Purified EDEM3 or EDEM1 alone produces determinant oligosaccharide structures from M8B in mammalian glycoprotein ERAD
Figures
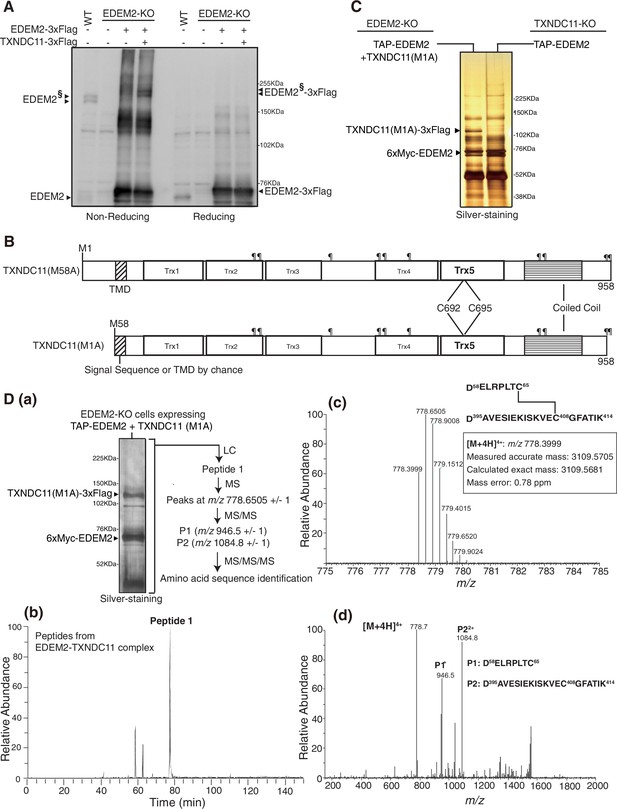
Identification of a disulfide-linked peptide in EDEM2-TXNDC11 complex by liquid chromatography (LC)/mass spectrometry (MS).
(A) Cell lysates were prepared from untransfected wild-type (WT) cells and EDEM2-knockout (KO) cells untransfected or transfected with (+) or without (-) plasmid to express EDEM2-3xFlag or TXNDC11-3xFlag, subjected to SDS-PAGE under non-reducing and reducing conditions, and analyzed by immunoblotting using anti-EDEM2 antibody. EDEM2§ denotes EDEM2 stably disulfide-bonded to TXNDC11. (B) Structures of the M58A and M1A mutants of human TXNDC11 containing the transmembrane domain (TMD), five Trx domains, and coiled coil domain are shown schematically. ¶ denotes potential N-glycosylation sites. (C) Eluates were obtained from EDEM2-KO cells overexpressing TAP-EDEM2 plus TXNDC11(M1A) and from TXNDC11-KO cells overexpressing TAP-EDEM2, subjected to SDS-PAGE under reducing conditions, and silver-stained. The positions of TXNDC11(M1A)–3xFlag and 6xMyc-EDEM2 are indicated. (D) (a) EDEM2 stably disulfide-bonded to TXNDC11 was purified at a larger scale and silver-stained after reducing SDS-PAGE. The eluate was analyzed sequentially by LC/MS, MS/MS, and MS/MS/MS as indicated. This experiment was conducted once. (b) Extracted ion chromatogram of the ion at m/z 778.40 (±0.01) from EDEM2-TXNDC11 complex is shown. (c) MS spectrum of Peptide 1 observed in (b) is shown. The six peaks other than m/z 778.3999 are isotopic (13C-containing) ion peaks. (d) Electron-transfer/higher-energy collisional dissociation-tandem mass spectrometry (EThcD-MS/MS) spectrum of the ion at m/z 778.6505 ± 1 (m/z 778.3999–779.4015) derived from Peptide 1 (c) is shown.
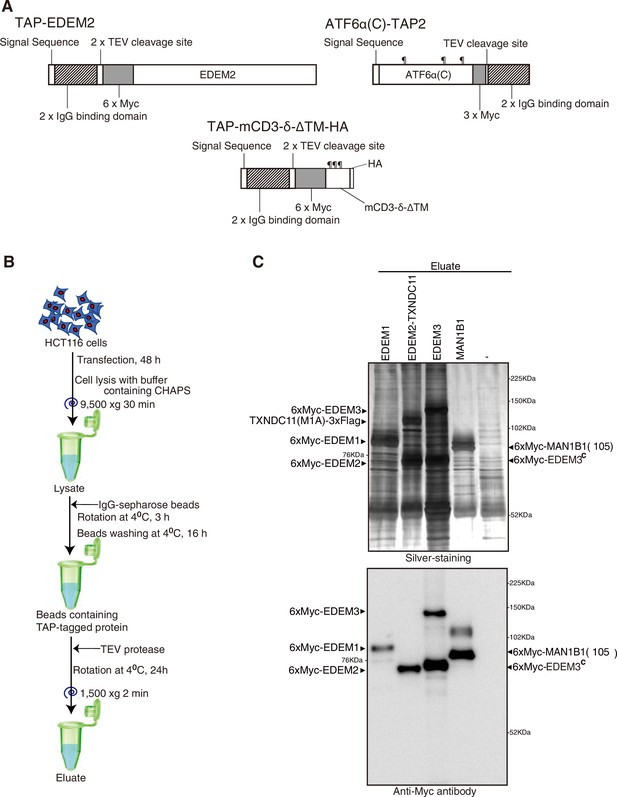
Purification strategy.
(A) Schematic structures of TAP-EDEM2, TAP-mCD3-δ-ΔTM-HA, and ATF6α(C)-TAP2 used for purification are shown. (B) Procedures of purification are shown. (C) Eluates obtained from wild-type (WT) cells untransfected (-) or transfected with plasmid to express TAP-EDEM1, TAP-EDEM2 plus TXNDC11(M1A), TAP-EDEM3, or TAP-MAN1B1(Δ105) were subjected to SDS-PAGE under reducing conditions, silver-stained, and then analyzed by immunoblotting using anti-Myc antibody. EDEM3C denotes cleaved EDEM3.
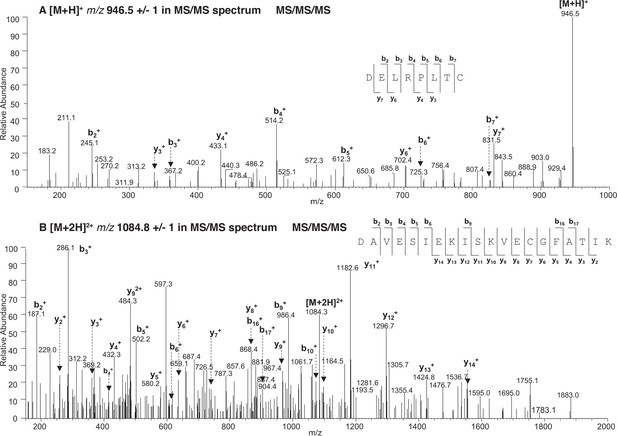
Higher-energy collisional dissociation (HCD)-mass spectrometry (MS)/MS/MS spectra.
(A) HCD-MS/MS/MS spectra of the product ion at m/z 946.5 ± 1 in EThcD-MS/MS spectrum revealed that the ion corresponds to D58ELRPLTC65 (P1). (B) HCD-MS/MS/MS spectra of the product ion at m/z 1084.8 ± 1 in EThcD-MS/MS spectrum revealed that the ion corresponds to D395AVESIEKISKVEC408GFATIK414 (P2).
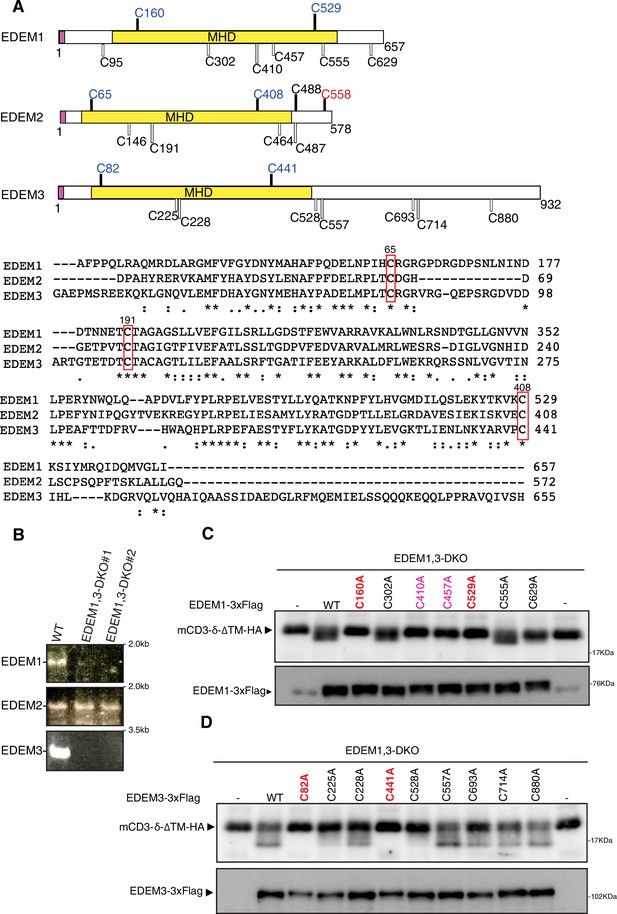
Effect of mutation of various cysteine residues in EDEM1 and EDEM3 on endoplasmic reticulum-associated degradation of misfolded glycoproteins (gpERAD).
(A) Structures of human EDEM1, EDEM2, and EDEM3 are schematically shown with cysteine residues (C) highlighted together with their positions (black bars underneath C indicate conserved cysteine residues, whereas white bars over C indicate non-conserved cysteine residues). The purple and yellow boxes denote the signal sequence and mannosidase homology domain (MHD), respectively. Sequence comparison around the three cysteine residues (C65, C191, and C408 of EDEM2) is shown below (asterisk and colon indicate identical and similar amino acids, respectively). (B) RT-PCR to amplify cDNA corresponding to full-length open reading frame in EDEM1/2/3 mRNA in wild-type (WT) and EDEM1, 3-double knockout (DKO) cells (two independent clones #1 and #2) is shown. (C) Cell lysates were prepared from EDEM1, 3-DKO cells expressing WT or one of various cysteine mutants of 3× Flag-tagged EDEM1 together with mCD3-δ-ΔTM-HA by transfection, and analyzed by immunoblotting using anti-HA and anti-EDEM1 antibodies. (D) Cell lysates were prepared from EDEM1, 3-DKO cells expressing WT or one of various cysteine mutants of 3× Flag-tagged EDEM3 together with mCD3-δ-ΔTM-HA by transfection, and analyzed by immunoblotting using anti-HA and anti-Flag antibodies.
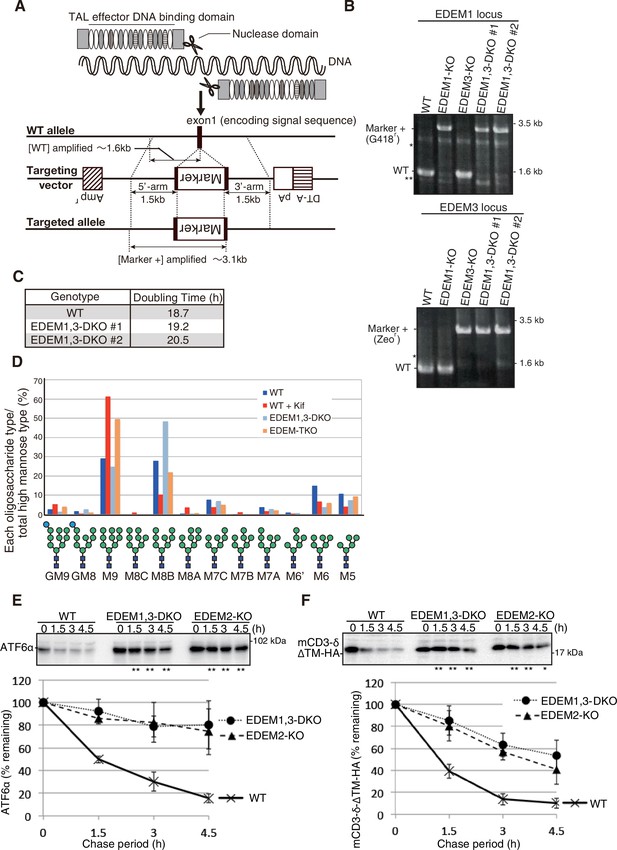
Construction and characterization of EDEM1, 3-double knockout (3-DKO) cells.
(A) Strategy for TALEN-mediated gene disruption is shown. Cleavage of exon 1 encoding the signal sequence of human EDEM1 or EDEM3 with the designed TALEN facilitates subsequent homologous recombination with the respective targeting vector containing both positive and negative selection markers. (B) Results of genomic PCR to confirm homologous recombination in HCT116 cells of various genotypes are shown. Asterisks denote non-specific bands. (C) Doubling time of wild-type (WT) and EDEM1, 3-DKO cells (two independent clones #1 and #2) is shown. (D) Isomer composition of N-glycans prepared from total cellular glycoproteins of WT, WT treated with kifunensine (Kif, 10 μg/ml, 12 hr), EDEM1, 3-DKO, and EDEM-triple knockout (TKO) cells is shown. This experiment was conducted once. (E) Cycloheximide chase was conducted to determine the degradation rate of endogenous ATF6α in WT, EDEM1, 3-DKO, and EDEM2-KO cells, and cells lysates were analyzed by immunoblotting using anti-ATF6α antibody (n = 3). Quantified data are shown below. (F) Cycloheximide chase was conducted to determine the degradation rate of transfected mCD3-δ-ΔTM-HA in WT, EDEM1, 3-DKO, and EDEM2-KO cells, and cells lysates were analyzed by immunoblotting using anti-HA antibody (n = 3). Quantified data are shown below.
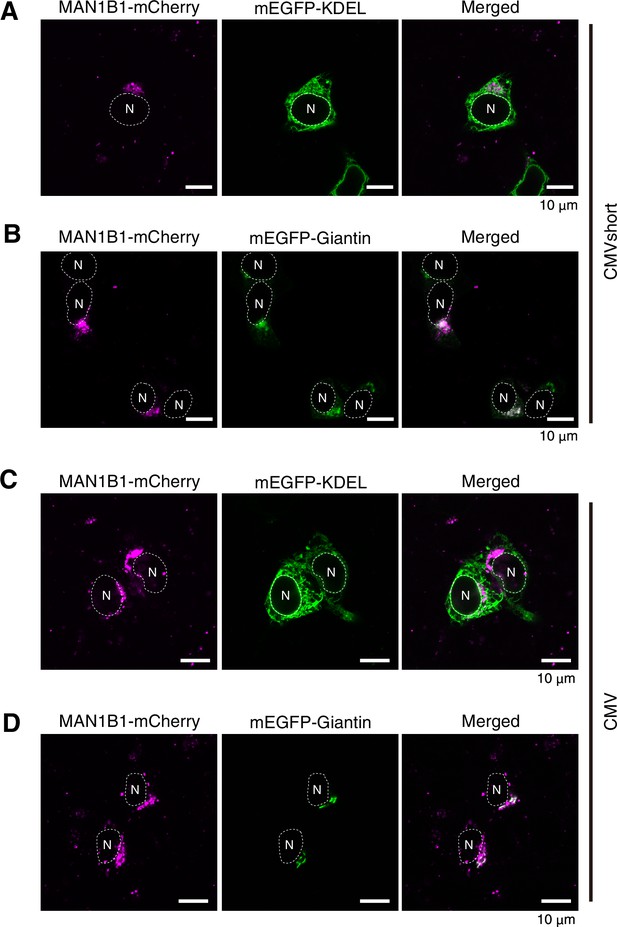
Localization of MAN1B1-mCherry in HCT116 cells.
(A, B) HCT116 cells were transfected with plasmid to express MAN1B1-mCherry under the control of the CMVshort promoter together with plasmid to express mEGFP-KDEL (A) or mEGFP-Giantin (B), and analyzed by confocal microscopy (Airyscan). Scale bar: 10 μm. (C, D) HCT116 cells were transfected with plasmid to express MAN1B1-mCherry under the control of the CMV promoter together with plasmid to express mEGFP-KDEL (C) or mEGFP-Giantin (D), and analyzed by confocal microscopy (Airyscan). Sale bar: 10 μm.
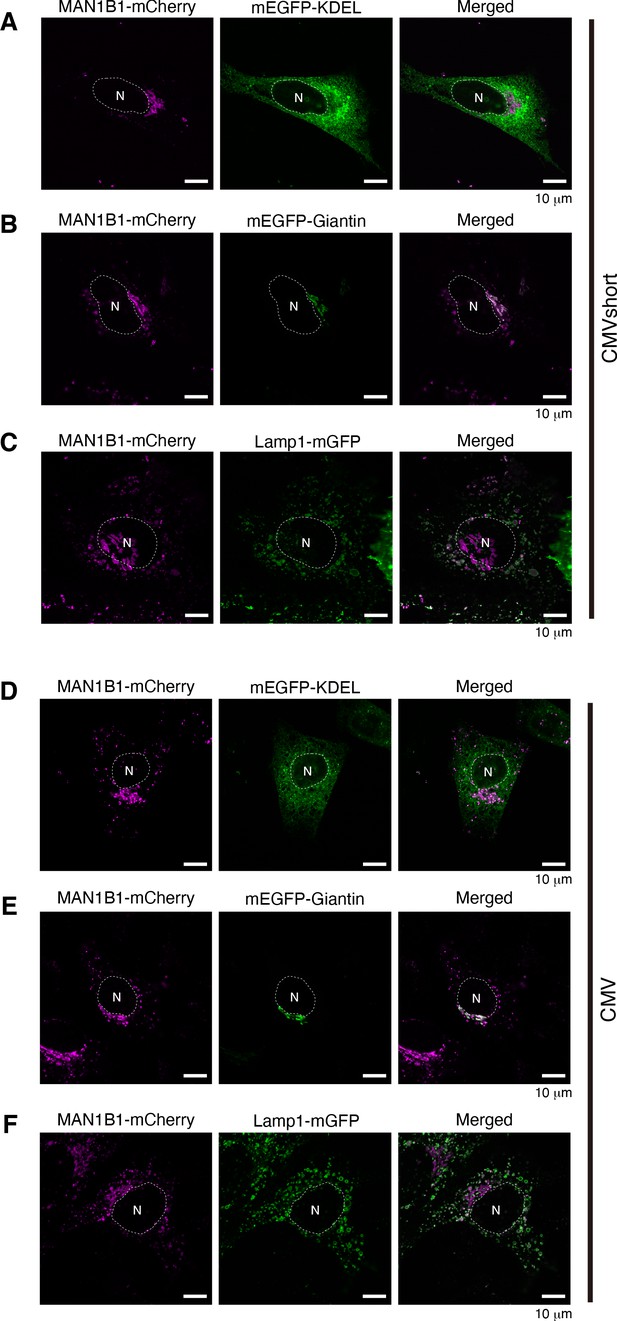
Localization of MAN1B1-mCherry in HeLa cells.
(A, B, C) HeLa cells were transfected with plasmid to express MAN1B1-mCherry under the control of the CMVshort promoter together with plasmid to express mEGFP-KDEL (A), mEGFP-Giantin (B), or Lamp1-mGFP (C), and analyzed by confocal microscopy (Airyscan). Scale bar: 10 μm. (D, E, F) HeLa cells were transfected with plasmid to express MAN1B1-mCherry under the control of the CMV promoter together with plasmid to express mEGFP-KDEL (D), mEGFP-Giantin (E), or Lamp1-mGFP (F), and analyzed by confocal microscopy (Airyscan). Scale bar: 10 μm.
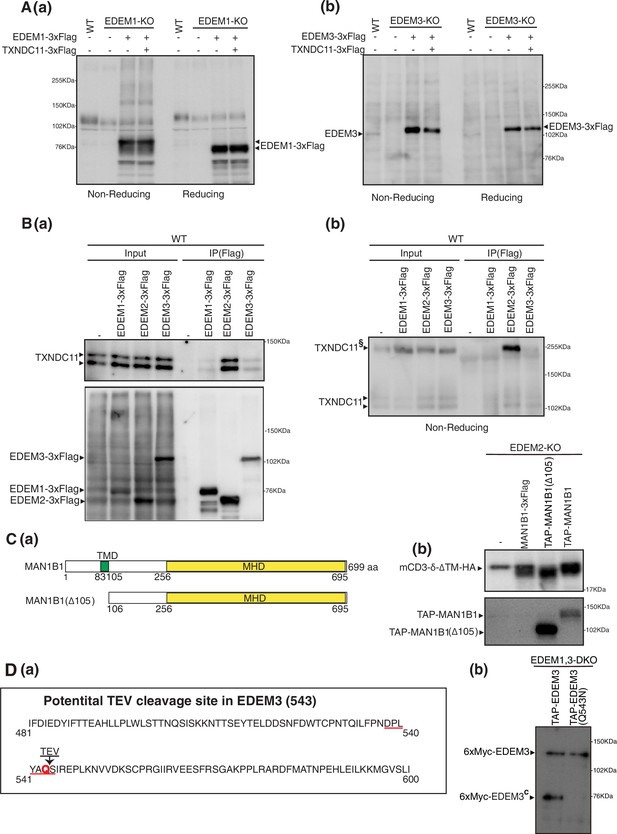
Effect of coexpression of TXNDC11 on EDEM1 and EDEM3.
(A) (a) Cell lysates were prepared from untransfected wild-type (WT) cells and EDEM1-knockout (KO) cells untransfected or transfected with (+) or without (-) plasmid to express EDEM1-3xFlag or TXNDC11-3xFlag, subjected to SDS-PAGE under non-reducing and reducing conditions, and analyzed by immunoblotting using anti-EDEM1 antibody. (b) Cell lysates were prepared from untransfected WT cells and EDEM3-KO cells untransfected or transfected with (+) or without (-) plasmid to express EDEM3-3xFlag or TXNDC11-3xFlag, subjected to SDS-PAGE under non-reducing and reducing conditions, and analyzed by immunoblotting using anti-EDEM3 antibody. (B) Cell lysates were prepared from WT cells untransfected or transfected with plasmid to express EDEM1-3xFlag, EDEM2-3xFlag, or EDEM3-3xFlag, and subjected to immunoprecipitation using anti-Flag antibody. Aliquots of cell lysates (Input) and immunoprecipitates (IP[Flag]) were subjected to SDS-PAGE under reducing (a) and non-reducing (b) conditions, and analyzed by immunoblotting using anti-TXNDC11 and anti-Flag antibodies. TXNDC11§ denotes TXNDC11 stably disulfide-bonded to EDEM2. (C) (a) Structures of MAN1B1 and MAN1B1(Δ105) are schematically shown. TMD denotes the transmembrane domain. (b) EDEM2-KO cells were transfected with plasmid to express mCD3-δ-ΔTM-HA together with or without plasmid to express MAN1B1-3xFlag, TAP-MAN1B1(Δ105), or TAP-MAN1B1. Cell lysates were then prepared and analyzed by immunoblotting using anti-HA and anti-Myc antibodies. (D) (a) Location of potential TEV cleavage site in EDEM3 is shown. Its consensus sequence is E-X-X-Y-X-Q-G/S. (b) EDEM1, 3-DKO cells expressing TAP-EDEM3 or TAP-EDEM3(Q543N) by transfection were subjected to small-scale purification as in Figure 1—figure supplement 1B. Eluates were analyzed by immunoblotting using anti-Myc antibody.
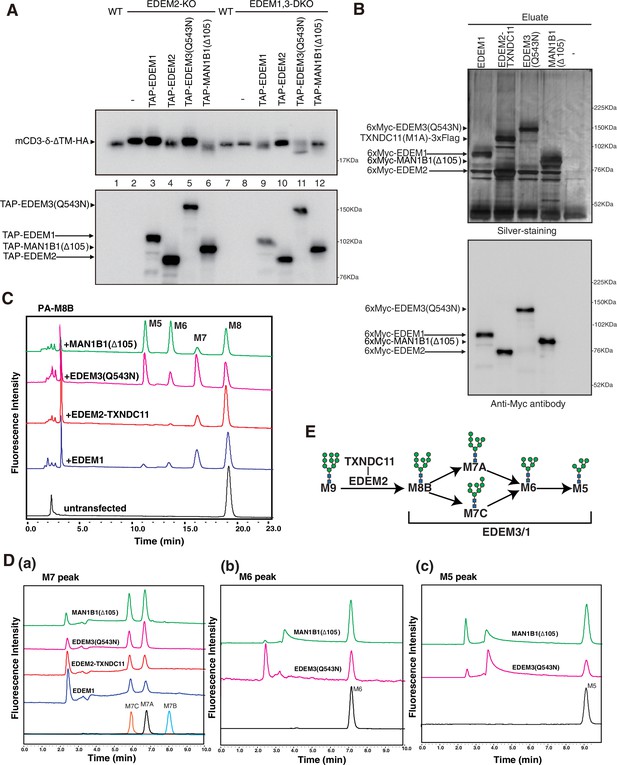
Effect of four purified α1,2-mannosidases on PA-M8B.
(A) Wild-type (WT) cells were transfected with plasmid to express mCD3-δ-ΔTM-HA. EDEM2-knockout (KO) cells and EDEM1, 3-double knockout (DKO) cells were transfected with plasmid to express mCD3-δ-ΔTM-HA together with or without plasmid to express TAP-EDEM1, TAP-EDEM2, TAP-EDEM3(Q543N), or TAP-MAN1B1(Δ105). Cell lysates were then prepared and analyzed by immunoblotting using anti-HA and anti-Myc antibodies. (B) Eluates obtained from WT cells untransfected (-) or transfected with plasmid to express TAP-EDEM1, TAP-EDEM2 plus TXNDC11(M1A), EDEM3(Q543N), or MAN1B1(Δ105) were subjected to SDS-PAGE under reducing conditions, silver-stained, and then analyzed by immunoblotting using anti-Myc antibody. PA-M8B was incubated with samples in (B) for 24 hr as indicated and then analyzed by HPLC (amide column) for mannose contents. (D) The M7 peak (a), M6 peak (b), and M5 peak (c) obtained in (C) were analyzed by HPLC (ODS column) for isomer identification. (E) Route of oligosaccharide processing in the mammalian endoplasmic reticulum-associated degradation of misfolded glycoproteins (gpERAD) is shown.
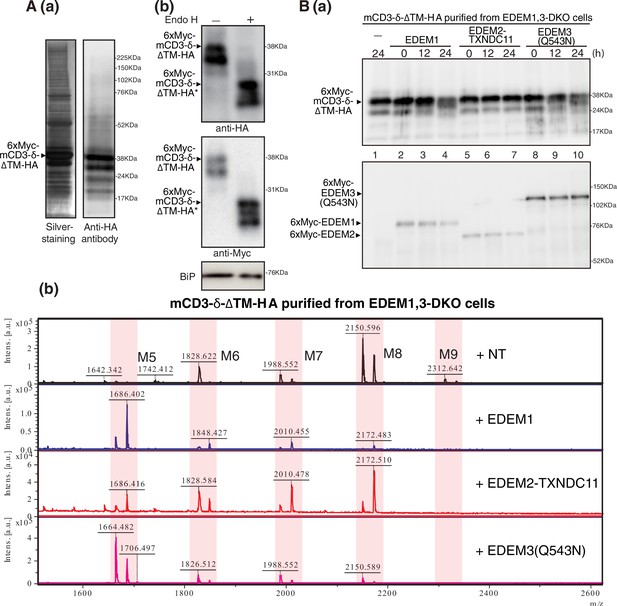
Effect of purified EDEM1/2/3 on M8B present in mCD3-δ-ΔTM-HA.
(A) (a) Eluate obtained from EDEM1, 3-double knockout (DKO) cells overexpressing TAP-mCD3-δ-ΔTM-HA was subjected to SDS-PAGE under reducing conditions, silver-stained, and then analyzed by immunoblotting using anti-HA antibody. (b) Eluate in (a) was untreated (-) or treated (+) with EndoH, subjected to SDS-PAGE under reducing conditions, and analyzed by immunoblotting using anti-HA, anti-Myc, and anti-GRP78 (which is identical to BiP) antibodies. (B) (a) Eluate in (A) was incubated with purified EDEM1, EDEM2-TXNDC11 complex, or EDEM3(Q543N) for the indicated time, and then analyzed by immunoblotting using anti-Myc antibody. (b) N-glycans prepared from samples in (a) after 24 hr incubation were analyzed by mass spectrometry (MS). This experiment was conducted once.
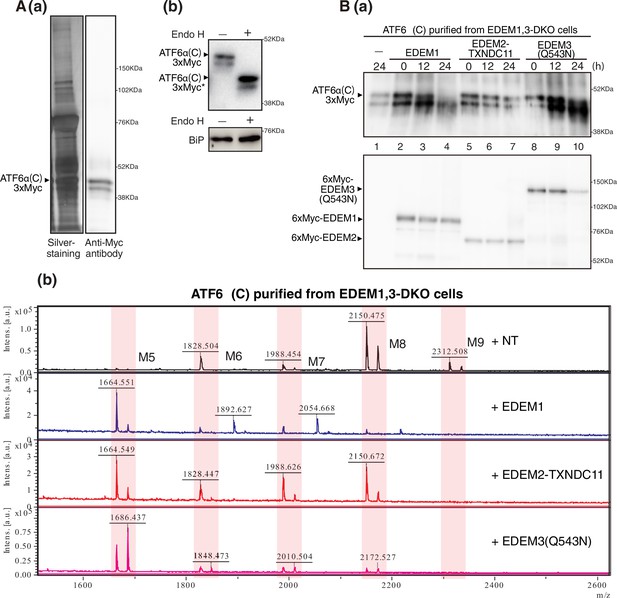
Effect of purified EDEM1/2/3 on M8B present in ATF6α(C).
(A) (a) Eluate obtained from EDEM1, 3-double knockout (DKO) cells overexpressing ATF6α(C)-TAP2 was subjected to SDS-PAGE under reducing conditions, silver-stained, and analyzed by immunoblotting using anti-Myc antibody. (b) Eluate in (a) was untreated (-) or treated (+) with EndoH, subjected to SDS-PAGE under reducing conditions, and analyzed by immunoblotting using anti-Myc and anti-GRP78 (which is identical to BiP) antibodies. (B) (a) Eluate in (A) was incubated with purified EDEM1, EDEM2-TXNDC11 complex, or EDEM3(Q543N) for the indicated time, and then analyzed by immunoblotting using anti-Myc antibody. (b) N-glycans prepared from samples in (a) after 24 hr incubation were analyzed by mass spectrometry (MS). This experiment was conducted once.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Homo sapiens) | Colorectal carcinoma | ATCC | HCT116 | This cell line has been authenticated and tested negative for mycoplasma. |
| Recombinant DNA reagent | p3xFlag-CMV-14 | Sigma-Aldrich | ||
| Recombinant DNA reagent | pmCherry-N1 | TAKARA | ||
| Recombinant DNA reagent | pSecTag2/Hygro | Thermo Fisher Scientific | ||
| Recombinant DNA reagent | pEGFP-C1 | CLONTECH | ||
| Antibody | Anti-TXNDC11(rabbit monoclonal) | Abcam | Cat#: ab188329 | WB (1:500) |
| Antibody | Anti-EDEM1 (rabbit polyclonal) | Sigma-Aldrich | Cat#: E8406 | WB (1:500) |
| Antibody | Anti-EDEM2 (rabbit polyclonal) | Novusbio | Cat#: NBP2-37921 | WB (1:500) |
| Antibody | Anti-EDEM3 (mouse monoclonal) | Sigma-Aldrich | Cat#: E0409 | WB (1:500) |
| Antibody | Anti-GRP78 antibody (rabbit polyclonal) | Thermo Fisher Scientific | Cat#: PA1-014A | WB (1:1000) |
| Antibody | Anti-HA (rabbit polyclonal) | Recenttec | Cat#: R4-TP1411100 | WB (1:1000) |
| Antibody | Anti-Flag (mouse monoclonal) | Sigma-Aldrich | Cat#: F3165 | WB (1:1000) IP (2.5 μl) |
| Antibody | Anti-Myc-direct-HRP antibody | MBL | Cat#: M047-7 | WB (1:1000) |





