Cyclin F drives proliferation through SCF-dependent degradation of the retinoblastoma-like tumor suppressor p130/RBL2
Figures
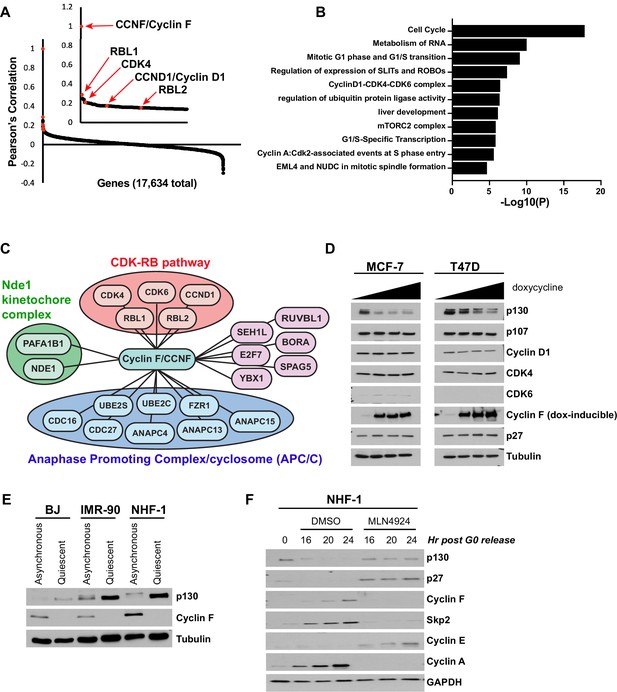
Analysis of the Cancer Dependency Map reveals that CCNF is highly correlated with the CDK-RB network.
(A) Cancer Dependency Map data from project Achilles were analyzed to identify the impact of gene loss-of-function on cellular fitness, and fitness correlation with that of CCNF, based on pooled CRISPR/Cas9 gene knockout screens performed in 789 cell lines. Pearson’s correlation coefficients are reported for all gene pairs (each dot corresponds to a single CCNF-gene X pair). The Pearson’s correlations for CCNF compared to 17,634 other genes are shown. The CDK-RB network members highlighted in red all score in the top 0.5% of genes whose impact on fitness is most highly correlated with CCNF. (B) Gene ontology (GO) analysis was performed for the top 0.05% of genes whose impact on fitness is most highly correlated with CCNF. The top 10 enriched GO terms and their corresponding p-value is shown. (C) The top 0.05% of genes whose impact on fitness is most highly correlated with CCNF were sorted by the GO term cell division (GO:0051301). A graphical representation of the remaining 21 genes is shown. Genes are grouped by their known associations with specific functional pathways or complex, including CDK-RB, Nde1-kinetochore, or APC/C. (D) MCF-7 and T47D cells were engineered to contain a TET-inducible cyclin F transgene. Cells were treated with 0 (vehicle control), 5, 25, or 100 ng/ml of doxycycline to induce cyclin F expression, and the indicated proteins were analyzed by immunoblot. No band was detected for CDK6 in T47D cells. Representative of n=3 experiments. (E) BJ, IMR-90, and NHF-1 human fibroblast cell lines were synchronized in G0 by 48 hr serum starvation (quiescent) or allowed to proliferate normally (asynchronous). Whole-cell extracts were collected for immunoblot analysis. Representative of n=3 experiments. (F) NHF-1 cells were synchronized in G0 by serum starvation for 48 hr. Cells were then released into the cell cycle upon the addition of serum-containing media supplemented with either MLN4924 or vehicle (DMSO) as a control. Cells were collected at the indicated time points after release. Protein levels were assessed by immunoblot. Data represent n=3 independent experiments.
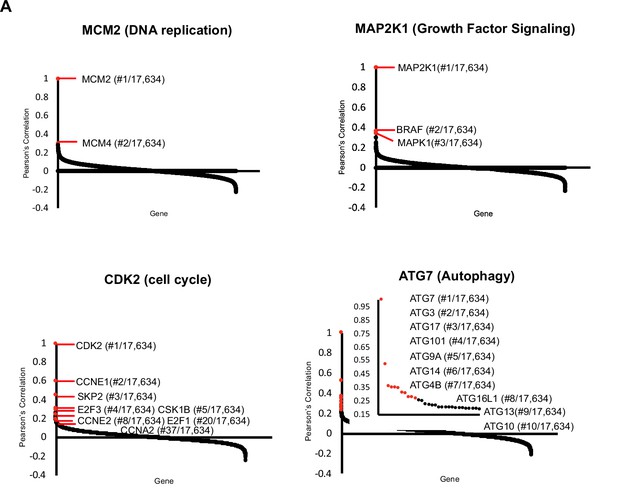
Analysis of Project Achilles Dependency Map reveals that highly correlated genes encode proteins known to interact.
(A) Cancer Dependency Map data from Project Achilles were analyzed to identify genes whose loss-of-function impact on cellular fitness is correlated with known proliferative regulators. Data are based on pooled CRISPR/Cas9 gene knockout screens performed in 789 cell lines. Pearson’s correlation coefficients for genes correlated with MCM2, CDK2, MAPK1, and ATG7. Known interactors are highlighted in red. Pearson’s correlation coefficients are reported for all gene pairs (each dot corresponds to a gene pair).
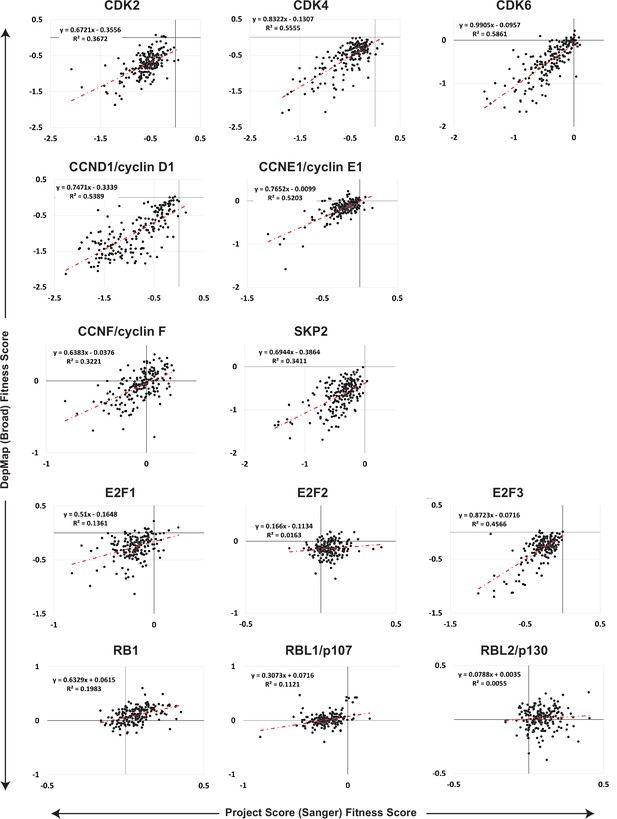
Gene dependency scores from the Broad Institute Project Achilles Dependency Map data set and the Sanger Project Score data set correlate highly.
Fitness scores for gene knockouts of CDK2, CDK4, CDK6, CCND1, CCNE1, E2F1, E2F2, E2F3, RB1, RBL1, and RBL2 were independently determined for Project Achilles (Broad Institute) and for Project Score (Sanger) in hundreds of cancer cell lines. Of the cell lines tested, 189 cell lines are included in both data sets. The fitness score from the Project Achilles Dependency Map (DEPMAP) data set was plotted versus the gene dependency scores independently determined and included in the Sanger Project Score data set. Linear fit and R2 values were calculated to compare the two data sets.
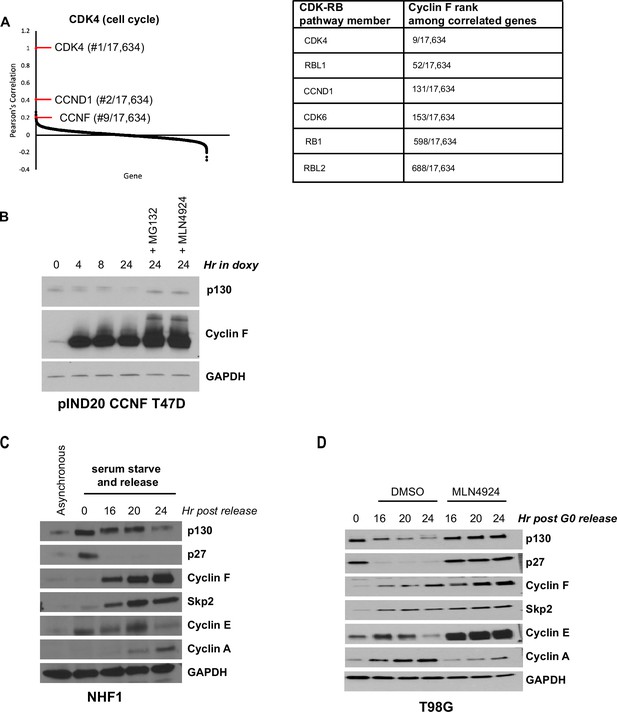
Analysis of Project Achilles Dependency Map reveals that CDK-RB network genes correlate highly with CCNF.
(A) Pearson’s correlation coefficients for genes whose fitness score correlates with CDK4. Genes of interest are highlighted in red (left). CCNF rank among genes correlated with CDK4, CDK6, CCND1, RB1, RBL1, and RBL2 (right). (B) T47D cells engineered to contain a TET-inducible cyclin F transgene were treated with 100 ng/ml doxycycline to induce cyclin F expression. Cells were collected at the indicated time points. Where indicated, the proteasome inhibitor, MG132, or the neddylation inhibitor, MLN4924, was added. Proteins were analyzed by immunoblot. Representative of n=2 experiments. (C) NHF-1 cells were synchronized in G0 by 48 hr serum starvation (time 0 hr) and then released into the cell cycle by re-introduction of serum-containing media. Cells were collected at the indicated times and immunoblotted. Untreated, asynchronous cells (lane 1) are shown as a control. Representative of n=3 experiments. (D) T98G cells were synchronized in G0 by serum starvation for 48 hr. Cells were then released into the cell cycle upon the addition of serum-containing media supplemented with either MLN4924 or vehicle (DMSO) as a control. Cells were collected at the indicated time points after release. Protein levels were assessed by immunoblot. Data represent n=2 independent experiments for T98G cells and n=3 independent experiments for NHF-1 cells.
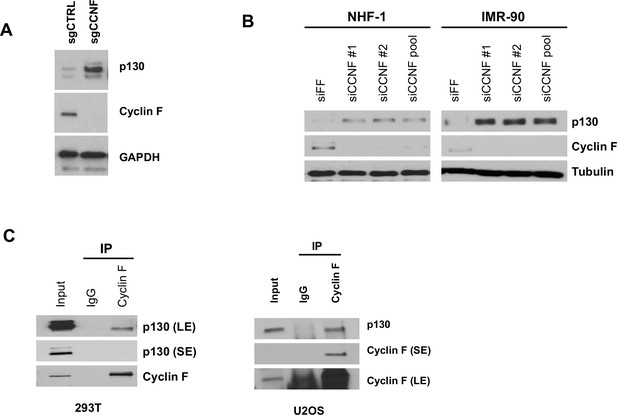
Cyclin F regulates and interacts with endogenous p130.
(A) Asynchronously proliferating CCNF CRISPR/Cas9 knockouts (sgCCNF) and control (sgCtrl) HeLa cells were blotted for levels of the indicated proteins. Representative of n=3 experiments. (B) NHF-1 and IMR-90 cells were transfected with two different siRNAs targeting CCNF or a control siRNA targeting Firefly Luciferase (siFF). Whole-cell lysates were immunoblotted for the indicated proteins. Representative of n=3 experiments. (C) Endogenous cyclin F was immunoprecipitated from asynchronously proliferating HEK293T cells (left) or asynchronously proliferating U2OS cells (right). Indicated proteins were immunoblotted. SE=short exposure; LE=long exposure; representative of n=3 experiments.
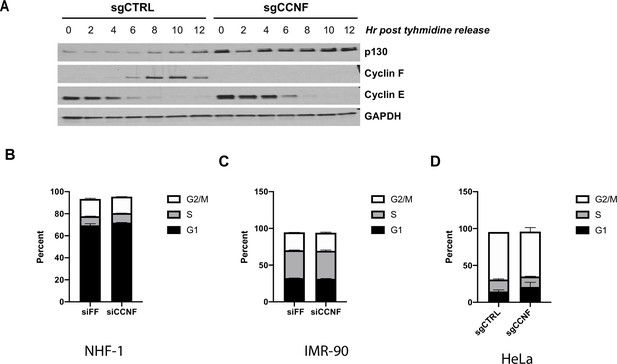
Cyclin F knockout and depletion across cell lines.
(A) CCNF CRISPR/Cas9 knockouts (sgCCNF) and control (sgCtrl) HeLa cells were synchronized in S-phase by double thymidine block and then released synchronously into the cell cycle upon the addition of drug-free media. Protein levels were monitored at the indicated time points by immunoblot. Representative of n=3 experiments. (B–D) Cells were fixed, stained with propidium iodide, and flow cytometry was performed to determine cell cycle distribution for NHF-1 cells treated with siFF or siCCNF (B), IMR-90 cells treated with siFF or siCCNF (C), and CCNF CRISPR/Cas9 knockouts (sgCCNF) and control (sgCtrl) HeLa cells (D). Data represent n=3 experiments.
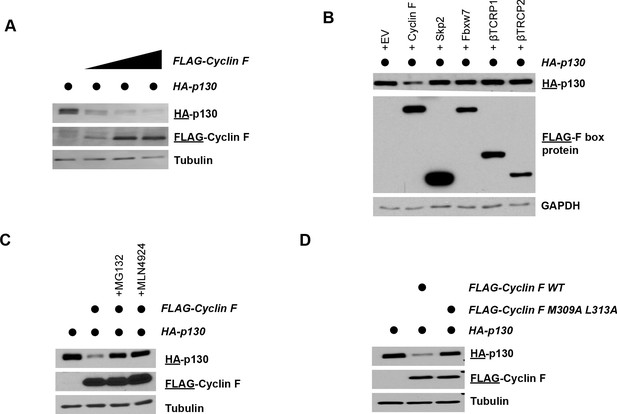
Cyclin F promotes p130 degradation.
(A) HEK293T cells transiently expressing HA-p130 with an empty FLAG vector control (lane 1) or together with increasing amounts of FLAG-cyclin F (lanes 2–4). Cells were collected and analyzed by immunoblot 24 hr post-transfection. The antigen being immunoblotted for is represented by the underline, here and in all experiments below. Representative of n=3 experiments. (B) HEK293T cells transiently expressing HA-p130 with an empty FLAG vector control (lane 1) or together with FLAG-cyclin F (lanes 2–4). MG132 (proteasome inhibitor) or MLN4924 (neddylation inhibitor) were added for 6 hr prior to harvesting. Cells were collected and analyzed by immunoblot 24 hr post-transfection. Representative of n=3 experiments. (C) HEK293T cells transiently expressing HA-p130 with an empty FLAG vector control (lane 1) or together with FLAG-cyclin F WT (lane 2) or FLAG-cyclin F(M309A L313A) (lane 3), as indicated. Cells were collected and analyzed by immunoblot 24 hr post-transfection. Representative of n=3 experiments. (D) HEK293T cells transiently expressing HA-p130 with an empty FLAG vector control (lane 1) or together with the indicated FLAG-tagged F-box proteins (lanes 2–6). Cells were collected and analyzed by immunoblot 24 hr post-transfection. Representative of n=3 experiments.
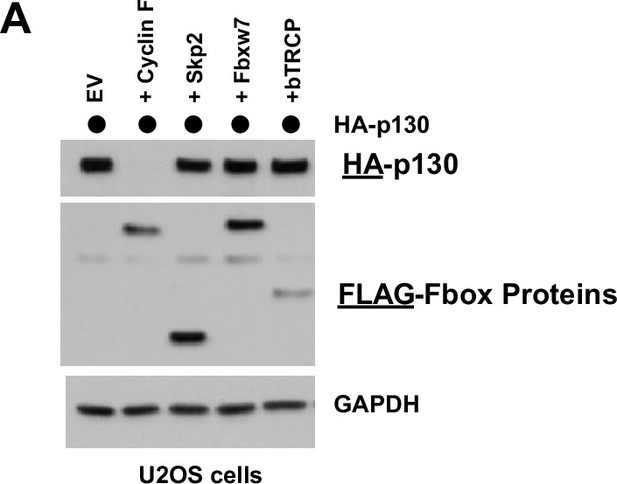
P130 levels following Fbox protein expression in U2OS cells.
(A) U2OS cells transiently expressing HA-p130 with an empty FLAG vector control (lane 1) or together with the indicated FLAG-tagged F-box proteins (lanes 2–5). Cells were collected and analyzed by immunoblot 24 hr post-transfection. Representative of n=3 experiments.
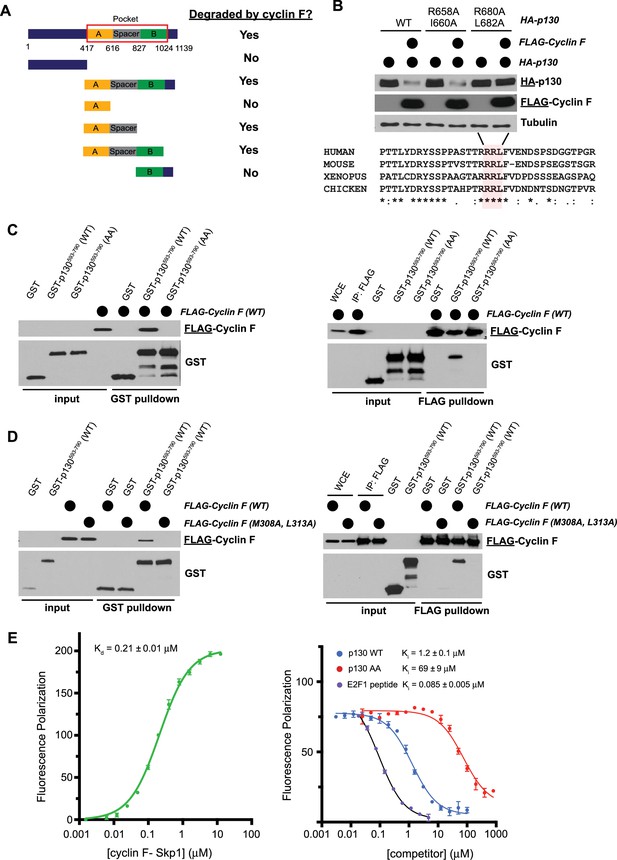
Cyclin F binds p130 directly and promotes p130 degradation through a conserved degron motif.
(A) Graphical depiction of p130 domain structure. Indicated p130 truncation mutants were screened for their ability to be degraded following co-overexpression with cyclin F. The data supporting these conclusions are shown in Figure 4—figure supplement 1. (B) The first and last amino acids in the two potential cyclin F binding sites in the p130 spacer domain (R658-I660 and R680-L682) were mutated to alanine (AxA). HEK293T cells transiently expressed HA-p130 alone (WT or AxA mutants, as indicated) or together with FLAG-cyclin F WT. Cells were collected and analyzed by immunoblot 24 hr post-transfection. Representative of n=3 experiments (top) and amino acid sequence alignment for human, mouse, frog, and chicken p130 (bottom). (C) GST-p130593–790 (WT) and GST-p130593–790 (AA) were produced in Escherichia coli and purified. FLAG-cyclin F was transiently expressed in HEK293T cells. GST pulldowns (left) and FLAG pulldowns (right) were used to determine the interaction of p130 and cyclin F. Interaction was assessed by immunoblot after pulldown (representative of n=3 experiments). (D) GST-p130(WT) and FLAG-cyclin F(WT) or FLAG-cyclin F(M309A L313A) were expressed as described in (C). Interaction and binding were assessed as in (C) (representative of n=3 experiments). (E) Fluorescence polarization anisotropy assay to detect direct association of p130 with cyclin F. (Left) 10 nM TAMRA-p130674–692 probe was titrated with increasing concentrations of purified GST-cyclin F25–546-Skp1. (Right) The p130 probe bound with 0.5 μM GST-cyclin F25–546-Skp1 was displaced with increasing concentrations of the indicated p130 protein construct or E2F184–99 peptide. Experiments were performed in triplicate, and the standard deviation is reported as the error.
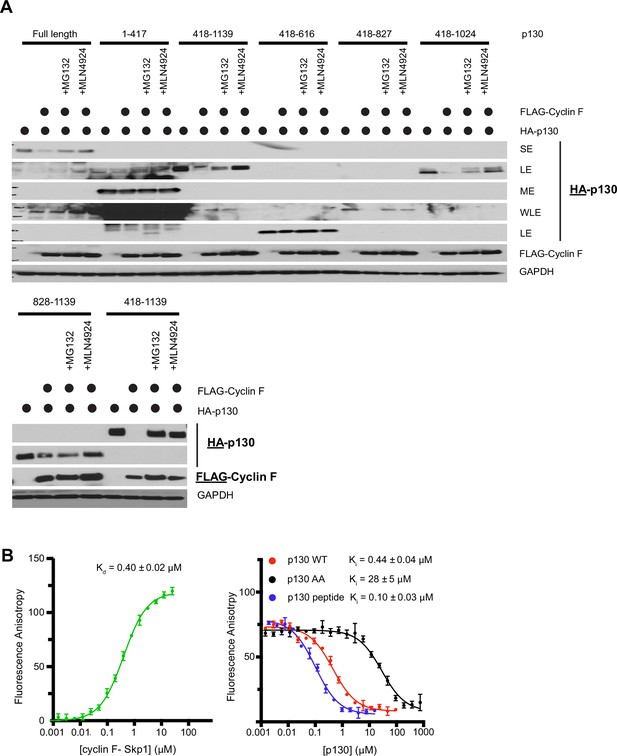
Requirements for cyclin F binding and degradation of p130 in vivo and in vitro.
(A) HEK293T cells were transiently transfected for 24 hr with the indicated HA-tagged p130 truncations either alone or in combination with FLAG-cyclin F. MG132 or MLN4924 were added for the last 6 hr where indicated. Cells were collected 24 hr post-transfection and analyzed by immunoblot. (Top) p130 full-length (amino acids 1–1139), p1301–417, p130418–1139, p130418–616, p130418–827, and p130418–1024 and (Bottom) p130828–1139 and p130418–1139. Blots are representative of n=3 replicate experiments. SE=short exposure; ME=medium exposure; LE=long exposure; WLE=wicked long exposure. (B) Additional fluorescence polarization anisotropy assay to detect direct association of E2F1 and p130 with cyclin F. (Left) 10 nM TAMRA-E2F184–99 was titrated with increasing concentrations of purified GST-cyclin F25–546-Skp1. (Right) The E2F1 probe bound with 0.5 μM GST-cyclin F25–546-Skp1 was displaced with increasing concentrations of the indicated p130 protein construct or synthetic peptide (residues 674–692). Experiments were performed in triplicate, and the standard deviation is reported as the error.
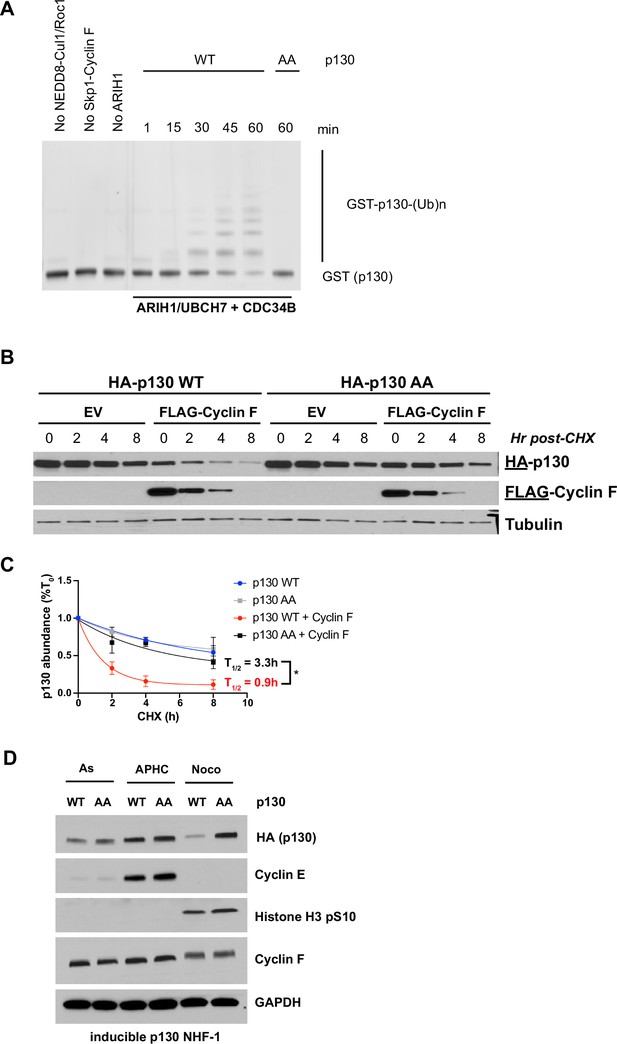
SCFCyclin F regulates the ubiquitination and stability of p130.
(A) Ubiquitination reactions were performed with SCFcyclin F, ARIH1/UBCH7, and CDC34B. GST-tagged p130593–790 WT and GST-p130593–790 were used as substrates and detected by immunoblot against GST. Data are representative of n=3 experiments. (B) FLAG-cyclin F, HA-p130(WT), and/or HA-p130(AA) were transiently expressed in HEK293T cells for 24 hr. The protein synthesis inhibitor cycloheximide (CHX) was added and cells were collected at the indicated time points. Protein levels were determined by immunoblot. Representative of n=3 experiments. (C) Quantification of (B). *=p<0.05 (Student’s t-test). Data are shown as mean ± SEM for n=3 experiments. (D) Inducible p130 NHF-1 cells were grown in media-containing 100 ng/ml doxycycline for 14 days to induce p130 expression, or in media-containing vehicle control. On indicated days, cells were pulsed with EdU for 30 min prior to harvest/fixation, DNA was stained with DAPI, and cells were analyzed by flow cytometry. Data for S- and G2/M phases are in Figure 6—figure supplement 1D. Data represent mean ± SEM for n=3 replicates.
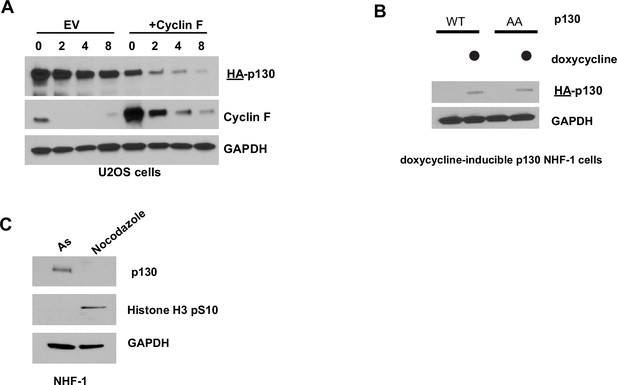
p130 stability in U2OS and NHF-1 cells.
(A) HA-p130(WT) with or without FLAG-cyclin F was transiently expressed in U2OS cells for 24 hr. The protein synthesis inhibitor cycloheximide (CHX) was added and cells were collected at the indicated time points. Protein levels were determined by immunoblot. Representative of n=2 experiments. (B) p130(WT) and p130(AA) expression is induced with 100 ng/ml doxycycline for 24 hr. No p130 expression is detected in vehicle control (water) inductions. (C) Inducible p130 NHF-1 cells were grown for 24 hr in media-containing 100 ng/ml doxycycline. Then, media was replaced with fresh media supplemented with doxycycline plus vehicle (control), aphidicolin (S-phase synchronization), or nocodazole (mitotic synchronization) for 24 hr. Total protein lysates were immunoblotted for the indicated proteins.
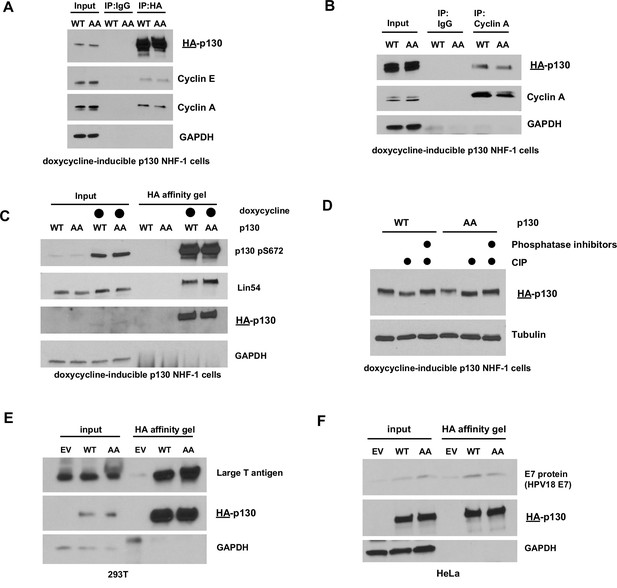
Both p130(WT) and p130(AA) can be phosphorylated and can associate with cyclin E, cyclin A, the DREAM complex, Large T antigen, and E7 protein.
(A–C) Inducible p130(WT) and p130(AA) NHF-1 cells were grown in 100 ng/ml doxycycline for 24 hr. (A) HA-tagged p130 was immunoprecipitated using an anti-HA antibody (or IgG control), and the indicated proteins were immunoblotted. Representative of n=2 independent experiments. (B) Endogenous cyclin A was immunoprecipitated, and the indicated proteins were immunoblotted. Representative of n=3 independent experiments. (C) HA-tagged p130 was isolated using anti-HA affinity resin, and the indicated proteins were immunoblotted. Representative of n=3 independent experiments. (D) Inducible p130(WT) and p130(AA) NHF-1 cells were grown in 100 ng/ml doxycycline for 24 hr. Cells were lysed in phosphatase-free lysis buffer. Total protein lysates were treated with control (buffer), Calf intestinal phosphatase (CIP), or CIP plus a phosphatase inhibitor cocktail for 1 hr at 37°C. Indicated proteins were immunoblotted. Representative of n=2 independent experiments. (E, F) HA-p130(WT) or HA-p130(AA) were transiently transfected into 293T (E) or HeLA (F) cells for 24 hr. The HA-p130 and any co-immunoprecipitating proteins were isolated using HA affinity gel, and indicated proteins were immunoblotted. Representative of n=2 experiments.
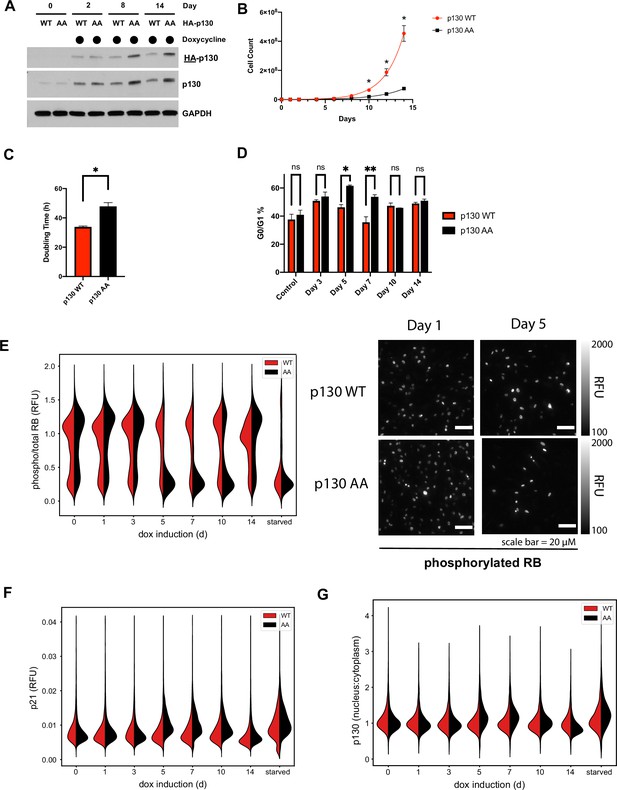
Cells expressing p130(AA) exhibit proliferation defects.
(A) NHF-1 cells were engineered to express a TET-inducible HA-p130(WT) or HA-p130(AA) transgene. Doxycycline (100 ng/ml) was used to induce p130 expression, and water was used as a vehicle control. HA-p130 levels were assessed by immunoblot after cells were grown in doxycycline for up to 14 days. (B) Inducible p130 NHF-1 cells were grown in 100 ng/ml doxycycline-containing media for 14 days to induce p130 expression. Cells were counted on the indicated days. Data represent mean ± SEM for n=3 independent experiments. (C) Doubling time was calculated from counting experiment in (B). Error bars are SEM for n=3 independent experiments. Representative of n=2 independent experiments. (D) NHF-1 cells were grown in media supplemented with nocodazole for 16 hr, then immunoblotted for the indicated proteins. Representative of n=2 independent experiments. (E–G) Inducible p130 NHF-1 cells were grown in 100 ng/ml doxycycline for indicated times. Cells were fixed and analyzed by iterative immunofluorescent staining and imaging. Distributions of single-cell measurements are shown for nuclear phosphorylated versus total RB (E, left) and representative images of phosphorylated RB are shown for indicated days (E, right). Distributions of single-cell measurements are also shown for cytoplasmic p21 (F) and nuclear versus cytoplasmic p130 (G), as indicated.
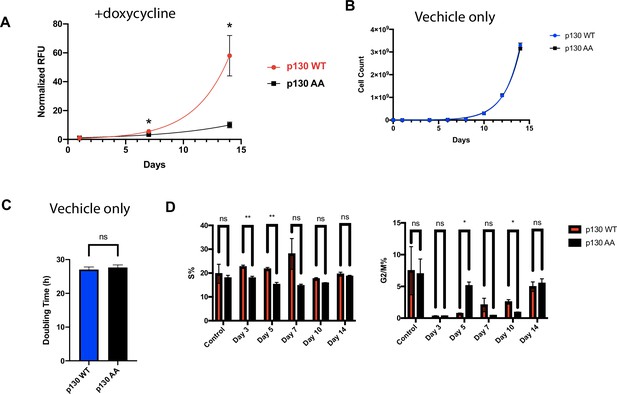
TET-inducible p130(WT) and p130(AA) NHF-1 cells proliferate rapidly prior to induction of p130 expression.
(A) Inducible p130 NHF-1 cells were grown in 100 ng/ml doxycycline-containing media for 14 days to induce p130 expression. Cell number/viability were assessed on the indicated days using the PrestoBlue assay. Data represent mean ± SEM for n=3 independent experiments. (B) Inducible p130(WT) and p130(AA) NHF-1 cells were grown for 2 weeks in media-containing vehicle (water) as control induction of p130. Cells were counted at the indicated times. Data represent mean of n=3 experiments ± SEM. (C) Doubling time was calculated from counting data in (B). Error bars are SEM. (D) Inducible p130 NHF-1 cells were grown in media-containing 100 ng/ml doxycycline for 14 days to induce p130 expression, or in media-containing vehicle control. On indicated days, cells were pulsed with EdU for 30 min prior to harvest/fixation, DNA was stained with DAPI, and cells were analyzed by flow cytometry. Data for G1-phase is in Figure 6D. Data represent the mean of n=3 independent experiments ± SEM.
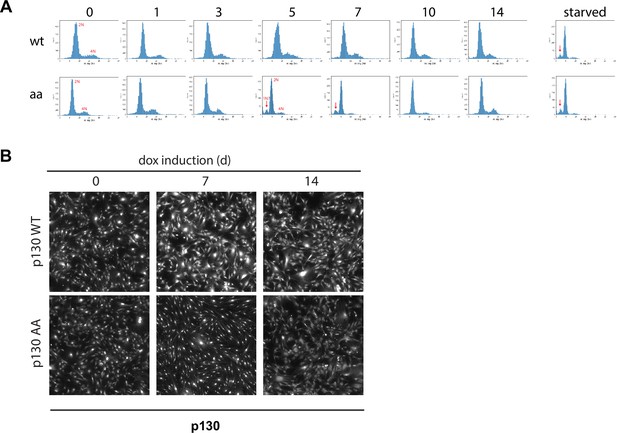
Cell cycle distribution and p130 staining from 4i experiments.
(A, B) Inducible p130 NHF-1 cells were grown in 100 ng/ml doxycycline for indicated time, then protein expression and protein localization were visualized by 4i staining. (A) DNA content assessed by DAPI staining. (B) p130 localization.
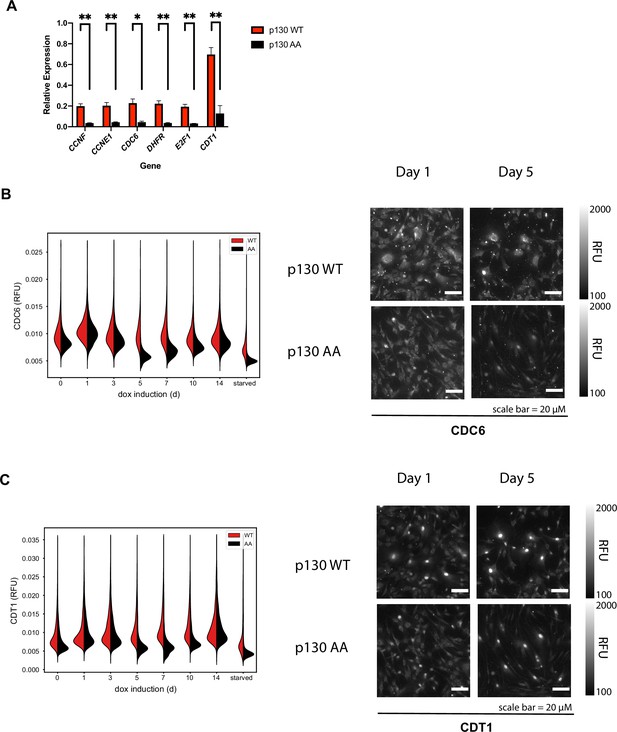
DREAM targets are downregulated in cells expressing p130(AA).
(A) Inducible p130(WT) and p130(AA) NHF-1 cells were grown for 8 days in media-containing 100 ng/ml doxycycline to induce p130 expression. RNA was extracted for rt-qPCR analysis. Gene expression is relative to GAPDH expression and normalized to the vehicle control. Data are mean of n=3 experiments, and error bars are SEM. (B, C) Inducible p130 NHF-1 cells were grown in 100 ng/ml doxycycline for indicated times, and protein levels of CDC6 (B) and Cdt1 (C) were analyzed by iterative immunofluorescent staining and fixed cell imaging. Distributions of single-cell measurements of Cytoplasmic CDC6 (B) and nuclear CDT1 (C) were plotted (left) and representative images are shown for the indicated days (right).
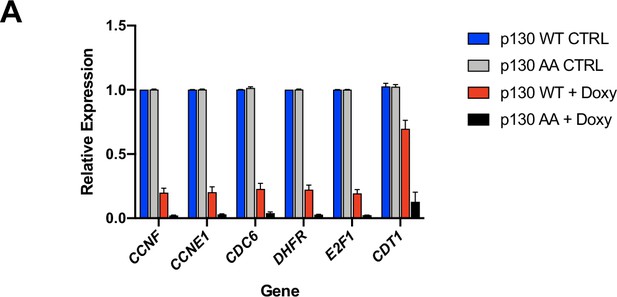
RT-pPCR for DREAM target genes in NHF-1 cells expressing control versus p130(WT) versus p130(A).
(A) Inducible p130(WT) and p130(AA) NHF-1 cells were grown in for 8 days in media-containing 100 ng/ml doxycycline to induce p130 expression (or containing water as a control). RNA was extracted for rt-qPCR analysis. Gene expression is relative to GAPDH expression and normalized to the vehicle control. Data are mean of n=3 experiments and error bars are SEM.
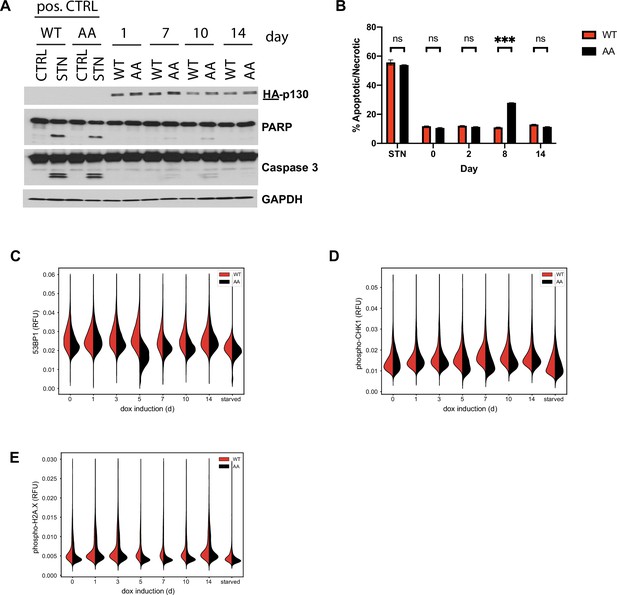
Apoptosis and DNA damage markers in cells expressing p130(WT) or p130(AA).
(A) Inducible p130(WT) and p130(AA) NHF-1 cells were grown in 100 ng/ml doxycycline-containing media for the indicated number of days. Cells were collected on the indicated days and analyzed by immunoblot for the indicated proteins. As a positive control for apoptosis, cells were treated with the broad-spectrum kinase inhibitor staurosporine (or DMSO as a control). Representative of n=2 independent experiments. (B) Inducible p130 (WT) and p130 (AA) NHF-1 cells were grown and treated as in (A). Cells were stained for Annexin V and DNA content (propidium idodide), and flow cytometry was performed to determine the presence of apoptosis/necrosis. Representative of n=3 independent experiments. (C–E) Inducible p130 NHF-1 cells were grown in 100 ng/ml doxycycline for the indicated time, then protein expression in single cells was visualized by 4i staining for (C) total 53BP1 levels; (D) total phospho-chk1 levels; and (E) total phospho-H2A.X levels.
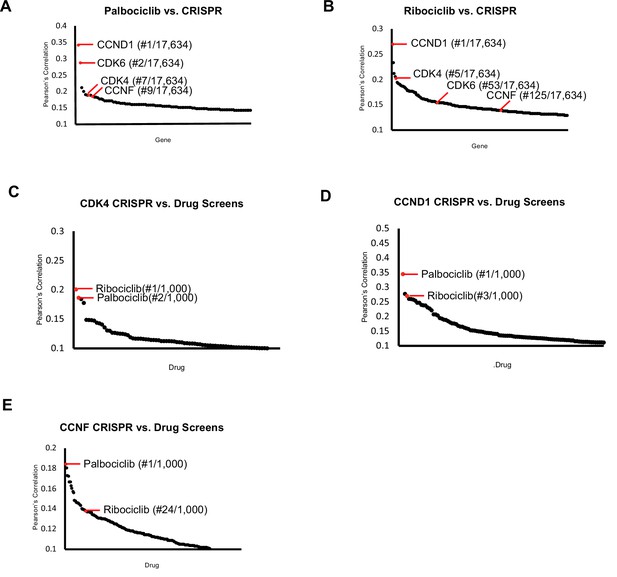
CDK4/6 inhibitors and CCNF knockout correlate highly.
Project Achilles Dependency Map data sets were analyzed to determine: (A) Pearson’s correlation coefficients for gene knockout correlated with Palbociclib treatment. (B) Pearson’s correlation coefficients for gene knockout correlated with Ribociclib treatment. (C) Pearson’s correlation coefficients for the correlation between CDK4 knockout and 1000 drug treatments. (D) Pearson’s correlation coefficients for the correlation between CCND1 knockout and 1000 drug treatments. (E) Pearson’s correlation coefficients for the correlation between CCNF knockout and 1000 drug treatments. For (A–E), only the top scoring, most statistically significant associations are shown.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Homo sapiens) | CCNF | GenBank | Gene ID: 899 | |
| Gene (H. sapiens) | RBL2 | GenBank | Gene ID: 5934 | |
| Antibody | CDK4 (rabbit monoclonal) antibody | CST | Cat. #: D9GE3; RRID:AB_2799229 | IB (1:1000) |
| Antibody | CDK6 (rabbit monoclonal) antibody | CST | Cat. #:D4S8S; RRID:AB_2721897 | IB (1:1000) |
| Antibody | Cyclin A1 (rabbit monoclonal) antibody | Abcam | Cat. #: ab53699; RRID:AB_879763 | IB (1:1000) |
| Antibody | Cyclin A1 (mouse monoclonal) antibody | Santa Cruz | Cat. #: SC-751; RRID:AB_631329 | IB (1:5000); IP (1 µg) |
| Antibody | Cyclin D1 (rabbit monoclonal) antibody | CST | Cat. #: 2978; RRID:AB_2259616 | IB (1:1000) |
| Antibody | Cyclin E1 (mouse monoclonal) antibody | CST | Cat. #: 4129; RRID:AB_2071200 | IB (1:5000); IP (1 µg) |
| Antibody | Cyclin E1 (rabbit monoclonal) antibody | CST | Cat. #: 20808; RRID:AB_2783554 | IB (1:1000) |
| Antibody | Cyclin F (rabbit polyclonal) antibody | Santa Cruz | Cat. #: SC-952; RRID:AB_2071212 | IB (1:5000) |
| Antibody | FLAG (HRP-conjugated mouse monoclonal) antibody | Sigma-Aldrich | Cat. #: A8592; RRID:AB_439702 | IB (1:10,000) |
| Antibody | GAPDH (mouse monoclonal) antibody | Santa Cruz | Cat. #: sc-47724; RRID:AB_627678 | IB (1:5000) |
| Antibody | GST (HRP-conjugated mouse monoclonal) antibody | GeneTex | Cat. #: GTX114099; RRID:AB_1949436 | IB (1:5000) |
| Antibody | HA (mouse monoclonal) antibody | Covance/BioLegend | Covance Cat. #: MMS-101P; RRID:AB_2314672 | IB (1:2000) |
| Antibody | Lin54 (rabbit polyclonal) antibody | Bethyl | Cat. #: A303-799A; RRID:AB_11218173 | IB (1:1000) |
| Antibody | Normal rabbit IgG (polyclonal) antibody | ProteinTech | Cat. #: 30000-0-AP; RRID:AB_2819035 | IP (1 µg) |
| Antibody | p107 (rabbit monoclonal) antibody | CST | Cat. #: D3P3C; RRID:AB_2800144 | IB (1:1000) |
| Antibody | p130 (rabbit monoclonal) antibody | CST | Cat. #: D9T7M; RRID:AB_2798274 | IB (1:1000) |
| Antibody | p130 pS672 (rabbit monoclonal) antibody | Abcam | Cat. #: ab76255; RRID:AB_2284799 | IB (1:5000) |
| Antibody | p27 (rabbit monoclonal) antibody | CST | Cat. #: 2552; RRID:AB_10693314 | IB (1:1000) |
| Antibody | Skp2 (rabbit monoclonal) antibody | CST | Cat. #: 2652; RRID:AB_11178941 | IB (1:5000) |
| Antibody | Tubulin (mouse monoclonal) antibody | Santa Cruz | Cat. #: 32293; RRID:AB_628412 | IB (1:5000) |
| Antibody | Goat anti-mouse IgG HRP-conjugated (goat polyclonal) antibody | Jackson ImmunoResearch | Cat. #: 115-035-003; RRID:AB_10015289 | IB (1:5000) |
| Antibody | Goat anti-rabbit IgG HRP-conjugated (goat polyclonal) antibody | Jackson ImmunoResearch | Cat. #: 111-035-003; RRID:AB_2313567 | IB (1:5000) |
| Antibody | Phospho-RB(S807/S811) (rabbit monoclonal) antibody | CST | Cat. #: 8516; RRID:AB_11178658 | 4i (1:1000) |
| Antibody | RB (mouse monoclonal) antibody | CST | Cat. #: 9309; RRID:AB_823629 | 4i (1:500) |
| Antibody | p21 (goat polyclonal) antibody | R&D Systems | Cat. #: AF1047; RRID:AB_2244704 | 4i (1:200) |
| Antibody | p130 (rabbit monoclonal) antibody | CST | Cat. #: 13610; RRID:AB_2798274 | 4i (1:100) |
| Antibody | Phospho-H2A.X(ser139) (mouse monoclonal) antibody | CST | Cat. #: 80312; RRID:AB_2799949 | 4i (1:200) |
| Antibody | Anti-phospho-Chk1 (rabbit monoclonal) antibody | CST | Cat. #: 12302; RRID:AB_2783865 | 4i (1:800) |
| Antibody | 53 BP1 (rabbit polyclonal) antibody | Abcam | Cat. #: ab36823; RRID:AB_722497 | 4i (1:250) |
| Antibody | CDT1 (rabbit monoclonal) antibody | CST | Cat. #: 8064; RRID:AB_10896851 | 4i (1:200) |
| Antibody | CDC6 (mouse monoclonal) antibody | Santa Cruz | Cat. #: sc-9964; RRID:AB_627236 | 4i (1:100) |
| Antibody | Donkey anti-rabbit AlexaFluor Plus 488 (Donkey polyclonal) antibody | Thermo Fisher Scientific | Cat. #: A32790; RRID:AB_2762833 | 4i (1:500) |
| Antibody | Donkey anti-goat AlexaFluor plus 647 (Donkey polyclonal) antibody | Thermo Fisher Scientific | Cat. #: A32758; RRID:AB_2762828 | 4i (1:500) |
| Antibody | Donkey anti-mouse AlexaFluor Plus 555 (Donkey polyclonal) antibody | Thermo Fisher Scientific | Cat. #: A32773; RRID:AB_2762848 | 4i (1:500) |
| Strain, strain background (Escherichia coli) | BL21 Competent Escherishia coli | NEB | Cat. #: C2530H | |
| Strain, strain background (E. coli) | DH5-ɑ Competent Escherishia coli | NEB | Cat. #: C2988J | |
| Commercial Assay or Kit | GeneJET Plasmid Miniprep Kit | Thermo Fisher Scientific | Cat. #: K0503 | |
| Commercial Assay or Kit | Q5 Site-Directed Mutagenesis Kit (without competent cells) | NEB | Cat. #: E0552S | |
| Commercial Assay or Kit | Rneasy Plus Mini Kit | QIAGEN | Cat. #: 74134 | |
| Commercial Assay or Kit | SuperScript III First-Strand Synthesis System | Thermo Fisher Scientific | Cat. #: 18080051 | |
| Commercial Assay or Kit | Dead Cell Apoptosis Kit with Annexin V FITC and PI for flow cytometry | Thermo Fisher Scientific | Cat. #: V13242 | |
| Chemical compound, drug | SSO Advanced Universal SYBR Green Supermix | Bio-Rad | Cat. #: 1725271 | |
| Chemical compound, drug | Bio-Rad Protein Assay Dye Reagent Concentrate | Bio-Rad | Cat. #: 5000006 | |
| Chemical compound, drug | PrestoBlue Cell Viability Reagent | Thermo Fisher Scientific | Cat. #: A13261 | |
| Chemical compound, drug | Calf Intestinal Phosphatase (CIP) | NEB | Cat. #: M0290 | |
| Chemical compound, drug | Clarity ECL Western Blot Substrate | Bio-Rad | Cat. #: 1705060 | |
| Chemical compound, drug | Cyclohexamide | MilliporeSigma | Cat. #: 1810 | |
| Chemical compound, drug | MG132 | Selleck Chemicals | Cat. #: S2619 | |
| Chemical compound, drug | MLN4924 | Active Biochem | Cat. #: A-1139 | |
| Chemical compound, drug | Glutathione Agarose Resin | GoldBio | Cat. #: G-250-5 | |
| Chemical compound, drug | EZView Red Anti-FLAG M2 Affinity Gel | MilliporeSigma | Cat. #: F2426 | |
| Chemical compound, drug | EZView Red Anti-HA Affinity Gel | MilliporeSigma | Cat. #: E6779 | |
| Chemical compound, drug | PierceTM Protein A/G agarose Beads | Thermo Fisher Scientific | Cat. #: 20421 | |
| Chemical compound, drug | Rnase A | MilliporeSigma | Cat. #: R6513 | |
| Chemical compound, drug | Lipofectamine RNAiMAX | Thermo Fisher Scientific | Cat. #: 13778150 | |
| Chemical compound, drug | PolyJet Transfection Reagent | SignaGen | Cat. #: SL100688 | |
| Chemical compound, drug | Lipofectamine 2000 | Thermo Fisher Scientific | Cat. #: 11668019 | |
| Cell line (H. sapiens) | 293T | ATCC | Cat. #: CRL-3216, RRID:CVCL_0063 | |
| Cell line (H. sapiens) | U2OS | ATCC | Cat. #: HTB-96; RRID:CVCL_0042 | |
| Cell line (H. sapiens) | HeLa | ATCC | Cat. #: CCL-2; RRID:CVCL_0030 | |
| Cell line (H. sapiens) | HeLa sgCTRL | PMID: 27653696 | ||
| Cell line (H. sapiens) | HeLa sgCCNF | PMID: 27653696 | ||
| Cell line (H. sapiens) | MCF7 pIND CCNF | This paper | See Figure 1, Figure 1—figure supplement 3 | |
| Cell line (H. sapiens) | T47D pIND CCNF | This paper | See Figure 1, Figure 1—figure supplement 3 | |
| Cell line (H. sapiens) | NHF-1 | William Kaufman Lab (UNC; retired) | ||
| Cell line (H. sapiens) | IMR-90 | Yue Xiong Lab (UNC; retired) | ||
| Cell line (H. sapiens) | T98G | Tissue Culture Facility, UNC | ||
| Cell line (H. sapiens) | NHF-1 doxy-inducible p130 WT | This paper | See Figure 6, Figure 7, Figure 5—figure supplements 1 and 26,7 | |
| Cell line (H. sapiens) | NHF-1 doxy-inducible p130 AA | This paper | See Figure 6, Figure 5—figure supplements 1 and 2 | |
| Transfected Construct (H. sapiens) | pBABE-p130 | Gift from Larisa Litovchick lab (VCU) | For lentiviral transfection | |
| Transfected Construct (H. sapiens) | pDEST-HA3-p130 | This paper | Transfected construct (human); See Figures 3—5, Figure 4—figure supplement 1 | |
| Transfected Construct (H. sapiens) | pDEST-HA3-p130 1–417 | This paper | Transfected construct (human); See Figure 4—figure supplement 1 | |
| Transfected Construct (H. sapiens) | pDEST-HA3-p130 418–1139 | This paper | Transfected construct (human); See Figure 4—figure supplement 1 | |
| Transfected Construct (H. sapiens) | pDEST-HA3-p130 418-616 | This paper | Transfected construct (human); See Figure 4—figure supplement 1 | |
| Transfected Construct (H. sapiens) | pDEST-HA3-p130 418-827 | This paper | Transfected construct (human); See Figure 4—figure supplement 1 | |
| Transfected Construct (H. sapiens) | pDEST-HA3-p130 418-1024 | This paper | Transfected construct (human); See Figure 4—figure supplement 1 | |
| Transfected Construct (H. sapiens) | pDEST-HA3-p130 828-1139 | This paper | Transfected construct (human); See Figure 4—figure supplement 1 | |
| Transfected Construct (H. sapiens) | pDEST-HA3-p130 R658A I660A | This paper | Transfected construct (human); See Figure 4—figure supplement 1 | |
| Transfected Construct (H. sapiens) | pDEST-HA3-p130 R680A L682A | This paper | Transfected construct (human); See Figure 4—figure supplement 1 | |
| Transfected Construct (H. sapiens) | pDEST-FLAG-Cyclin F | PMID: 27653696 | Transfected construct (human) | |
| Transfected Construct (H. sapiens) | Cyclin F M309A L131A | PMID: 20596027 | Transfected construct (human) | |
| Transfected Construct (H. sapiens) | pDEST-FLAG-Cyclin F M309A L313A | This paper | Transfected construct (human) | |
| Transfected Construct (H. sapiens) | pGEX-GST-p130 593-790 | PMID: 9188854 | For protein expression in Escherichia coli | |
| Transfected Construct (H. sapiens) | pGEX-GST-p130 593-790 R680A L682A | This paper | For protein expression in Escherichia coli | |
| Transfected Construct (H. sapiens) | pINDUCER20 | PMID: 21307310 | Addgene #44012; RRID:Addgene_44012 | For lentiviral transfection |
| Transfected Construct (H. sapiens) | pINDUCER20 CCNF | This paper | For lentiviral transfection | |
| Transfected Construct (H. sapiens) | pLV[Exp]-CMV> Tet3G/Hygro | VectorBuilder | ID:VB180123-1018bxq | For lentiviral transfection |
| Transfected Construct (H. sapiens) | pLV[TetOn]-Neo-TRE3G > HA/{p130 AA} | this paper, VectorBuilder | ID:VB200319-6469hpy | For lentiviral transfection |
| Transfected Construct (H. sapiens) | pLV[TetOn]-Neo-TRE3G > HA/{p130 WT} | this paper, VectorBuilder | ID:VB200319-6451nqd | For lentiviral transfection |
| Sequence-based reagent | p130 from AA1 w/attb site Forward | PCR primers | 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTAAT GCCGTCGGGAGGTGACCAG | |
| Sequence-based reagent | p130 from AA417 w/attb site Reverse | PCR primers | 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTA CTACACACAAGGGCTATTCTCCTT | |
| Sequence-based reagent | p130 from AA418 w/attb site Forward | PCR primers | 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTT AACTCCAGTTTCTACAGCTACG | |
| Sequence-based reagent | p130 from AA 1139 w/attb site Reverse | PCR primers | 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTA TCAGTGGGAACCACGGTCATT | |
| Sequence-based reagent | p130 from AA 616 w/attb site Reverse | PCR primers | 5′-GGGGACCACTTTGTACAAGAAAGCTGGG TACTAAACTCTGTTTTCATTGTCTCT | |
| Sequence-based reagent | p130 from AA 827 w/attb site Reverse | PCR primers | 5′-GGGGACCACTTTGTACAAGAAAGCTGGG TACTAACTACTGCTGGTTACAGACTG | |
| Sequence-based reagent | p130 from AA 1024 w/attb site Reverse | PCR primers | 5′-GGGGACCACTTTGTACAAGAAAGCTGGG TACTAGTACTTCATGGCAAATGTCTT | |
| Sequence-based reagent | p130 from AA 828 w/attb site Forward | PCR primers | 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTT AAATAGACCCAGGAAGACCAGC | |
| Sequence-based reagent | p130 R658A I660A Forward | PCR primers | 5′-CGCCACATCTCCAACCACATTATAC | |
| Sequence-based reagent | p130 R658A I660A Reverse | PCR primers | 5′-CTGGCTCCAAGTCCTCCAGTATC | |
| Sequence-based reagent | p130 R680A L682A Forward | PCR primers | 5′-GGCCTTTGTTGAGAATGATAGCCCCTC | |
| Sequence-based reagent | p130 R680A L682A Reverse | PCR primers | 5′-CGGGCTCTGGTAGTGCTGGCTGG | |
| Sequence-based reagent | Cyclin F from AA 1 w/attb site Forward | PCR primers | 5′-GGGGACAAGTTTGTACAAAAAAGCAGGC TTAATGGGGAGCGGCGGCGTGGTCC | |
| Sequence-based reagent | Cyclin F from end w/attb site Reverse | PCR primers | 5′-GGGGACCACTTTGTACAAGAAAGCTGG GTATTACAGCCTCACAAGGCCCAGG | |
| Sequence-based reagent | CCNE1 RT-qPCR Forward | PCR primers | 5′-AGACATACTTAAGGGATCAGC | |
| Sequence-based reagent | CCNE1 RT-qPCR Reverse | PCR primers | 5′-CACACCTCCATTAACCAATC | |
| Sequence-based reagent | CDC6 RT-qPCR Forward | PCR primers | 5′-ATGTAAATCACCTTCTGAGC | |
| Sequence-based reagent | CDC6 RT-qPCR Reverse | PCR primers | 5′-GTCATCCTGTTACCATCAAC | |
| Sequence-based reagent | DHFR RT-qPCR Forward | PCR primers | 5′-TTCCAGAAGTCTAGATGATGC | |
| Sequence-based reagent | DHFR RT-qPCR Reverse | PCR primers | 5′-CTTCCTTATAAACAGAACTGCC | |
| Sequence-based reagent | E2F1 RT-qPCR Forward | PCR primers | 5′-CTGATGAATATCTGTACTACGC | |
| Sequence-based reagent | E2F1 RT-qPCR Reverse | PCR primers | 5′-CTTTGATCACCATAACCATCTG | |
| Sequence-based reagent | GAPDH RT-qPCR Forward | PCR primers | 5′-GGCCTCCAAGGAGTAAGACC | |
| Sequence-based reagent | GAPDH RT-qPCR Reverse | PCR primers | 5′-AGGGGTCTACATGGCAACTG | |
| Sequence-based reagent | Cyclin F RT-qPCR Forward | PCR primers | 5′-AGGACAAGCGCTATGGAGAA | |
| Sequence-based reagent | Cyclin F RT-qPCR Reverse | PCR primers | 5′-TCTGTCTTCCTGGAGGCTGT | |
| Sequence-based reagent | CDT1 RT-qPCR Forward | PCR primers | 5′-CCTGGGGAAATGGAGAAG | |
| Sequence-based reagent | CDT1 RT-qPCR Reverse | PCR primers | 5′-TTGTCCAGCTTGACGTAG | |
| Sequence-based reagent | Cyclin F subcloning into pfastbac Forward | PCR primers | 5′-GCTAGGGTCGGATCCAGGAGGCCCCGAAACCTGACC | |
| Sequence-based reagent | Cyclin F subcloning into pfastbac Reverse | PCR primers | 3′-GCTAGGCATAGCGGCCGCACCTTAGCTGT CTTGTGTCACTCCTAATGCAGC | |
| Sequence-based reagent | Skp1 subcloning into PGEX-4T-1 Forward | PCR primers | 5′-GCTAGGGTCGGATCCATGCCTT CAATTAAGTTGCAGAGTTCTGATGG | |
| Sequence-based reagent | Skp1 subcloning into PGEX-4T-1 Reverse | PCR primers | 3′-GCTAGGCATAGCCTCGAGTTACTT CTCTTCACACCACTGGTTCTC | |
| Sequence-based reagent | CCNF #1 | siRNA | 5′-UAGCCUACCUCUACAAUGAUU | |
| Sequence-based reagent | CCNF #2 | siRNA | 5′-GCACCCGGUUUAUCAGUAAUU | |
| Sequence-based reagent | siFF | siRNA | 5′-CGUACGCGGAAUACUUCGAUU | |
| Other | EdU | Sigma-Aldrich | Cat. #: T511285 | For flow cytometry-–10 µM to cell media for 30 min prior to fixation |
| Other | DAPI | Thermo Fisher Scientific | Cat. #: D1306 | For flow cytometry (1 µg/ml) |
| Other | Alexa-Fluor 488 Azide | Thermo Fisher Scientific | Cat. #: A10266 | For flow cytometry (0.2 µM) |
Additional files
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/70691/elife-70691-transrepform1-v2.pdf
-
Source code 1
Violin plotting.
- https://cdn.elifesciences.org/articles/70691/elife-70691-supp1-v2.zip
-
Source data 1
Immunoblots.
This source data file includes all uncropped blots used to generate data for the main figures and figure supplements. Additionally, copies of the uncropped images are shown a second time where blot strips shown in figures are highlighted with a red square and the protein that was blotted for is noted.
- https://cdn.elifesciences.org/articles/70691/elife-70691-supp2-v2.zip
-
Source data 2
Cycloheximide Chase.
This source data file contains quantified band densities and background used to generate the cycloheximide protein degradation plot and for the protein half-life calculations.
- https://cdn.elifesciences.org/articles/70691/elife-70691-supp3-v2.xlsx
-
Source data 3
Flow cytometry.
This source data file contains percentages of cells in various populations for the flow cytometry experiments.
- https://cdn.elifesciences.org/articles/70691/elife-70691-supp4-v2.zip
-
Source data 4
Cell counting and PrestoBlue.
This source data file contains the raw cell counts for the courting experiment and the raw fluorescence measurements for the PrestoBlue experiment.
- https://cdn.elifesciences.org/articles/70691/elife-70691-supp5-v2.zip
-
Source data 5
Rt-qPCR.
This source data file contains the output from the rt-qPCR machine that was used to determine gene expression for the rt-qPCR blot.
- https://cdn.elifesciences.org/articles/70691/elife-70691-supp6-v2.zip





