Group II truncated haemoglobin YjbI prevents reactive oxygen species-induced protein aggregation in Bacillus subtilis
Figures
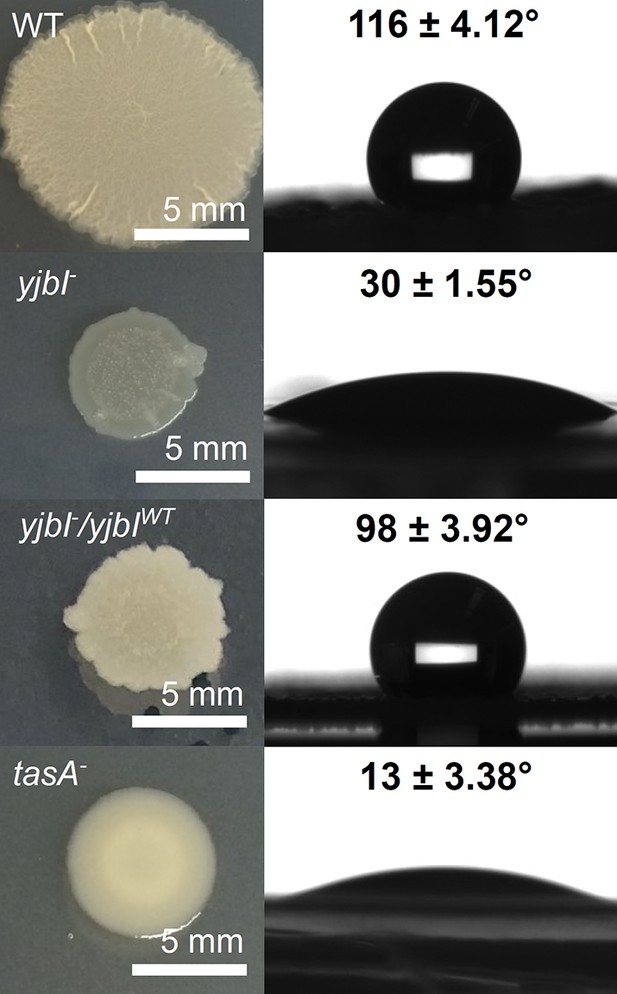
YjbI is needed for normal biofilm formation.
Images showing the morphology (left panels) and surface repellency (right panels) of the biofilms of the wild-type (WT), yjbI-, yjbI-/yjbIWT, and tasA-deficient (tasA-) strains of Bacillus subtilis. The water contact angles indicated in the right panels represent the mean ± SD of three independent experiments (n=3).
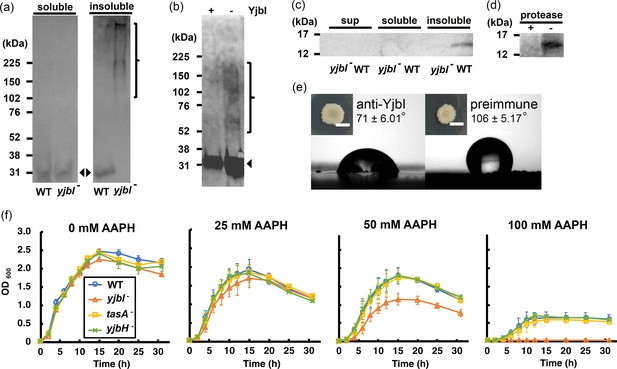
yjbI is required to prevent oxidative TasA aggregation and for the normal antioxidant properties of Bacillus subtilis cells.
(a) Detection of His-tagged TasA (TasA-His) in the soluble (left panel) and insoluble (right panel) fractions of the pellicles of the wild-type (WT) strain or yjbI-deficient mutant (yjbI-) strain carrying pHtasA1, which constitutively expresses TasA-His. After culturing in liquid MSgg medium, lysates of these pellicles were further analysed by western blotting using an anti-His-tag antibody. The positions of the molecular mass marker proteins are shown on the left. The arrowheads and bracket indicate monomeric and aggregated TasA-His, respectively. Similar results were obtained from three independent experiments. (b) In vitro reactive oxygen species (ROS)-induced formation of aggregates of purified TasA (pEtasA2-derived TasA28-261-His) in the presence (+) or absence (-) of purified recombinant YjbI, as evidenced by western blotting analysis using an anti-His-tag antibody. The arrowhead and bracket indicate monomeric and aggregated TasA28-261-His, respectively. Protein aggregation was induced with 1 mM H2O2 and 10 μM FeCl2. Similar results were obtained from two independent experiments. (c) Localisation of YjbI in the insoluble biofilm fraction. The soluble and insoluble fractions of the WT and yjbI- pellicles and culture supernatants (sup) were analysed by western blotting using an anti-YjbI antiserum. Similar results were obtained from two independent experiments. (d) Localisation of YjbI to the cell surface, as evidenced by western blot analysis of the intact WT pellicles with (+) or without (-) protease digestion. YjbI was detected using an anti-YjbI antiserum. Similar results were obtained from two independent experiments. (e) Images showing biofilm surface repellency. Two microliters of rabbit anti-YjbI serum (left panel) or pre-immune serum (right panel) were mixed with 2 µL of pre-cultured Bacillus subtilis cells and inoculated on MSgg solid medium. The morphologies of the biofilms are shown in the insets. The water contact angles indicated in the right panels represent the mean ± SD of three independent experiments. (f) Sensitivity of planktonically grown B. subtilis strains to 2,2′-azobis(2-amidinopropane) dihydrochloride (AAPH)-induced oxidative stress. B. subtilis cells (WT, yjbI-, tasA-, or yjbH-) were inoculated at OD600=0.02 in LB medium containing 0–100 mM AAPH and grown at 37°C with shaking. Data are shown as the mean ± SD of three independent experiments.
-
Figure 2—source data 1
PVDF membrane after western blotting treatment of soluble and insoluble fractions of Bacillus subtilis extracts with anti-TasA antibody.
- https://cdn.elifesciences.org/articles/70467/elife-70467-fig2-data1-v2.zip
-
Figure 2—source data 2
Gel image of Fenton reaction-treated TasA after electrophoresis and silver staining.
- https://cdn.elifesciences.org/articles/70467/elife-70467-fig2-data2-v2.zip
-
Figure 2—source data 3
PVDF membrane after western blotting treatment of each fraction of Bacillus subtilis using anti-YjbI antiserum.
- https://cdn.elifesciences.org/articles/70467/elife-70467-fig2-data3-v2.zip
-
Figure 2—source data 4
PVDF membrane after western blotting treatment of protease-treated Bacillus subtilis extracts using anti-YjbI antiserum.
- https://cdn.elifesciences.org/articles/70467/elife-70467-fig2-data4-v2.zip
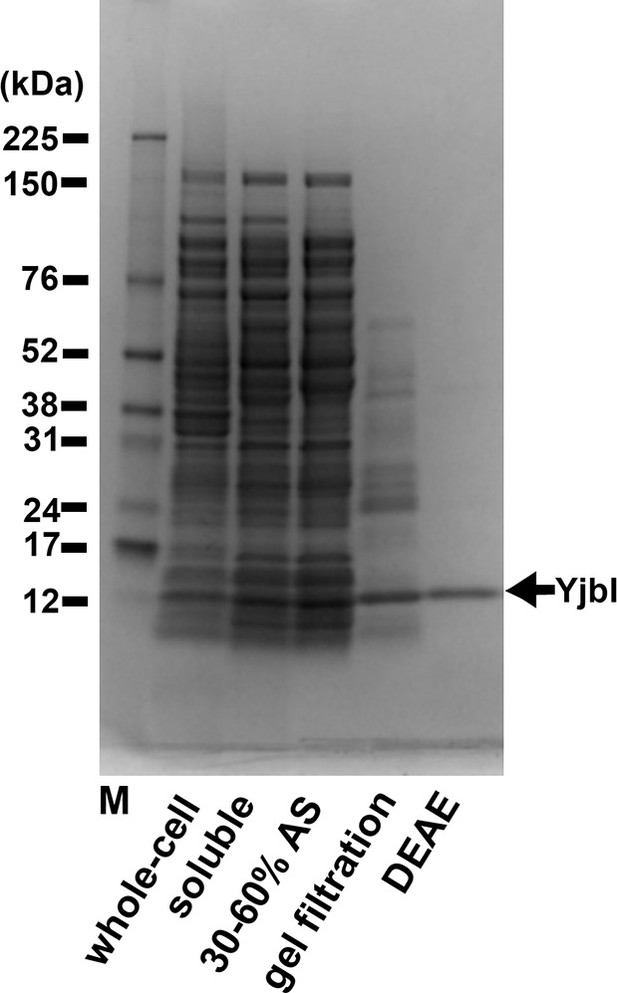
Purification of YjbI.
YjbI was produced in Escherichia coli BL21(DE3)/pEyjbI2 cells grown in lysogeny broth (LB) medium. Proteins from each purification step (i.e., whole-cell and soluble crude extracts); 30–60% saturation ammonium sulphate (AS) fractionation; Sephacryl S-100 gel filtration; and Toyopearl DEAE-650M chromatography were resolved by SDS-PAGE and then stained with Coomassie Brilliant Blue.
-
Figure 2—figure supplement 1—source data 1
Gel image of column-purified recombinant YjbI after electrophoresis.
- https://cdn.elifesciences.org/articles/70467/elife-70467-fig2-figsupp1-data1-v2.zip
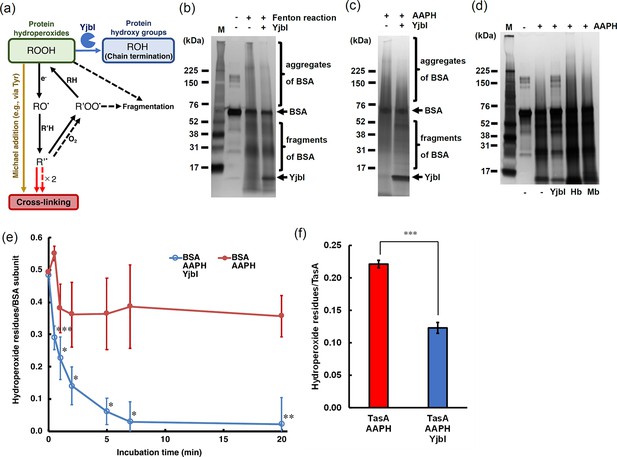
YjbI exhibits protein hydroperoxide peroxidase activity.
(a) Schematic drawing summarising the proposed protein cross-linking and fragmentation induced by protein radicals derived from protein hydroperoxides and prevention of these reactions by the protein hydroperoxide peroxidase activity of YjbI. The solid and dashed arrows indicate the already known reactions (Davies, 2016) occurring at protein side chains and backbones, respectively. Protein cross-linking proceeds via spontaneous radical coupling (red arrows), and the Michael addition occurs at protein side chains (yellow arrow). YjbI catalyses the protein hydroperoxide peroxidase reaction (blue arrow) to convert protein hydroperoxides (ROOH) to hydroxy groups (ROH) at protein side chains and prevents further protein cross-linking and fragmentation. Side chain alkoxyl radicals (RO·) can be formed through one-electron reduction (e.g., homolysis) of ROOH, followed by hydrogen atom abstraction reactions with other protein side chains or backbones (R'H). The carbon-centred radicals (R'·) of side chains or backbones lead to protein cross-linking to yield protein aggregates (indicated with solid and dashed red arrows). The side chain or backbone R'· also reacts with oxygen to produce protein peroxy radicals (R'OO·) under aerobic conditions. The side chain R'OO· reacts with the side chains or backbones of other amino acid residues (RH) in its vicinity to yield protein ROOH moieties. The unstable backbone R'OO· and ROOH moieties cause protein fragmentation. (b) BSA was treated with (+) or without (-) H2O2 and FeCl2 to produce BSA-OOH by Fenton reaction. BSA-OOH was incubated in the presence (+) or absence (-) of YjbI and analysed by SDS-PAGE and silver staining. YjbI and monomeric BSA are indicated by arrows, and BSA aggregates and fragments are indicated by brackets. The sizes of the molecular mass marker proteins (M) are shown on the left. Similar results were obtained from two independent experiments. (c) BSA-OOH prepared by 2,2′-azobis(2-amidinopropane) dihydrochloride (AAPH) treatment was incubated in the presence (+) or absence (-) of YjbI and analysed by SDS-PAGE and silver staining. The labels on the left or right of the images are the same as those in (b). Similar results were obtained from two independent experiments. (d) YjbI prevents AAPH-induced BSA-OOH aggregation. BSA was incubated for 3 hr with (+) or without (-) AAPH before adding YjbI, haemoglobin from bovine blood (Hb) (Sigma-Aldrich), myoglobin from equine heart (Mb) (Sigma-Aldrich), or none (-) and then analysed by SDS-PAGE and silver staining. The positions of the molecular mass marker proteins (M) are shown on the left. Similar results were obtained from two independent experiments. (e) Time course of the YjbI-induced reduction of the hydroperoxide groups in BSA-OOH. Peroxide-treated BSA (30 μM) in 50 mM Tris-acetate buffer (pH 7.0) was incubated with (blue circles) or without (red circles) YjbI (3.3 μM) at 37°C, and the reaction was terminated by adding 4 volumes of cold acetone at the indicated incubation times (0, 0.5, 1, 2, 5, 7, and 20 min). The number of hydroperoxide groups per BSA subunit was determined for each sample. The data are the mean ± SD of three independent experiments (*p<0.05; **p<0.01; and ***p<0.005; t-test, one-tailed). (f) TasA-OOH (30 μM) in 50 mM Tris-acetate buffer (pH 7.0) was incubated with YjbI (3.3 μM) at 37°C, and the reaction was terminated by adding 4 volumes of cold acetone after incubation for 20 min. The number of hydroperoxide groups per TasA was determined for each sample. Data are shown as the mean ± SD of three independent experiments (***p<0.005; t-test, one-tailed).
-
Figure 3—source data 1
Gel image of Fenton reaction-treated BSA after electrophoresis and silver staining.
- https://cdn.elifesciences.org/articles/70467/elife-70467-fig3-data1-v2.zip
-
Figure 3—source data 2
Gel image of 2,2′-azobis(2-amidinopropane) dihydrochloride (AAPH)-treated BSA after electrophoresis and silver staining.
- https://cdn.elifesciences.org/articles/70467/elife-70467-fig3-data2-v2.zip
-
Figure 3—source data 3
Gel image of 2,2′-azobis(2-amidinopropane) dihydrochloride (AAPH)-treated BSA and haem proteins after electrophoresis and silver staining.
- https://cdn.elifesciences.org/articles/70467/elife-70467-fig3-data3-v2.zip

Purification of TasA and YjbI (Y63F).
Mature TasA was produced in Escherichia coli BL21(DE3)/pEtasA3 cells grown in lysogeny broth (LB) medium. Proteins from each purification step, that is, soluble crude extracts (lane 1), elution fraction from immobilised metal affinity chromatography (IMAC) resin charged with cobalt (lane 2), and wash fraction from IMAC after treatment with SUMO-protease (lane 3), were resolved by SDS-PAGE and stained with Coomassie Brilliant Blue.
-
Figure 3—figure supplement 1—source data 1
Gel image of column-purified recombinant TasA after electrophoresis.
- https://cdn.elifesciences.org/articles/70467/elife-70467-fig3-figsupp1-data1-v2.zip
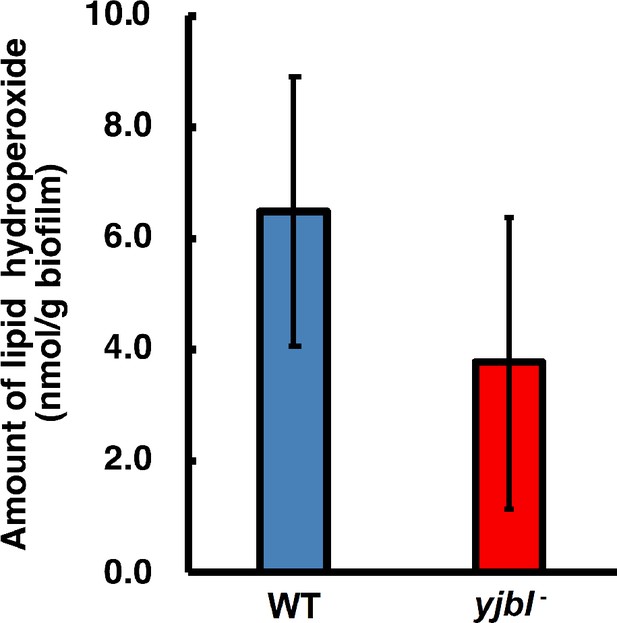
Lipid hydroperoxide (LOOH) levels were not affected in yjbI- strain biofilms.
Bacterial colony biofilms were obtained by inoculating a 3 µL aliquot of pre-cultured Bacillus subtilis on solid MSgg medium, followed by cultivation at 37°C for 48 hr. The biofilms were collected and suspended in phosphate-buffered saline by using a spatula. These wild-type (WT) and yjbI-deficient mutant biofilm suspensions were analysed with a colorimetric LOOH assay kit (LPO assay kit, Cayman Chemical Company) according to the manufacturer’s instructions. The data are shown as the mean ± SD of three independent experiments (p=0.083; t-test, one-tailed).
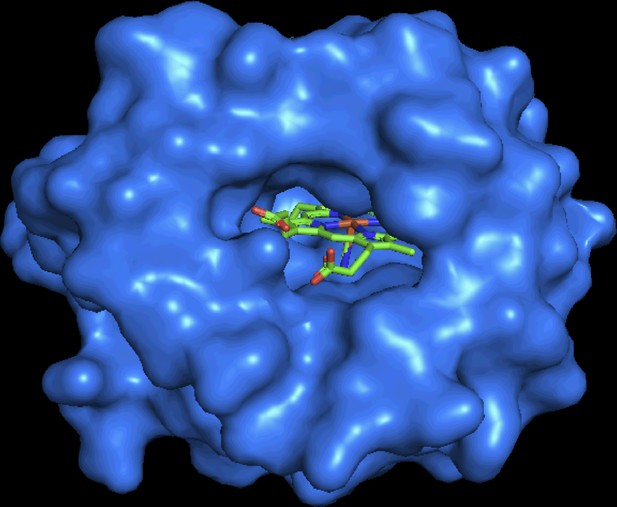
X-ray crystal structure of Bacillus subtilis YjbI (PDB ID: 1UX8) (Giangiacomo et al., 2005), showing an opening on the molecular surface.
The protein surface is shown in blue, and the haem molecule is shown with a stick model. The image was drawn using PyMOL.
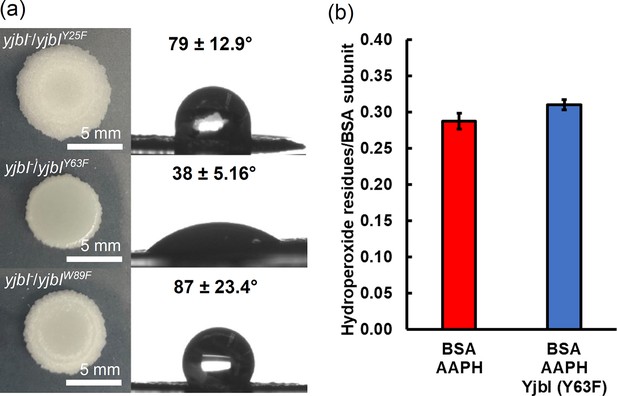
Gene complementation study using the yjbI-deficient strain of Bacillus subtilis with plasmids expressing YjbI derivatives (Y25F, Y63F, and W89F).
a) The morphology and surface repellence of the biofilms of the yjbI-deficient strains carrying each plasmid are shown. The water contact angles indicated in the right panels represent the mean ± SD of three independent experiments (n=3). (b) BSA-OOH (30 μM) in 50 mM Tris-acetate buffer (pH 7.0) was incubated with the YjbI derivative Y63F (3.3 μM) at 37°C, and the reaction was terminated by adding 4 volumes of cold acetone after incubation for 20 min. The number of hydroperoxide groups per BSA subunit was determined for each sample. Data are shown as the mean ± SD of three independent experiments.

Y63F derivative of YjbI was produced in Escherichia coli BL21(DE3)/pEyjbI3 cells grown in lysogeny broth (LB) medium.
Proteins from each purification step, soluble crude extracts (lane 1), elution fraction from immobilised metal affinity chromatography (IMAC) (lane 2), and wash fraction from IMAC after treatment with SUMO-protease (lane 3) were resolved by SDS-PAGE and stained with Coomassie Brilliant Blue. The positions of the molecular mass marker proteins (M) are shown on the left.
-
Figure 4—figure supplement 1—source data 1
Gel image of column-purified recombinant YjbI (Y63F) after electrophoresis.
- https://cdn.elifesciences.org/articles/70467/elife-70467-fig4-figsupp1-data1-v2.zip
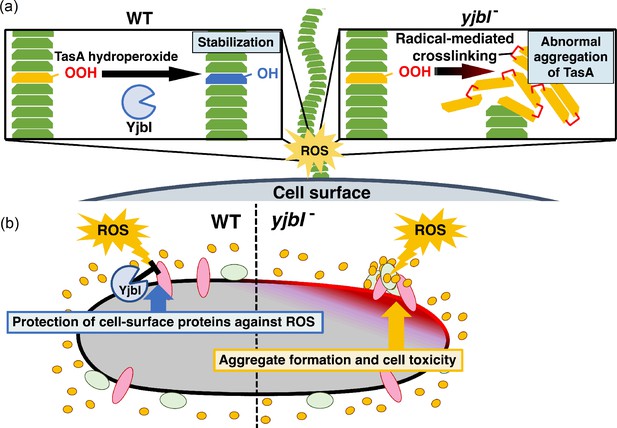
YjbI protects proteins and the cell from reactive oxygen species (ROS) by removing hydroperoxide groups from proteins.
(a) YjbI repairs the oxidatively damaged TasA-OOH in biofilms. In the wild-type (WT) strain (left panel), the normal fibrous TasA (green trapezoids) is damaged by ROS to generate TasA-OOH (yellow trapezoid) on the cell surface. YjbI converts TasA-OOH to TasA-OH (blue trapezoid) to stabilise the normal TasA fibre. In contrast, TasA-OOH aggregates via radical-mediated protein cross-linking in the yjbI- strain (right panel). (b) A proposed role of YjbI in the general protection of cell-surface proteins from ROS-induced oxidative damage. Protein hydroperoxide-modified residues generated in various cellular surface proteins (red, green, and yellow ellipses) are generally reduced to protein hydroxy residues by YjbI to protect from further damage (i.e., aggregation) (left half). yjbI disruption accumulates protein aggregates via hydroperoxidation and cross-linking of various proteins, resulting in cell toxicity (right half).
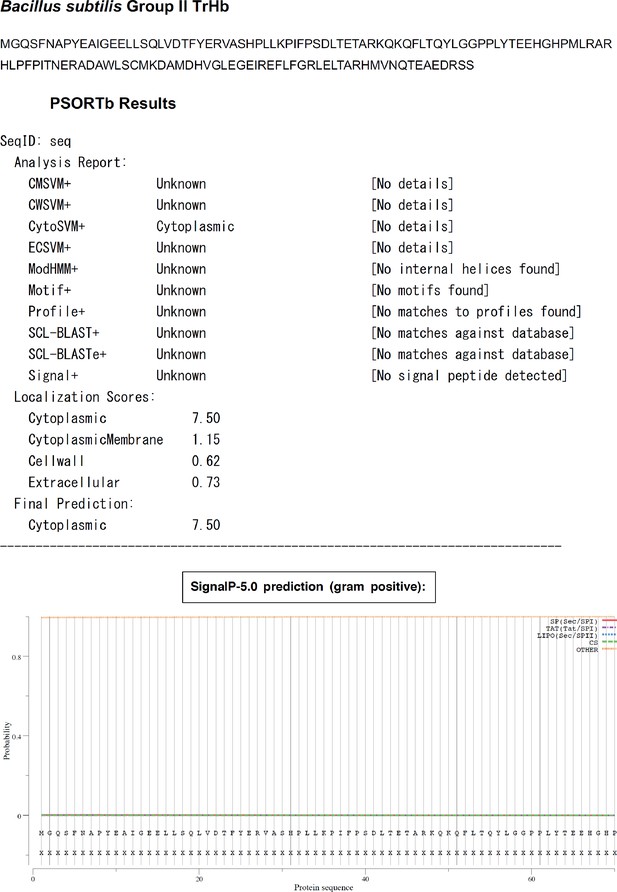
Bacillus subtilis YjbI does not contain a predicted signal sequence.
The results of bioinformatic analyses by using PSORTb (https://www.psort.org/psortb/) or SignalP-5.0 (http://www.cbs.dtu.dk/services/SignalP/).
Tables
Minimum bactericidal concentration following exposure to hypochlorous acid (HClO) (n=2).
| HClO (mM) | 62.5 | 31.3 | 15.6 | 5.00 | 2.50 | 1.25 | 0.625 | 0.313 | 0.156 |
|---|---|---|---|---|---|---|---|---|---|
| WT | − | + | + | + | + | + | + | + | + |
| yjbI- | − | − | − | − | − | − | − | + | + |
-
(+) visible growth, (−) no visible growth.
Minimum bactericidal concentration following exposure to hypochlorous acid in the yjbI-/yjbIWT and point-mutated strains (HClO) (n=2).
| HClO (mM) | 62.5 | 31.3 | 15.6 | 5.00 | 2.50 | 1.25 | 0.625 | 0.313 | 0.156 |
|---|---|---|---|---|---|---|---|---|---|
| yjbI-/yjbIWT | − | − | − | − | − | + | + | + | + |
| yjbI-/yjbIY25F | − | − | − | − | − | − | − | + | + |
| yjbI-/yjbIY63F | − | − | − | − | − | − | − | + | + |
| yjbI-/yjbIW89F | − | − | − | − | − | ± | + | + | + |
-
(+) visible growth, (−) no visible growth, (±) visible growth in one of two cases.





