold Ct value obtained following real time PCR for each primer. Error bar represents mean ± SEM. (C) Agarose gel profile showing the amplification of Input DNA (left panel) and BG4 pull down DNA (right panel). ‘M’ denotes 100 bp ladder. Refer also Figure 5—figure supplement 1.
-
Figure 5—source data 1
BG4 ChIP to show the binding of BG4 to mitochondrial G-quadruplex forming regions.
- https://cdn.elifesciences.org/articles/69916/elife-69916-fig5-data1-v3.zip

Evaluation of existence of G-quadruplex in mitochondrial DNA.
Related to Figure 5. Schematic showing the position of primers used for mito IP studies. GR1-GR5 represents primers that can amplify G-quadruplex forming motifs and CR1-CR10 represents the random control regions.
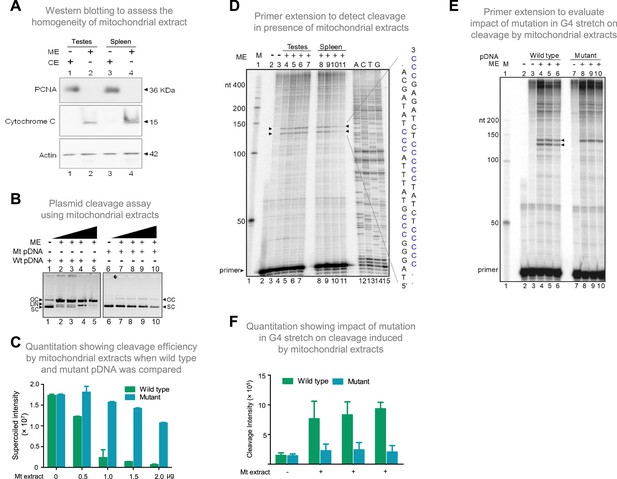
Evaluation of mitochondrial extract induced cleavage at G4 DNA formed at Region I of the mitochondrial genome.
(A) Purification and characterization of mitochondrial extracts. Mitochondrial extract was prepared from rat testis and spleen, and its purity was evaluated using specific markers by western blotting. Purity of CE (cytosolic extract) and ME (mitochondrial extract) were determined using antibodies against PCNA (nuclear and cytoplasmic marker), cytochrome C (mitochondrial marker) and Actin (loading control). (B) In vitro nicking assay on a plasmid containing wild type (WT pDNA) mitochondrial Region I (pDI1) and mutant (Mt pDNA) of Region I (pDI2). Both wild type and mutant plasmids were incubated with mitochondrial extract from rat testes (ME) and resolved on a 0.8% agarose gel. Lanes 1 and 6 served as the control without any extract in the reaction, whereas lanes 2–5 and lanes 7–10 are with increasing concentration (0.25, 0.5, 1, and 2 µg) of mitochondrial extract for pDI1 and pDI2, respectively. ‘OC’ is open circular, ‘LIN’ is linear, and ‘SC’ is supercoiled. (C) Quantification showing the efficiency of mitochondrial extracts (Mt extracts) mediated cleavage when a plasmid containing mutant and wild type G4 motif derived from Region I was compared. The values in Y-axis are expressed in PSLU (photo-stimulated luminescence units) representing the extent of pause. (D) Primer extension assay on plasmid containing mitochondrial Region I (pDI1) to determine the position and location of cleavage. pDI1 was incubated with either mitochondrial extract prepared from testes or spleen in an increasing concentration (0.25, 0.5, 1, and 2 μg), reaction products were purified and used for primer extension assay. Pause sites are indicated, which correspond to G4 DNA motif. Sequencing ladder was used as marker. Complementary sequence corresponding to G-quadruplex forming motif is indicated in blue. ‘M’ is 50 nt ladder. (E) Primer extension assay on pDI1 and pDI2 (G4 mutant) following incubation with mitochondrial extract prepared from rat testis (1 μg). ‘M’ is 50 nt ladder. ‘+’ indicates addition of 1 μg mitochondrial extract shown in triplicates. (F) Quantification showing the cleavage efficiency of mitochondrial extracts (testes) when a plasmid containing mutant and wild type G4 motif derived from Region I was incubated. Cleavage intensity is shown in Y-axis. Error bar represents ± SEM. Refer also Figure 6—figure supplements 1 and 2.
-
Figure 6—source data 1
Gel profiles showing the mitochondrial induced cleavage at mitochondrial region I.
- https://cdn.elifesciences.org/articles/69916/elife-69916-fig6-data1-v3.zip
-
Figure 6—source data 2
Cleavage assay on plasmid bearing wildtype and mutant G4 sequence.
- https://cdn.elifesciences.org/articles/69916/elife-69916-fig6-data2-v3.zip
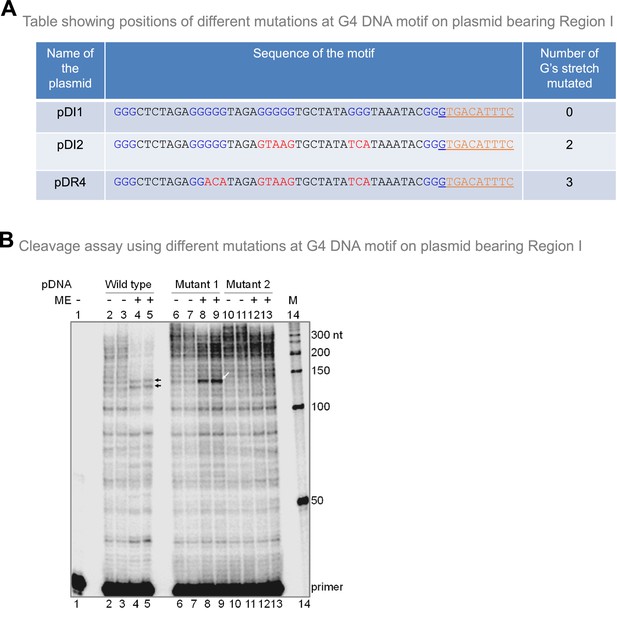
Mitochondrial extract mediated cleavage assay on plasmids bearing different mutations at G4 DNA motif containing plasmid.
Related to Figure 6. (A) Table showing the positions of different mutations at G4 DNA motif on plasmid bearing Region I. (B) Cleavage assay using different mutations at G4 DNA motif on plasmid bearing Region I.
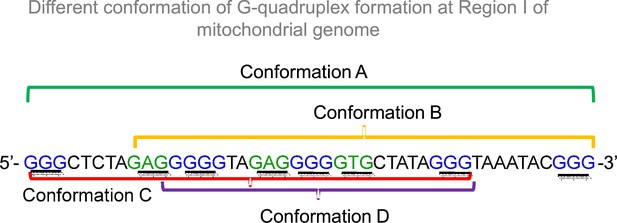
DNA sequences that support formation of different conformation of G-quadruplex DNA structures in Region I.
Related to Figure 6. The presence of five G-stretches (shown in blue) and three GNG stretches (shown in green) allows this region to fold into four different conformations of G-quadruplex structure.
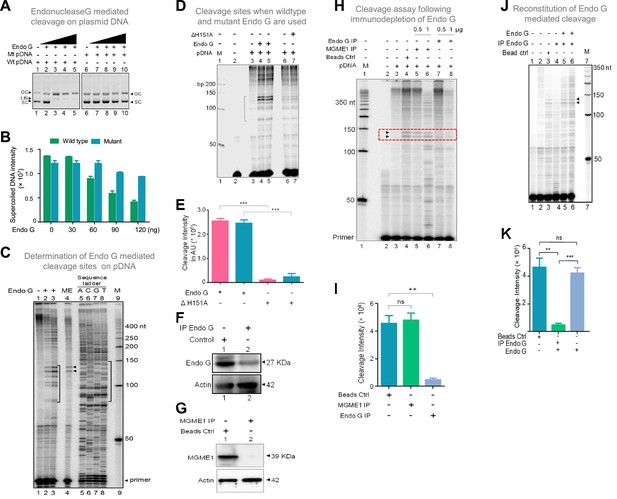
Studies to identify mitochondrial nuclease responsible for cleavage at G4 DNA formed at Region I of the mitochondrial genome.
(A) In vitro nicking assay using purified Endonuclease G on wild type and mutant plasmids containing mitochondrial Region I (pDI1 and pDI2). Both wild type and mutant plasmids were treated with increasing concentration of purified Endonuclease G and resolved on a 0.8% agarose gel. Lanes 1 and 6 served as the control without any protein in the reaction whereas lanes 2–5 and lanes 7–10 are with increasing concentration (30, 60, 90, 120 ng) of purified Endonuclease G. (B) Quantification showing the efficiency of Endonuclease-G-mediated cleavage when a plasmid containing mutant and wild type G4 motif derived from Region I was compared. (C) Cleavage assay was performed on a plasmid containing mitochondrial Region I, pDI1 following incubation with purified Endonuclease G. Primer extension assay was carried out using [γ-32P] radiolabeled VKK11 primer and resolved on 8% denaturing polyacrylamide gel. Lanes 2 and 3 represent 30 and 60 ng of Endonuclease G incubated samples. Lane 1 is without protein, lane 4 is with mitochondrial extract (ME). and lanes 5–8 are A, C, G, T represents the sequencing ladder. ‘M’ is 50 bp ladder. Marked regions represent the specific cleavage products. (D) Cleavage assay was performed on a plasmid containing mitochondrial Region I, pDI1 following incubation either with purified Endonuclease G or mutant Endonuclease G (ΔH151A). Primer extension assay was carried out using γ-32P radiolabeled VKK11 primer and resolved on 8% denaturing polyacrylamide gel. Lane 2 is primer alone, Lanes 3 and 6 are without protein, lanes 4 and 5 are with 30 and 60 ng of Endonuclease G, lanes 7 and 8 are with 30 and 60 ng of mutant Endonuclease G incubated samples. ‘M’ is 50 bp ladder. (E) Quantification showing the efficiency of wild type and mutant Endonuclease G (ΔH151A) mediated cleavage when a plasmid containing wild type G4 motif derived from Region I was compared. (F) Western blotting showing immunodepletion of Endonuclease G from rat testicular mitochondrial extracts. Protein A/G beads were incubated with anti-Endonuclease G and then with the extracts. Actin served as a loading control. (G) Immunodepletion of another endonuclease present in mitochondria, MGME1 from rat testicular mitochondrial extracts as described in panel F. (H) Endonuclease G or MGME1-depleted extract was incubated with pDI1 and used for the primer extension using VKK11 primer. Extract without the addition of antibody served as bead control (lane 4). Lane 3 is no protein control, lanes 5 and 6 corresponds to increasing concentrations of MGME1 depleted extracts, while in lanes 7 and 8, increasing concentrations of Endonuclease G depleted extracts were added. ‘M’ is 50 nt ladder. Cleavage positions are indicated by arrow and boxed (red). (I) Bar diagram depicting quantitation showing the impact of immunoprecipitation of Endonuclease G based on multiple experiments. (J) Reconstitution assay was performed by the addition of purified Endonuclease G following its immunodepletion. Lane 2 represents the primer alone; Lane 3 represents the beads control; lane 4 represents the Endonuclease G depleted extract while lanes 5 and 6 represent the addition of purified Endonuclease G to the depleted extracts (performed in duplicate reaction). (K) Bar diagram representing the cleavage intensity after reconstitution assay as shown in panel J. In panels, C, D, H and J, ‘M’ represents 50 nt ladder. In panels E, I and K, quantitation is based on three biological repeats and data is shown with error bar calculated as mean ± SEM (ns not significant, *p<0.05, **p<0.005, ***p<0.0001). Refer also Figure 7—figure supplement 1.
-
Figure 7—source data 1
Gel profiles showing the Endonuclease-G-induced cleavage at mitochondrial region I.
- https://cdn.elifesciences.org/articles/69916/elife-69916-fig7-data1-v3.zip
-
Figure 7—source data 2
Gel profiles showing the purification of different endonucleases.
- https://cdn.elifesciences.org/articles/69916/elife-69916-fig7-data2-v3.zip
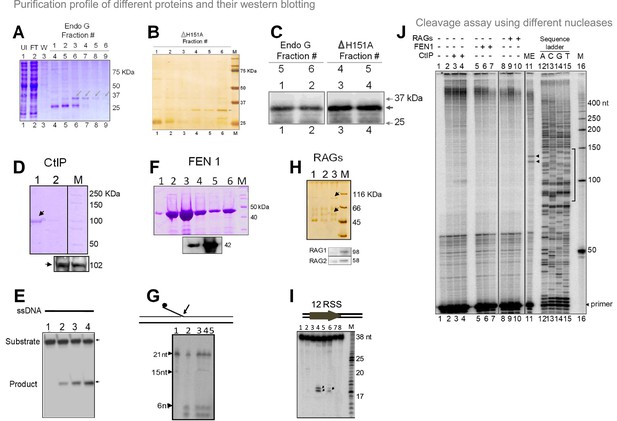
Overexpression, purification and activity assay of different endonucleases.
Related to Figure 7. (A, B) Overexpression and purification of Endonuclease G protein (wild type and mutant). The purity and identity of the protein was confirmed using SDS-PAGE for wild type (A) and mutant (B). (C) The identity and purity of the protein was confirmed using the western blotting for both wild type and mutant. Fractions 5 and 6 (wild type) and 4 and 5 (ΔH151A) were used for the western blot in both the cases. (D). SDS gel profile showing the overexpression and purification of CtIP. Western blotting (bottom panel) was performed for confirmation of the protein. (E) Activity assay showing resection of the substrate (ssDNA) upon addition of increasing concentrations (30, 60 and 90 ng) of purified CtIP (lanes 2–4). (F) SDS gel profile showing the overexpression and purification of FEN1. Western blotting confirming the identity of the protein is also shown. (G) Activity assay showing the cleavage of the substrate with flap DNA upon the addition of increasing concentrations (50, 100, and 500 ng) of purified FEN1 (lanes 2–5). (H) Silver-stained gel profile showing the overexpression and purification of RAGs from mammalian cells. Western blotting confirming the presence of RAG1 and RAG2. (I) Activity assay showing the cleavage of the RSS substrate upon addition of different fractions of purified RAGs (lanes 2–8). (J) Cleavage assay for different nucleases was performed on a plasmid containing mitochondrial Region I, pDI1. Following incubation with different purified nucleases CtIP, FEN1, and RAGs, primer extension assay was carried out using γ-32P radiolabeled VKK11 primer and resolved on 8% denaturing polyacrylamide gel along with sequencing ladder. Lane 1 is primer alone, lanes 2, 5, 8 are without protein, lane 11 is with mitochondrial extract and lanes 12–15 (A, C, G, T) represents the sequencing ladder. Lanes 3,4 represents (CtIP); lanes 6,7 (FEN1); lanes 9, 10 (RAGs) incubated samples. In each case, 30 and 60 ng of protein was used. ‘M’ is 50 bp ladder.
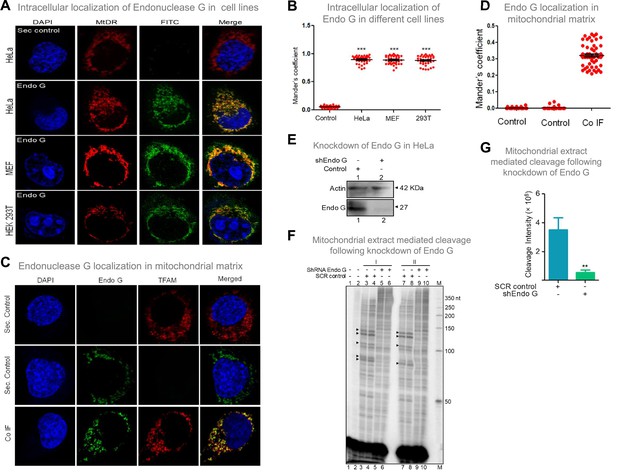
Evaluation of expression of Endonuclease G within different mammalian cells and Endonuclease-G-mediated cleavage at mtDNA following shRNA mediated knockdown within cells.
(A) Localization of Endonuclease G in mitochondria in different cell lines. Representative images of localization of Endonuclease G to mitochondria in HeLa, MEF and HEK293T cells. FITC-conjugated secondary antibodies were used for detecting Endonuclease G proteins. MtDR is Mitotracker Red, a mitochondrial marker. DAPI is used as nuclear stain. (B) Colocalization analyses of Endonuclease G and Mitotracker signals using JaCoP in ImageJ software based on immunofluorescence studies performed in multiple cell lines (see Figure 8—figure supplement 1). Minimum of 50 cells were used for analysis of colocalization of red and green signals and plotted as the colocalization value of green overlapping red. Y-axis depicts the Mander’s colocalization coefficient value calculated for green over red and plotted in the form of dot plot. The significance was calculated using GraphPad Prism 5.0 with respect to secondary control and shown as mean ± SEM (ns not significant, *p<0.05, **p<0.005, ***p<0.0001). (C) Representative images of colocalization of Endonuclease G to mitochondrial matrix (TFAM) in HeLa cells. Conjugated secondary antibodies were used for detecting Endonuclease G (Alexa Fluor 488) and mitochondrial matrix protein, TFAM (Alexa Fluor 568). DAPI is used as nuclear stain. (D) Colocalization analyses of Endonuclease G and TFAM signals using JaCoP in ImageJ software. Minimum of 50 cells were used for analysis of colocalization of red and green signals and plotted. Y-axis depicts the Mander’s colocalization coefficient value calculated for green over red and plotted in the form of dot plot. Control represents the panel where only one of the primary antibodies was used. (E) Knockdown of Endonuclease G from HeLa cell using PEI mediated transfection. shRNA against Endonuclease G cloned plasmid was used for transfection. Cells were harvested after 48 h and mitochondrial extracts were prepared. Western blotting was performed to confirm the knockdown of Endonuclease G from the HeLa cells. Actin served as a loading control. (F) The knockdown extract was incubated with the plasmid and used for the primer extension using VKK11 primer (‘I’ and ‘II’ are two biological repeats). Extract prepared from the sample transfected with scrambled plasmid served as a control (SCR control). Lanes 3, 4, 7, and 8 serve as scrambled controls while lanes 5, 6, 9, and 10 are for knockdown extracts. I and II represent two independent batches of experiments. ‘M’ is a 50 nt ladder. (G) Bar diagram representing the cleavage intensity of the extracts prepared after transfection with scrambled plasmid and shEndo G plasmid. In panels F and G, a minimum of three biological repeats were performed and the data is shown with the error bar calculated as SEM (ns: not significant, *p<0.05, **p<0.005, ***p<0.0001). Refer also Figure 8—figure supplement 1.
-
Figure 8—source data 1
Localization of Endonuclease G to mitochondria.
- https://cdn.elifesciences.org/articles/69916/elife-69916-fig8-data1-v3.zip
-
Figure 8—source data 2
Immunofluorescence showing the Localization of Endonuclease G to mitochondria.
- https://cdn.elifesciences.org/articles/69916/elife-69916-fig8-data2-v3.zip
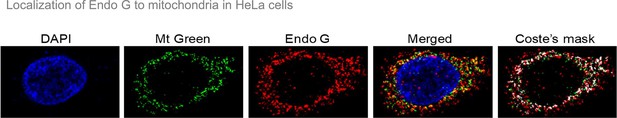
Analysis of Endonuclease G localization in mitochondria.
Related to Figure 8.
Representative image showing colocalization of Endonuclease G using MitoTracker Green FM (Mt Green) in HeLa cells. Alexa-568 conjugated secondary antibody was used for the detection of Endonuclease G. DAPI is used to stain the nucleus.
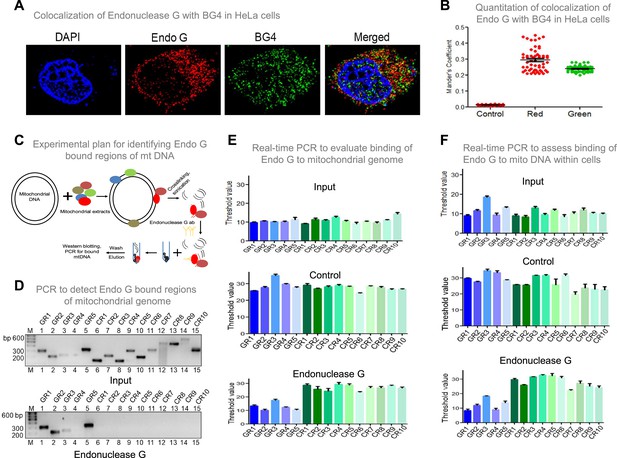
Investigation of binding efficacy of Endonuclease G to G4 DNA at Region I of mitochondrial genome.
(A) Representative image showing colocalization of Endonuclease G with BG4 in HeLa cells. Alexa Fluor 568 and Alexa Fluor 488 conjugated secondary antibodies were used for detection of Endonuclease G and BG4 proteins, respectively. DAPI was used as nuclear stain. (B) The quantitation showing colocalization of Endonuclease G and BG4. The colocalization was quantified using Mander’s colocalization coefficient (ImageJ software) by analyzing a minimum of 100 cells and presented as a dot plot. Red plot represents the overlapping of Endonuclease G over BG4 while green plot represents the overlapping of BG4 over Endonuclease G. (C) Schematic showing the pull-down assay used for evaluation of binding of Endonuclease G present in the rat testicular mitochondrial extracts to the mitochondrial genome. Bound regions were pulled out using anti-Endonuclease G and protein A/G beads. Regions of interest were detected by either semi-quantitative PCR or real-time PCR using appropriate primers. (D) Agarose gel profile showing the amplification through semi-quantitative PCR of Input DNA (upper panel) and Endonuclease G pull down DNA (lower panel). Primers specific to 5 G-quadruplex forming regions (GR1-GR5) and 10 random regions (CR1-CR10) were also used for the amplification. (E) Real-time PCR of 5 G-quadruplex forming regions (blue) and 10 random regions (green) following pull-down assay. Input DNA served as template control. Antibody control served as a negative control. Error bar represents three independent biological repeats. (F) Evaluation of binding of Endonuclease G to different regions of the mitochondrial genome within cells by mito IP. Cells were crosslinked and then mitochondria were isolated. Endonuclease G bound DNA was obtained and was amplified for different regions of mitochondria, as explained in panel E. Graph is plotted for the Ct values obtained following real-time PCR as described above. The error bar represents three independent biological repeats. Refer also Figure 9—figure supplement 1, Figure 2.
-
Figure 9—source data 1
ChIP assay showing the binding of Endonuclease G with the mitochondrial G-quadruplex regions within cells.
- https://cdn.elifesciences.org/articles/69916/elife-69916-fig9-data1-v3.zip
-
Figure 9—source data 2
ChIP assay showing the binding of Endonuclease G with the mitochondrial G-quadruplex regions when purified Endonuclease G was used.
- https://cdn.elifesciences.org/articles/69916/elife-69916-fig9-data2-v3.zip
-
Figure 9—source data 3
P1 nuclease assay showing the binding of Endonuclease G to mitochondrial G quadruples regions.
- https://cdn.elifesciences.org/articles/69916/elife-69916-fig9-data3-v3.zip
Binding of Endonuclease G to G-quadruplex regions of the mitochondrial genome.
Related to Figure 9. (A) Representative immunofluorescence images showing the colocalization of Endonuclease G and BG4. The ‘Merged’ image shown in left is a colocalization of DAPI, Endonuclease G and BG4 foci, while ‘Merged’ in right shows colocalization of Endonuclease and BG4 foci. (B) Schematic showing the binding of purified Endonuclease G to the mitochondrial genome and the pull-down of a bound region using Endonuclease G antibody and protein A/G beads. These regions were then used for semi-quantitative and real-time PCR using appropriate primers. (C) Schematic showing the position of primers used for mito-IP studies. The upper panel shows the primer positions for G-quadruplex forming regions and lower panels for control regions. (D) 5 G-quadruplex forming regions and 10 random regions were used for amplification using mito-IP DNA as template (lower panel). Upper panel shows the amplification for input DNA. (E) Bar graph was plotted for the threshold cycle against different primers following real-time PCR. Input DNA served as template control. ‘Antibody’ control in which no antibody was added served as negative control. Blue bars represent G-quadruplex regions, while green bars represent the control regions. Error bar represents three independent biological repeats.
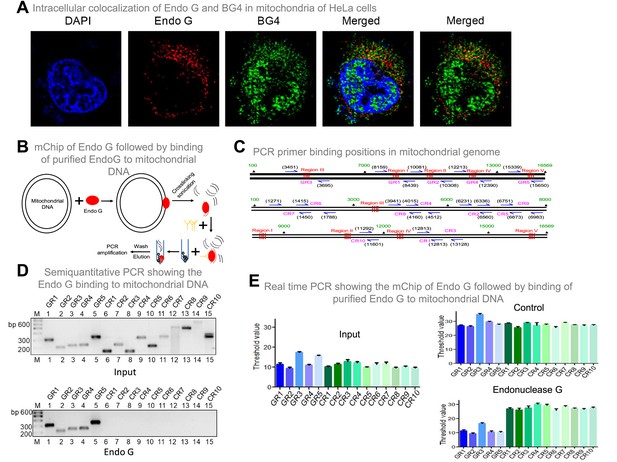
Binding of Endonuclease G to G-quadruplex regions of the mitochondrial genome.
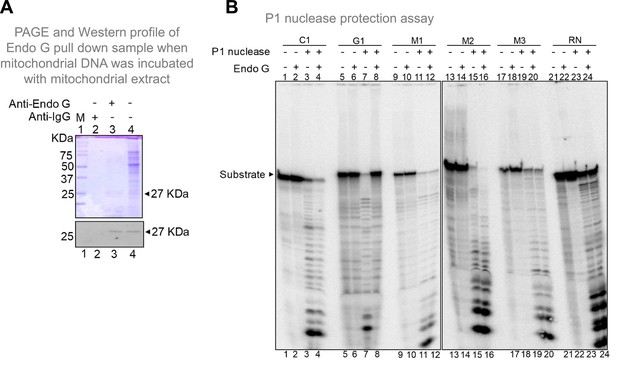
Related to Figure 9. (A) SDS profile and western blotting of the pulldown sample when mitochondrial DNA was incubated with mitochondrial extracts. As described in the methodology, mitochondrial extract was allowed to bind to mitochondrial DNA and Endonuclease G bound DNA was pulled down using Endonuclease G antibody. After the pull down, the extract was loaded into a SDS PAGE with Endonuclease G (lane 3) or IgG antibody (lane 2). For reference, mitochondrial extract was also loaded (lane 4). M is the protein marker. For other details refer main Figure 9. (B) P1 nuclease foot-printing to investigate the binding of Endonuclease G to G-quadruplex forming Region I. Radiolabelled oligomers were incubated with Endonuclease G protein and subjected to P1 nuclease and electrophoresed on 18% denaturing PAGE. In each case, lanes 1, 5, 9, 13, 17, 21 are substrate alone, lanes 2, 6, 10, 14, 18, 22 are Endonuclease G alone. Lanes 3, 7, 11, 15, 19, 23 are P1 nuclease alone treated samples and lanes 4, 8, 12, 16, 20, 24 are Endonuclease G plus P1 nuclease treated samples. C1 is C-strand, G1 is G strand, M1, M2 and M3 are mutants while random sequence (RN) is the oligomer with equal G-C content as G1 in a random manner. 50 ng of purified Endonuclease G and 0.03 U of P1 nuclease was used for the assay.
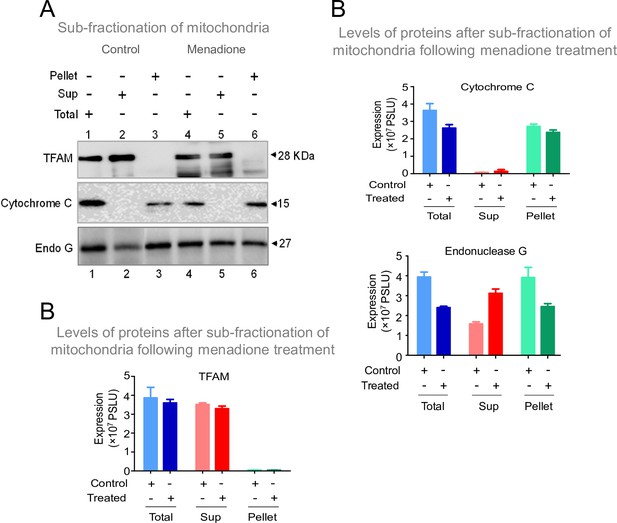
Investigation of stress conditions that favor the transport of Endonuclease G to mitochondrial matrix.
(A) Western blot showing the presence of Endonuclease G, Cytochrome C and TFAM in either total or supernatant (Sup) and pellet fraction with or without menadione treatment (25 µM) following sub-fractionation of mitochondrial compartments. Lanes 1–3 are for control samples and Lanes 4–6 are menadione treated samples. (B) Bar graph showing the quantitation of presence of Endonuclease G, Cytochrome C and TFAM in either total mitochondria or supernatant (Sup) and pellet fraction with or without menadione treatment (25 µM). Quantitation is based on three biological repeats and the data is shown with the error bar calculated as SEM. The values in Y-axis are expressed in PSLU (photo-stimulated luminescence units) representing the expression.
-
Figure 10—source data 1
Sub localization of Endonuclease G with or without induction of stress.
- https://cdn.elifesciences.org/articles/69916/elife-69916-fig10-data1-v3.zip
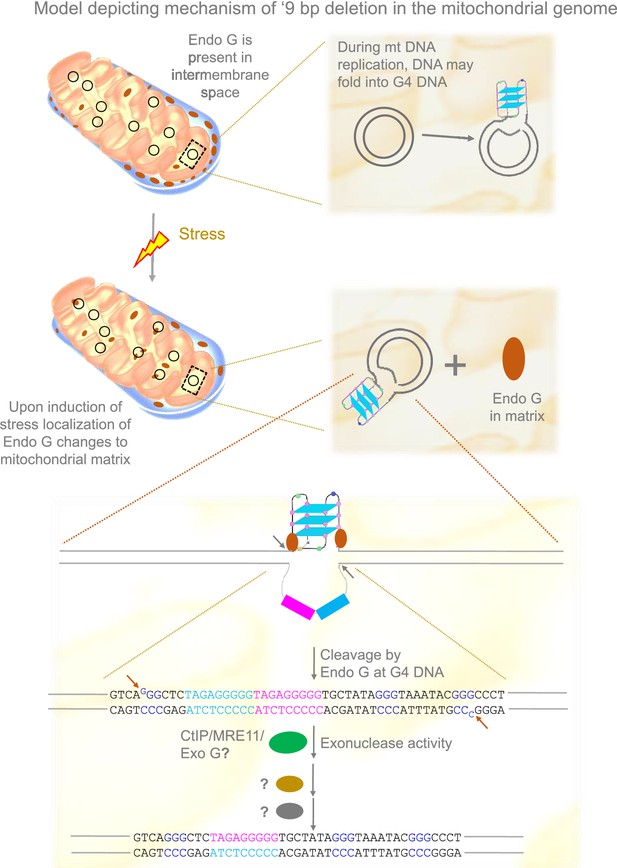
Model depicting mechanism of generation of ‘9 bp deletion’ seen in the mitochondrial genome.
When mitochondria are under stress, Endonuclease G releases into matrix from inner membrane space. In the matrix, Endonuclease G binds and induces cleavage at single-double stranded junctions of G4 DNA as indicated by arrows. Exonuclease action (CtIP/MRE11/Exo G) exposes the direct repeats/microhomology region, which is then paired with the help of unknown proteins, most likely using direct repeats. Ligation of broken ends can result in a ‘9 bp deletion’ at the Region I of the mitochondrial genome.
Tables
Oligomers used in the study.
| Oligomer Name | Sequence | Region |
|---|---|---|
| VKK11 | 5’- GCTGTGTCGACTACTACGGTCAATGCTCTG –3’ | GR1 |
| VKK12 | 5’- CTGAGGTCGACTGGGTGATGAGGAATAGTG - 3’ | |
| RBK46 | 5’- TAATCAACACCCTCCTAGCC –3’ | GR2 |
| VKK14 | 5’- GATAGTGTCGACGGCTCATGGTAGGGGTAA –3’ | |
| SD10 | 5'- TTCGCTGACGCCATAAAACT –3' | GR3 |
| SD11 | 5'- ATCAGGGCGTAGTTTGA –3' | |
| SD12 | 5'- GCTCACAAGAACTGCTAA –3' | GR4 |
| SD13 | 5'- TGGATGCGACAATGGAT –3' | |
| SD14 | 5'- TCTTGCACGAAACGGGAT –3' | GR5 |
| SD15 | 5'- TAGGATGAGGATGGATAGT –3' | |
| RBK15 | 5’- CTACTCCTGCTCGCATCTGC –3’ | CR2 |
| RBK16 | 5’- GAAGGTGGTGTTGAGGTTGC –3’ | |
| RBK17 | 5’- GCATTGTTCGTTACATGGTCC –3’ | CR3 |
| RBK18 | 5’- GTGGAAGCGGATGAGTAAGAAG –3’ | |
| RBK19 | 5’- CTCACCACTACAATCTTCCTAG –3’ | CR4 |
| RBK20 | 5’- CAAAGATGGTAGAGTAGATGACG –3’ | |
| RBK21 | 5’- CTAACCATCTTCTCCTTACACCTAG –3’ | CR5 |
| RBK22 | 5’- GTTTGCTAATACAATGCCAGTCAGG –3’ | |
| RBK23 | 5’- CGAAGGTGGATTTAGCAGTAAACTG –3’ | CR6 |
| RBK24 | 5’- CGGTACTATATCTATTGCGCCAGG –3’ | |
| RBK41 | 5’- GTATCATCAACTGATGAGCAAG –3’ | CR1 |
| SD2 | 5’- TCAGCAAACCCTGATGAA –3’ | CR7 |
| SD3 | 5'- CACTCTACTCTCAGTTTACT –3' | |
| SD4 | 5'- ACATCGAATACGCCGCA –3' | CR8 |
| SD5 | 5'- AGTTGGTCGTAGCGGAATCG –3' | |
| SD6 | 5'- TAGGGTTTATCGTGTGAG –3' | CR9 |
| SD7 | 5'- AGTGTGGCGAGTCAGCT –3' | |
| SD8 | 5'- TACTCACTCTCACTGCCCAA –3' | CR10 |
| SD9 | 5'- TGTTTGTCGTAGGCAGAT-3' | |
| VKK21 | 5'- GGATCCATGCGGGCGCTGCGG –3' | |
| VKK22 | 5'- GCGGCCGCTCACTTACTGCCCG –3' | |
| DI12 | 5'- GCAAACCACAGTTTCATGCCCATC –3' | |
| DI13 | 5'- GCCTATAATCACTGCGCCCGCTC –3' | |
| VKK1 | 5’- CCCGTATTTACCCTATAGCACCCCCTCTACCCCC –3’ | C1 |
| VKK2 | 5’- GGGGGTAGAGGGGGTGCTATAGGGTAAATACGGG –3’ | G1 |
| VKK5 | 5’- GTCAGTAGAGGGGGTGCTATAGGGTAAATACGGG –3’ | M1 |
| VKK6 | 5’- GTCAGTAGAGAATGTGCTATAGGGTAAATACGGG –3’ | M2 |
| VKK7 | 5’- GTCAGTAGAGGGGGTGCTATATCATAAATACGGG –3’ | M3 |
| SD 54 | 5'- GGCCAGGGCCCCGCGGTCGAAGCCACTGCC-3' | |
| SD 57 | 5'- TCACCTGGCCGCCGCCGCCAACCAC-3' | |
| DK27 | 5’-TGGGCTCTAGAGGACATAGAGTAAGTGCT-3’ | |
| DK28 | 5’-AGCACTTACTCTATGTCCTCTAGAGCCCA-3’ | |
| KD14 | 5’-CAAGCTCGAAATTAACCCTCAC-3’ | |
| KD13 | 5’-CCCAGTCACGACGTTGTAAAAC-3’ | |
| DI8 | 5’-CTTACAGTGGGCTCTAGAGGGGGTAGATAATAT GCTATAGGGTAAATACTCACTAAAAATCTTTGAA-ATAGGG –3’ | |
| DI9 | 5’-CTAAAAATCTTTGAAATAGGGTGAGTATTTA CCCTATAGCATATTATCTACCCCCTCTAGAGCCCA-CTGTAAG –3’ |





