Kap-β2/Transportin mediates β-catenin nuclear transport in Wnt signaling
Figures
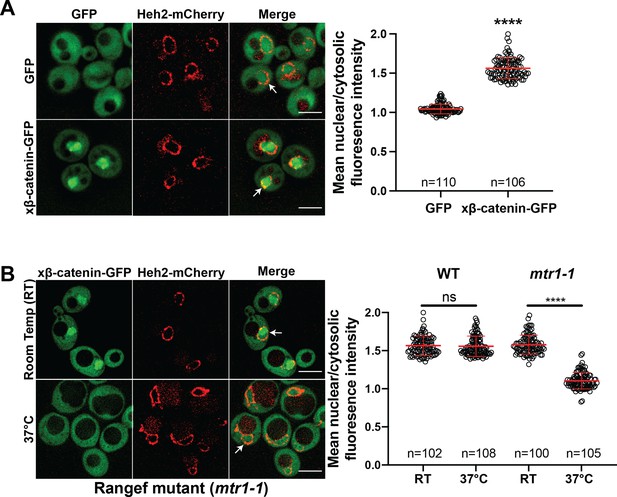
β-catenin requires a functional Ran cycle to accumulate in the nucleus of Saccharomyces cerevisiae.
(A) Representative deconvolved fluorescence images of xβ-catenin-GFP in a wildtype yeast strain that expresses Heh2-mCherry to label the nucleus (left). White arrows indicate the nuclear compartment. Plot showing the quantification of mean nuclear to cytosolic fluorescence intensity from 30 to 40 cells from three independent replicates (right). (B) Representative deconvolved fluorescence images of xβ-catenin-GFP in the RanGEF mutant (mtr1-1) strain at room temperature or 37°C that co-expresses Heh2-mCherry as a nuclear envelope marker (left). The ratio of mean nuclear to cytosolic fluorescence intensity was measured in the wildtype or mtr1-1 strain from 30 to 35 cells from three independent replicates (right). Scale bar is 5 μm in (A) and (B). Red bar indicates the mean value with the SD. p-Values are from unpaired two-tailed t-test where ns is p>0.05, and ****p<0.0001 for both (A) and (B). The data is uploaded as Figure 1—source data 1.
-
Figure 1—source data 1
Source data related to Figure 1.
- https://cdn.elifesciences.org/articles/70495/elife-70495-fig1-data1-v2.xlsx
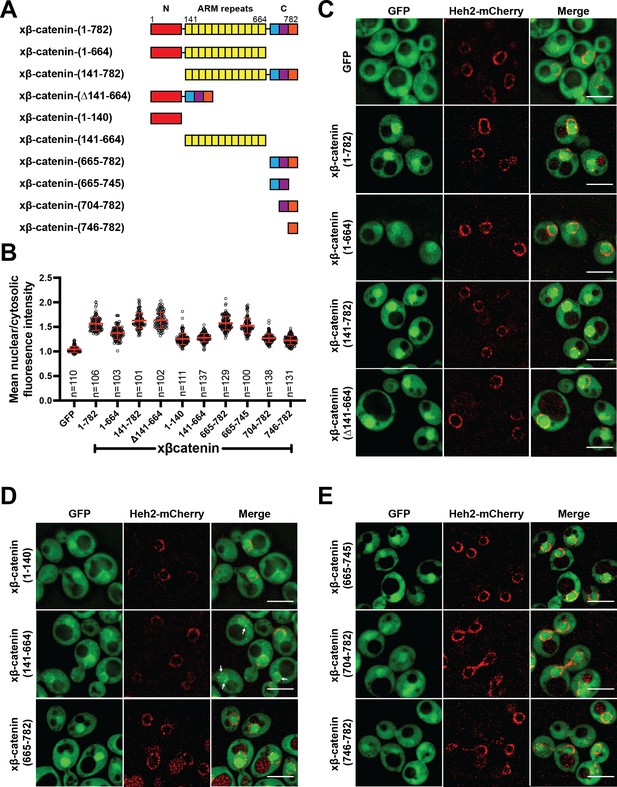
The C-terminus of β-catenin contains a nuclear localization signal (NLS).
(A) Schematic of Xenopus β-catenin truncation constructs tested in this study. (B) Plot of the ratio of mean nuclear to cytosolic fluorescence intensity of Xenopus β-catenin GFP truncation constructs tested in a wildtype yeast strain from 30 to 40 cells from three independent replicates. Red bar indicates the mean value with the SD. (C) Deconvolved fluorescence images of the N-terminal deletion (141-782), ARM-repeats deletion (Δ141-664) and C-terminal deletion (1-664) of Xenopus β-catenin GFP in the wildtype strain. GFP and full-length Xenopus β-catenin-GFP were used as controls. (D) Deconvolved fluorescence images of the indicated fragments of Xenopus β-catenin GFP in the wildtype strain. White arrows indicate nuclear rim localization. (E) Deconvolved fluorescence images of indicated C-terminus fragments of Xenopus β-catenin GFP in the wildtype strain. Heh2-mCherry was co-expressed to label the nuclear envelope in (C), (D), and (E). Scale bar is 5 µm in (C), (D), and (E). The data is uploaded as Figure 2—source data 1.
-
Figure 2—source data 1
Source data related to Figure 2.
- https://cdn.elifesciences.org/articles/70495/elife-70495-fig2-data1-v2.xlsx
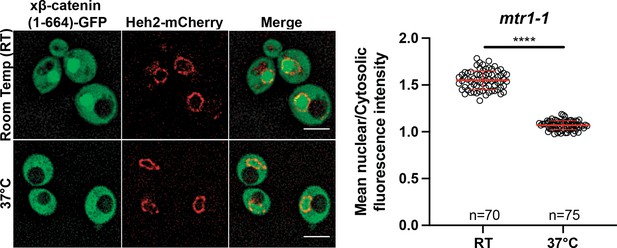
β-catenin (1-664) localizes to the nucleus in a RanGTPase dependent manner in Saccharomyces cerevisiae.
Representative deconvolved fluorescence image of xβ-catenin (1-664)-GFP in the RanGEF mutant (mtr1-1) strain at room temperature or 37°C that co-expresses Heh2-mCherry as a nuclear envelope marker (left). The ratio of mean nuclear to cytosolic fluorescence intensity was measured in the mtr1-1 strain from three independent replicates (right). Red bar indicates the mean value with the SD. p-Values are from unpaired two-tailed t-test where ns is p>0.05 and **** is p<0.0001. Scale bar is 5 µm. The data is uploaded as Figure 2—figure supplement 1—source code 1.
-
Figure 2—figure supplement 1—source code 1
Source data related to Figure 2—figure supplement 1.
- https://cdn.elifesciences.org/articles/70495/elife-70495-fig2-figsupp1-code1-v2.zip
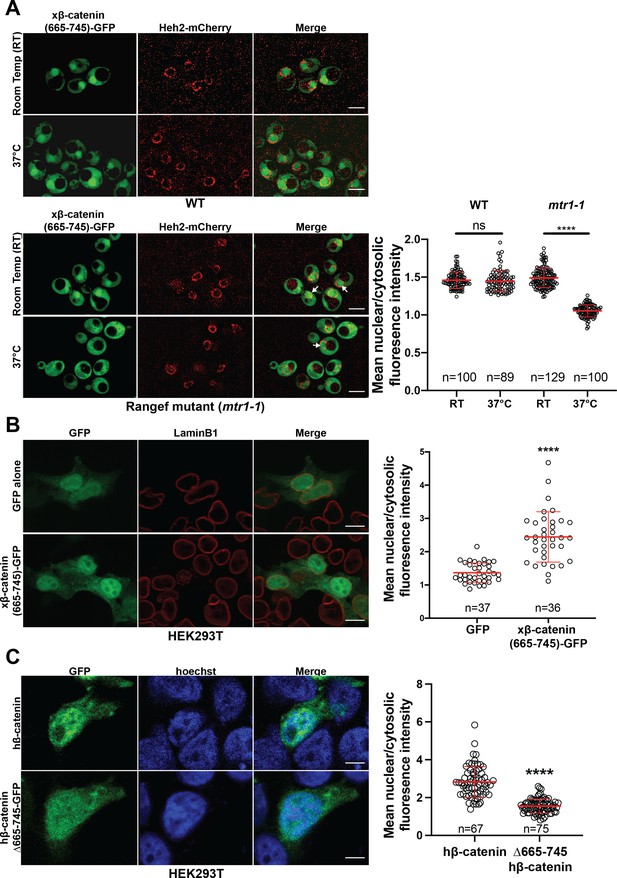
Ran dependence of the β-catenin (665-745)-GFP in the Saccharomyces cerevisiae and its requirement for nuclear localization in HEK293T cells.
(A) Deconvolved fluorescence image of Xenopus β-catenin-(665-745)-GFP in the wildtype (top left) and RanGEF mutant (mtr1-1) (bottom left) strain at room temperature (RT) or 37°C that co-expresses Heh2-mCherry as a nuclear envelope marker. White arrows indicate the nuclear compartment. The ratio of mean nuclear to cytosolic fluorescence intensity from a single experiment (right). Scale bar is 5 µm. (B) Representative image of HEK293T cells expressing Xenopus β-catenin (665-745)-GFP. LaminB1 was labeled to locate the nuclear envelope. GFP was used as a control. Ratio of nuclear to cytoplasmic intensities from two independent replicates (right). Scale bar is 10 µm. (C) Representative image of HEK293T expressing full-length human β-catenin GFP and human β-catenin (Δ665-745)-GFP. Ratio of nuclear to cytoplasmic intensities from three independent replicates (right). Hoechst was labeled to locate the nuclear compartment. Scale bar is 6 µm. p-Values are from unpaired two-tailed t-test where ns is p>0.05 and ****p<0.0001. The data is uploaded as Figure 2—figure supplement 2—source data 1.
-
Figure 2—figure supplement 2—source data 1
Source data related to Figure 2—figure supplement 2.
- https://cdn.elifesciences.org/articles/70495/elife-70495-fig2-figsupp2-data1-v2.xlsx
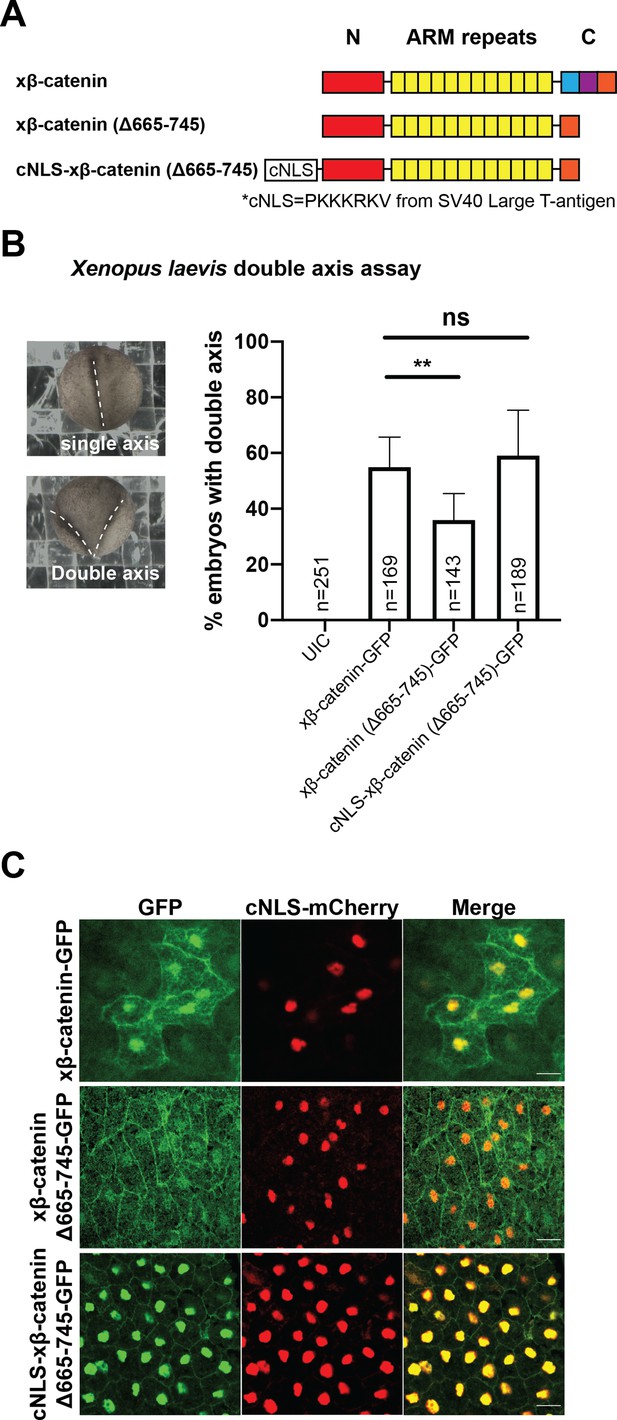
Residues 665–745 of β-catenin are required to induce secondary axes in Xenopus laevis.
(A) Schematic diagram of Xenopus β-catenin constructs. (B) Double axes were scored in st 19 embryos and viewed dorsally with anterior to the top (left). Data from three independent replicates depicted in histogram (right). p-values are from Fisher’s exact test where ns is p>0.05, p<0.05 (*), and 0.0021 (**). (C) Subcellular localization of xβ-catenin-GFP, xβ-catenin (Δ665-745)-GFP, or cNLS-xβ-catenin (Δ665-745)-GFP in the dorsal blastopore lip of stage 10 X. laevis embryos. cNLS-mCherry mRNA was co-injected to mark the nucleus. Scale bar is 30 µm.
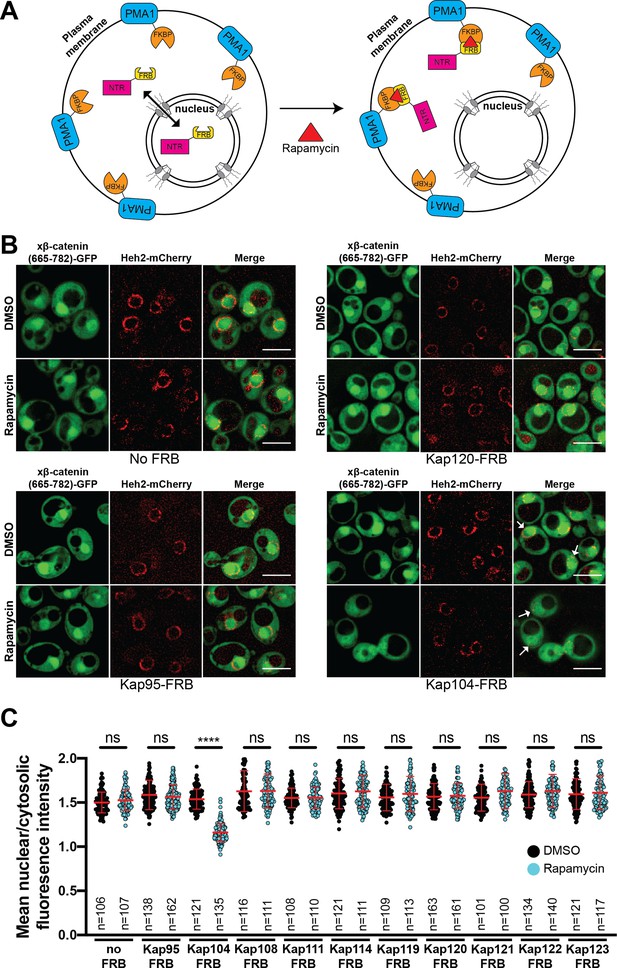
Kap104 is specifically required for β-catenin nuclear accumulation in Saccharomyces cerevisiae.
(A) Schematic of the Anchor-Away assay mediated by the rapamycin-induced dimerization of nuclear transport receptor (NTR)-FKBP-rapamycin binding (FRBP and Pma1-FKBP12). Pma1 is a plasma membrane ATPase. (B) Deconvolved fluorescence images of cells with indicated FRB fusions expressing Xenopus β-catenin (665-782)-GFP treated with DMSO (vehicle) or rapamycin for 15 min. Heh2-mCherry was used as a nuclear envelope marker. White arrows indicate the nucleus. Scale bar is 5 µm. (C) Plot showing the ratio of mean nuclear to cytosolic fluorescence intensity of Xenopus β-catenin (665-782)-GFP in the 10 NTR-FRB strains treated with DMSO or rapamycin from 30 to 40 cells from three independent replicates. Red bar indicates the mean value with the SD. Experiments were performed three times. p-Values are from unpaired two-tailed t-test where ns is p>0.05 and ****p<0.0001. The data is uploaded as Figure 3—source data 1.
-
Figure 3—source data 1
Source data related to Figure 3.
- https://cdn.elifesciences.org/articles/70495/elife-70495-fig3-data1-v2.xlsx
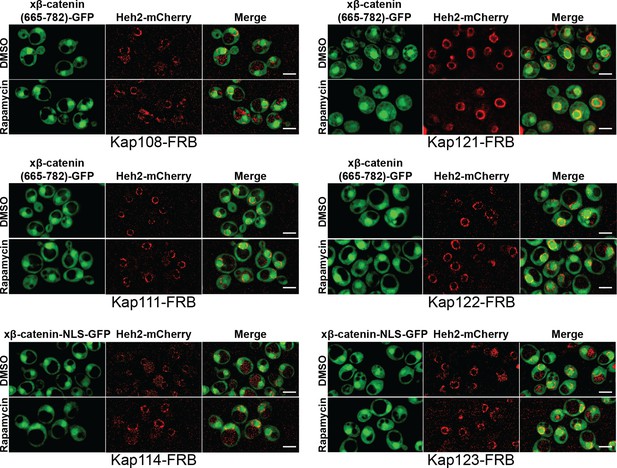
Sub-cellular localization of Xenopus β-catenin (665-782)-GFP in Anchor-Away strains in Saccharomyces cerevisiae.
Representative deconvolved fluorescence image of xβ-catenin (665-782)-GFP treated with DMSO (carrier) or rapamycin in the indicated nuclear transport receptor (NTR)-FKBP-rapamycin binding (FRB) strain. Heh2-mCherry was used as a nuclear membrane marker. Scale bar is 5 µm.
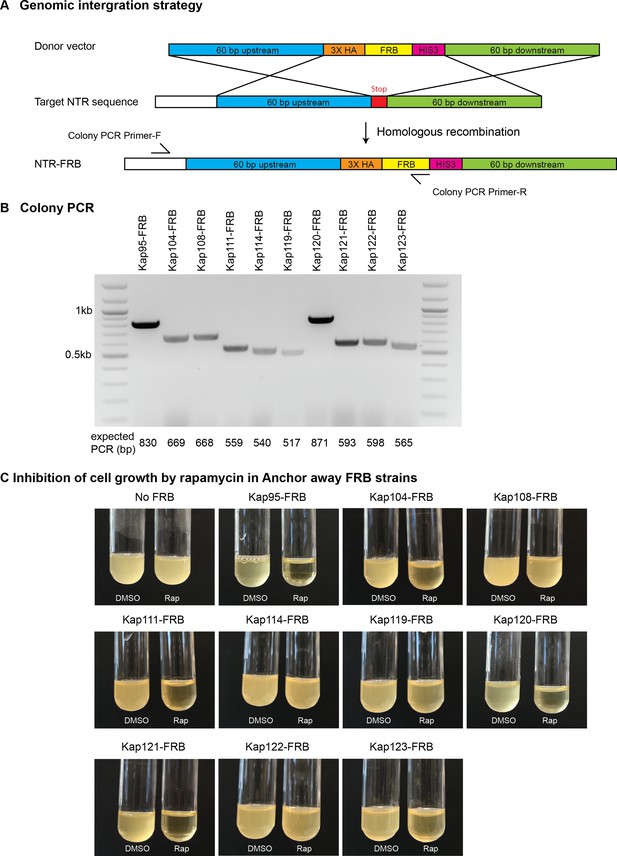
Anchor-Away cloning strategy in Saccharomyces cerevisiae.
(A) Schematic diagram of FKBP-rapamycin binding (FRB) tagging to individual endogenous nuclear transport receptors (NTRs) by homologous recombination. (B) Screening of NTR-FRB strains by colony PCR. Primers are listed in Supplementary file 3. (C) Positive NTR-FRB strains from the colony PCR in (B) were further tested for cell growth as some NTRs are essential for survival. No FRB and DMSO were used a negative controls.
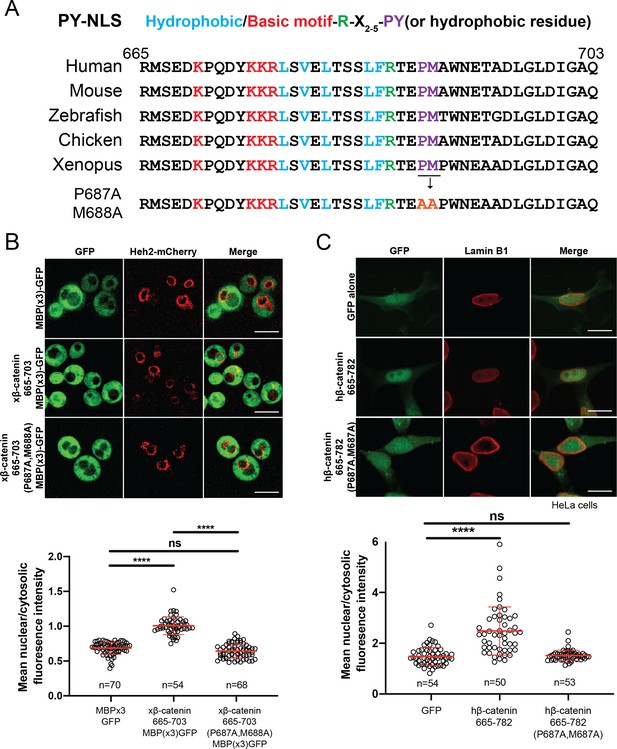
β-catenin contains a PY-NLS that is required for nuclear import.
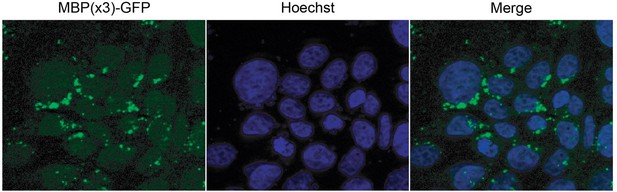
(A) Conservation of amino acid sequences that conform to the PY-NLS consensus (top, from Soniat and Chook, 2015) in the C-terminus of β-catenin. (B) Deconvolved fluorescence images of wildtype yeast cells expressing MBP(x3)-GFP tagged with the Xenopus β-catenin nuclear localization signal (NLS) (665-703) and also an NLS that contains the PM to AA mutation (top). Untagged MBP(x3)-GFP was used as a control. Plot of the ratio of mean nuclear to cytoplasmic fluorescence intensity from a single experiment (bottom). Scale bar is 5 µm. (C) Representative fluorescence image of HeLa cells expressing human β-catenin (665-782) or the PM to AA mutant version (top). LaminB1 was labeled to locate the nuclear envelope. GFP alone was used as a control. Plot of the ratio of mean nuclear to cytoplasmic fluorescence intensity from three experiments (bottom). Scale bar is 15 µm. p-Values are from unpaired two-tailed t-test where ns is p>0.05 and ****p<0.0001 for both (B) and (C). The data is uploaded as Figure 4—source data 1.
-
Figure 4—source data 1
Source data related to Figure 4.
- https://cdn.elifesciences.org/articles/70495/elife-70495-fig4-data1-v2.xlsx
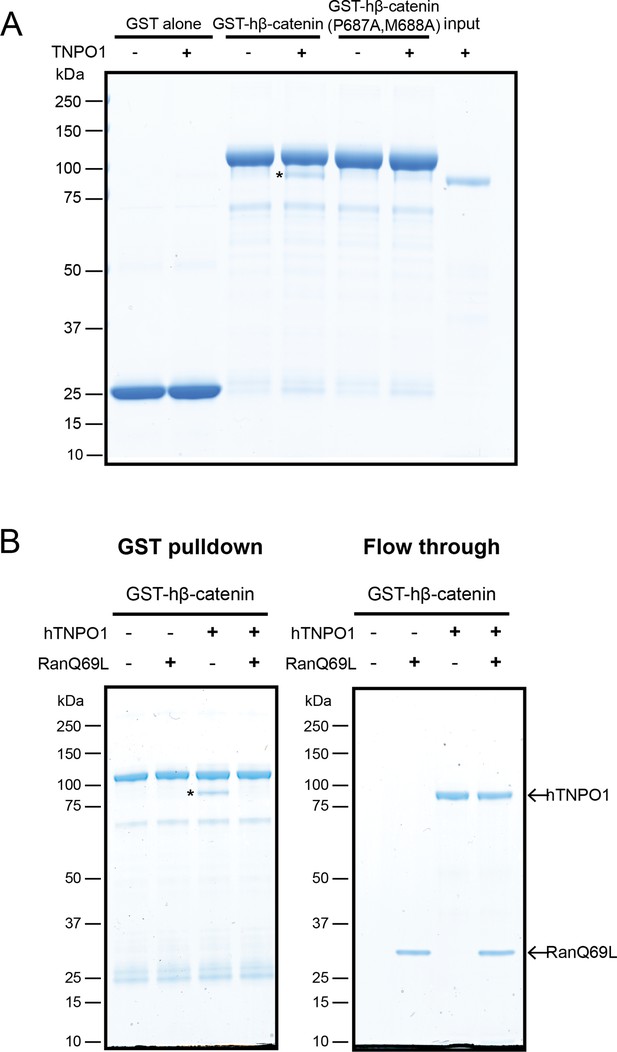
Direct binding of β-catenin and TNPO1 is destabilized by Ran-GTP.
(A) In vitro binding assay of purified recombinant TNPO1 and GST fusions of human β-catenin and human β-catenin containing the PM to AA mutations. GST alone was used as a negative control. (B) In vitro binding assay of purified recombinant TNPO1 to GST fusions of human β-catenin in the presence of GTP hydrolysis deficient Ran mutant loaded with GTP (RanQ69L). Proteins were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and stained with Coomassie blue in (A) and (B). * indicates TNPO1 bound to GST-hβ-catenin.
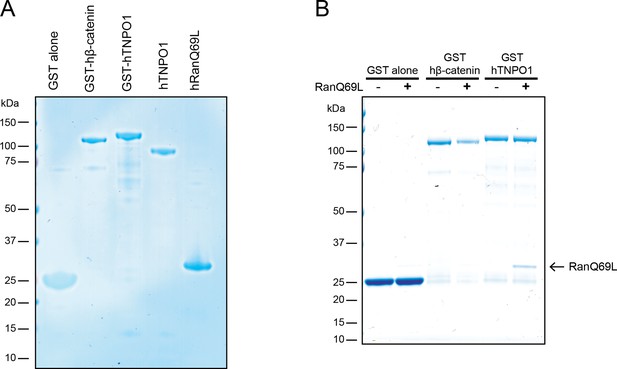
TNPO1 selectively binds to RanGTP in vitro.
(A) Generation of recombinant GST fusions of human β-catenin and TNPO1 and RanQ69L in vitro. (B) In vitro binding assay of purified recombinant RanQ69L to GST fusions of human TNPO1. GST fusions of human β-catenin and RanQ69L buffer were used as a control. Proteins were separated by SDS-PAGE and stained with Coomassie blue in (A) and (B).
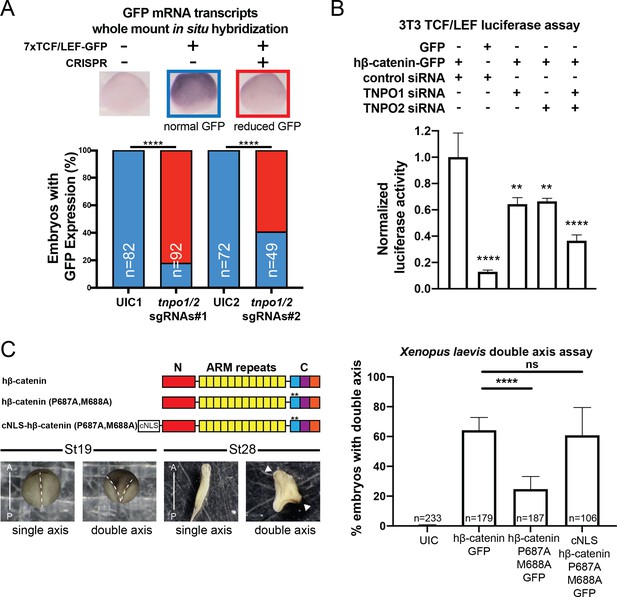
TNPO1/2 and the β-catenin nuclear localization signal (NLS) are required for Wnt signaling in vivo.
(A) Depletion of tnpo1 and tnpo2 using two different pairs of non-overlapping sgRNAs represses gfp expression in Xenopus tropicalis Tg(pbin7Lef-dGFP) embryos at stage 10. Key used to quantify embryos with whole mount in situ hybridization (WMISH) signal (blue – normal gfp signal nd red – reduced gfp signal). Uninjected control (UIC) embryos were used as a negative control. (B) siRNA mediated TNPO1 and/or TNPO2 knockdown reduces luciferase activity in mouse embryonic fibroblasts that harbor a stable integration of luciferase under the control of TCF/LEF promoters. Wnt signaling was activated by human β-catenin-GFP overexpression. Control siRNA and GFP were used as negative controls. Experiments were performed in triplicate. (C) Schematic diagram of three β-catenin constructs used in the double axis assay in Xenopus laevis. ** indicates P687A, M688A substitutions (top left). Dorsal views of X. laevis embryos with anterior to the top (bottom left). Dotted lines indicate the embryonic axis, and the white arrows indicate the head. Histogram of the percent of embryos with secondary axes from three independent replicates. p-Values are from Fisher’s exact test (A) and (C) and unpaired two-tailed t-test (B) where ns is p>0.05, p<0.05 (*), 0.0021 (**), 0.0002 (***), and p<0.0001 (****).The data is uploaded as Figure 6—source data 1.
-
Figure 6—source data 1
Source data related to Figure 6.
- https://cdn.elifesciences.org/articles/70495/elife-70495-fig6-data1-v2.xlsx
Sequence alignment of Tnpo1 and Tnpo2 across species.
Amino acid sequences of transportin 1 and transportin 2 were compared across four different species (Saccharomyces cerevisiae, human, mouse, and Xenopus tropicalis). Each residue in the alignment is colored using Jalview software (Waterhouse et al., 2009).
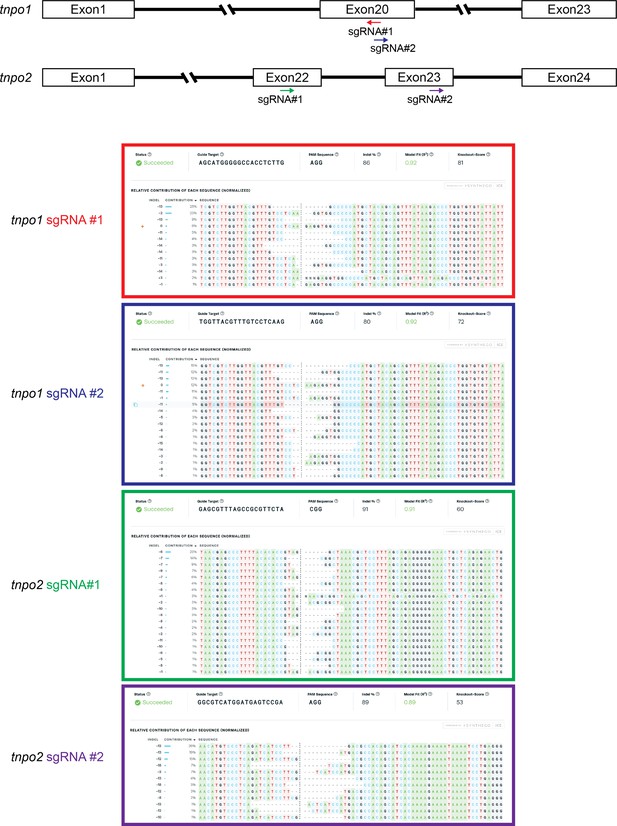
X. tropicalis tnpo1 and tnpo2 gene depletion by CRISPR/Cas9.
Schematic diagram of tnpo1 and tnpo2 sgRNA target sites (top). Inference of CRISPR Edits (ICE) analysis of indel mutations at predicted target sites (bottom).
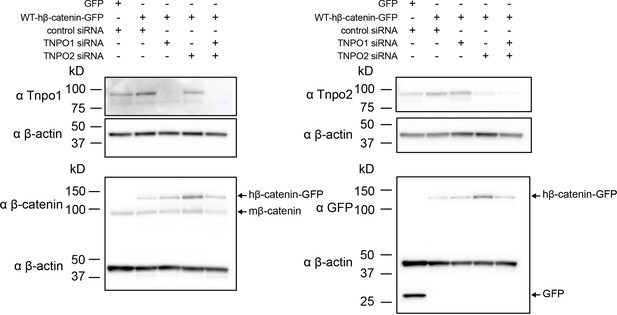
Western blots of Tnpo1/2 and β-catenin from 3T3 TCF/LEF luciferase assays.
Western blot demonstrating the efficacy of siRNA mediated Tnpo1 and Tnpo2 depletion in mouse embryonic fibroblast Wnt reporter cell lines.
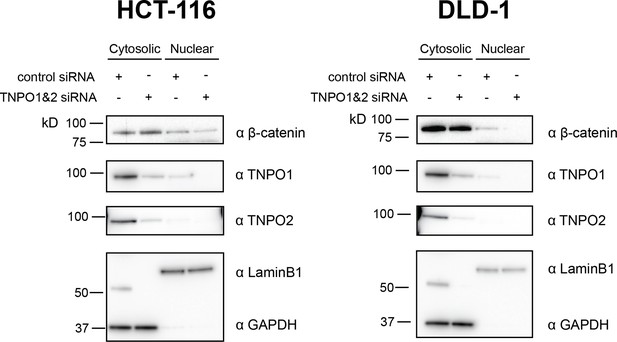
TNPO1/2 regulates nuclear β-catenin levels in colorectal cancer cells.
Western blot demonstrating the efficacy of siRNA mediated Tnpo1 and Tnpo2 depletion on β-catenin nuclear and cytoplasmic levels in two colorectal cancer cell lines, HCT-116 and DLD-1.
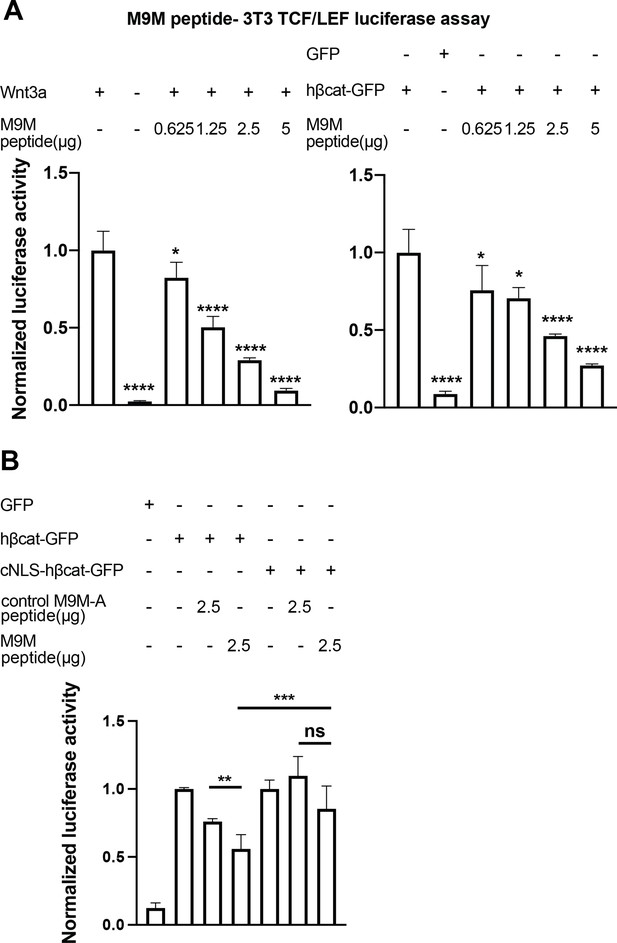
The M9M peptide inhibits Wnt signaling.
Wnt signaling was activated by Wnt3a (A, left), human β-catenin-GFP overexpression (A, right and B), or cNLS-human β-catenin-GFP (B). No Wnt3a or GFP overexpression were used as negative controls. Experiments were performed in triplicate (A) or duplicate in two independent experiments (B). p-Values are from unpaired two-tailed t-test where ns is p>0.05, p<0.05 (*), 0.0021 (**), 0.0002 (***), and p<0.0001 (****). The data is uploaded as Figure 7—source data 1.
-
Figure 7—source data 1
Source data related to Figure 7.
- https://cdn.elifesciences.org/articles/70495/elife-70495-fig7-data1-v2.xlsx
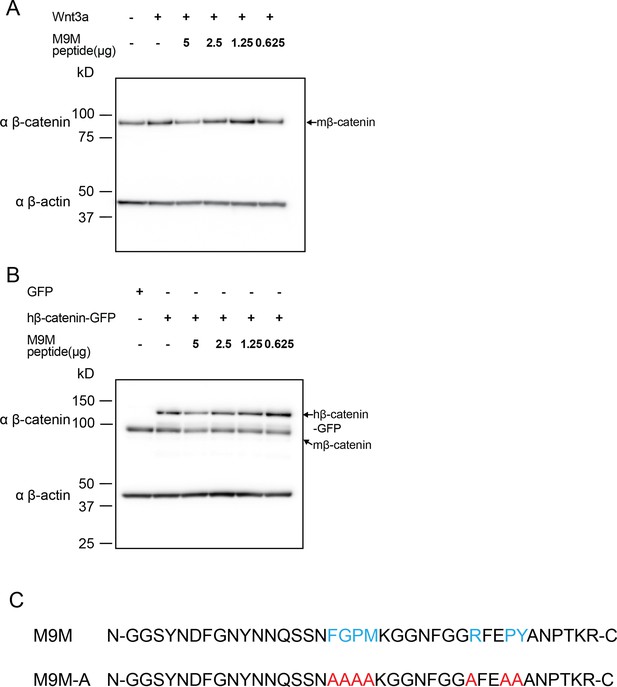
Western blots of β-catenin from 3T3 TCF/LEF luciferase assays with M9M peptide treatment.
Western blot data for M9M peptide treatment in mouse embryonic fibroblast Wnt reporter cell lines. Wnt signaling is activated by (A) Wnt3a or (B) human β-catenin. (C) PY-NLS residues (blue) in M9M peptide were mutated to alanine (red) to create M9M-A peptide.
Tables
List of human nuclear transport receptor (NTR) and Saccharomyces cerevisiae orthologs.
| Human NTR | S. cerevisiae orthologs |
|---|---|
| Kap β1 | Kap95 |
| Kap α | Kap60/Srp1 |
| Transportin 1 | Kap104 |
| Importin-5/Kap β3 | Kap121/Pse1 |
| Importin-4/RanBP5 | Kap123 |
| Importin-7/RanBP7 | Kap119/Nmd5 |
| Importin-8/RanBP8 | Kap108/Sxm1 |
| Importin-9 | Kap114 |
| Importin-11 | Kap120 |
| Transportin-SR/TNPO3 | Kap111/Mtr10 |
| Importin-13 | Kap122/Pdr6 |
| CRM1/Exportin-1 | Xpo1 |
| Exportin-t | Los1 |
| Exportin-5 | Kap142/Msn5 |
| CAS | Cse1 |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain and strain background (Xenopus tropicalis) | Tg(pbin7LEF-dGFP) | National Xenopus Resources at MBL | NXR_1094 | |
| Strain and strain background (Xenopus laevis) | X. laevis | NASCO | LM00535 and LM00715 | |
| Strain and strain background (Escherichia coli) | BL21 Gold (DE3) | Agilent | 230132 | |
| Strain and strain background (E. coli) | XL-10 Gold | Agilent | 200314 | |
| Strain and strain background (E. coli) | DH5-alpha | NEB | C2987 | |
| Cell line (M. musculus) | Leading Light Wnt Reporter Cell line-TCF/ LEF luciferase 3T3 mouse fibroblast | Enzo Life Sciences | ENZ-61001–0001 | |
| Cell line (Homo-sapiens) | Human embryonic kidney 293 (HEK293T) | ATCC | CRL-3216 | |
| Cell line (Homo-sapiens) | HeLa | ATCC | CCL-2 | |
| Cell line (Homo-sapiens) | Human colorectal cancer (HCT 116) | ATCC | CCL-247 | |
| Cell line (Homo-sapiens) | Human colorectal cancer (DLD-1) | ATCC | CCL-221 | |
| Transfected construct (M. musculus and human) | siRNA to TNPO1 & 2 | Thermo Fisher | ||
| Antibody | Anti-β-catenin (mouse monoclonal) | Santa Cruz | sc-7963 HRP, RRID:AB_626807 | WB (1:1000) |
| Antibody | Anti-β-actin (mouse monoclonal) | Santa Cruz | sc-47778 HRP, RRID:AB_2714189 | WB (1:10000) |
| Antibody | Anti-GFP (mouse monoclonal) | Santa Cruz | sc-9996 HRP, RRID:AB_627695 | WB (1:1000) |
| Antibody | Anti-Transportin-1 (mouse monoclonal) | Abcam | ab10303, RRID:AB_2206878 | WB (1:1000) |
| Antibody | Anti-Transportin-2 (Rabbit polyclonal) | Proteintech | 17831–1-AP, RRID:AB_10598481 | WB (1:3000) |
| Antibody | Anti-LaminB1 (Rabbit polyclonal) | Abcam | Ab16048, RRID:AB_443298 | IF (1:500) WB (1:1000) |
| Antibody | Anti-GAPDH (mouse monoclonal) | Santa Cruz | sc-47724 HRP; RRID:AB_627678 | WB (1:3000) |
| Sequence-based reagent | tnpo1 CRISPR 1 | This paper | Oligonucleotides | ttctaatacgactcactataGGCATGGGGGCCACCTCTTGgttttagagctagaa |
| Sequence-based reagent | tnpo1 CRISPR 2 | This paper | Oligonucleotides | ttctaatacgactcactataGGGTTACGTTTGTCCTCAAGgttttagagctagaa |
| Sequence-based reagent | tnpo2 CRISPR 1 | This paper | Oligonucleotides | ttctaatacgactcactataGGGCGTTTAGCCGCGTTCTAgttttagagctagaa |
| Sequence-based reagent | tnpo2 CRISPR 2 | This paper | Oligonucleotides | ttctaatacgactcactataGGCGTCATGGATGAGTCCGAgttttagagctagaa |
| Sequence-based reagent | siRNA: negative control | Thermo Fisher | 4390843 | Silencer Select |
| Sequence-based reagent | siRNA: mouse TNPO1 | Thermo Fisher | s108857 | Silencer Select |
| Sequence-based reagent | siRNA: mouse TNPO2 | Thermo Fisher | s102754 | Silencer Select |
| Sequence-based reagent | siRNA: human TNPO1 | Thermo Fisher | s7934 | Silencer Select |
| Sequence-based reagent | siRNA: human TNPO2 | Thermo Fisher | s26881 | Silencer Select |
| Peptide, recombinant protein | M9M-A | LifeTein | Custom | N-GGSYNDFGNYNNQSSNAAAAKGGNFGGAFEAAANPTKR-C |
| Peptide, recombinant protein | M9M | LifeTein | Custom | N-GGSYNDFGNYNNQSSNFGPMKGGNFGGRFEPYANPTKR-C |
| Commercial assay or kit | Luciferase Assay System | Promega | E1500 | |
| Commercial assay or kit | NE-PER Nuclear Cytoplasmic Extraction Reagents | Thermo Scientific | 78833 | |
| Commercial assay or kit | jetPRIME | Polyplus transfection | 114–15 | |
| Commercial assay or kit | ProteoJuice Protein Transfection | Millipore Sigma | 71281 | |
| Commercial assay or kit | MycoAlert Detection Kit | Lonza | LT07-118 | |
| Chemical compound and drug | Rapamycin | Fisher scientific | AAJ62473MF | |
| Chemical compound and drug | Glutathione Sepharose 4B | Millipore Sigma | GE17-0756-01 | |
| Chemical compound and drug | Protease Inhibitor Cocktail mix | Millipore Sigma | P8340-5ML | |
| Chemical compound and drug | ProTEV Plus | Promega | V6101 | |
| Chemical compound and drug | NEBExpress Ni-NTA Magnetic Beads | NEB | S1423S | |
| Chemical compound and drug | Isopropyl β-d-1-thiogalacto pyranoside (IPTG) | Thermo Fisher | 15529019 | |
| Software and algorithm | Fiji | ImageJ | https://imagej.net/Fiji | |
| Software and algorithm | Prism 9 | Graphpad | https://www.graphpad.com/ |
Additional files
-
Supplementary file 1
List of genotypes and origins of all Saccharomyces cerevisiae strains used in this study.
- https://cdn.elifesciences.org/articles/70495/elife-70495-supp1-v2.xlsx
-
Supplementary file 2
List of all plasmids used in this study.
- https://cdn.elifesciences.org/articles/70495/elife-70495-supp2-v2.xlsx
-
Supplementary file 3
List of primers used for the Anchor-Away assay in this study.
- https://cdn.elifesciences.org/articles/70495/elife-70495-supp3-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/70495/elife-70495-transrepform1-v2.pdf
-
Source data 1
Source data related to original gels and western blots.
- https://cdn.elifesciences.org/articles/70495/elife-70495-data1-v2.zip