Transcriptional profiling of sequentially generated septal neuron fates
Figures
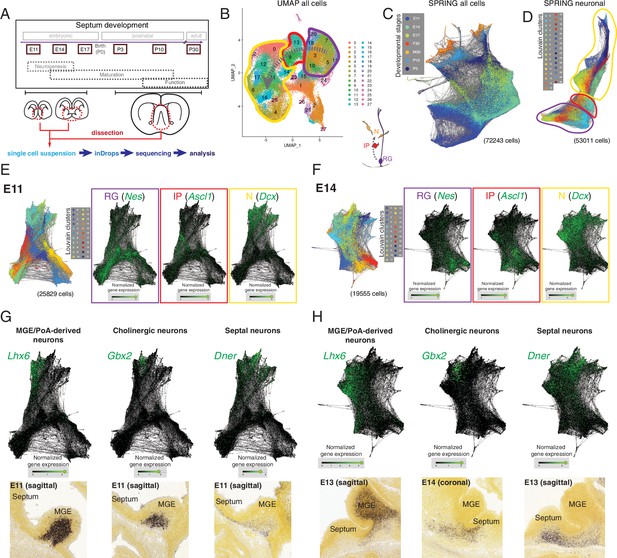
Single-cell RNA sequencing of the developing septum reveals early emergence of neuronal identities.
(A) Experimental approach: samples were collected at the indicated developmental stages and submitted to single-cell RNA sequencing using inDrops. (B) UMAP plot of all cells, where the transition from radial glia (RG, purple) to neurons (N, yellow) via intermediate progenitors (IP, red) schematized in the cartoon at the bottom right can be visualized (gray dashed arrows). (C) SPRING plot shows developmental stage-dependent organization of all sequenced cells. (D) SPRING plot of cells belonging to neuronal trajectory (colors indicate cell groups determined by Louvain clustering). (E, F) SPRING plots of all cells at embryonic stages E11 (E) and E14 (F) display differentiation-dependent alignment of cells, illustrated by the relative enrichment of the genes Nes, Ascl1, and Dcx (cell-type markers for RG, IP, and N, respectively) in adjacent areas of the graphs. (G, H) Analysis of each protrusion within the neuronal portion of E11 (G) and E14 (H) SPRING plots shows enrichment in marker genes for MGE/PoA-derived (Lhx6), cholinergic (Gbx2), and septal (Dner) newborn neurons; the presence of these neuronal types in the embryonic septum at similar stages is confirmed by in situ hybridization (bottom panels).
Image credit (bottom panels): Allen Institute – Allen Developing Mouse Brain Atlas.
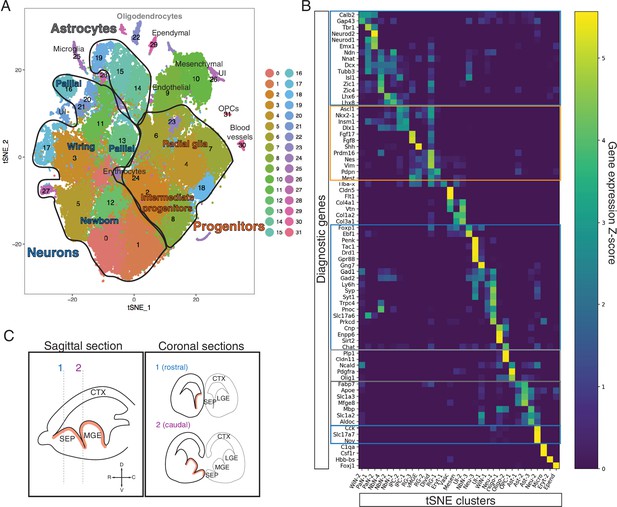
Cluster identity assignment in the scRNA-Seq dataset.
(A) t-SNE plot of all cells in the dataset. Assigned identities are written over the corresponding clusters. The three major groups of clusters encircled in black outlines contain progenitor cells (orange text), neurons (blue text), and astrocytes (dark gray text). (B) Heatmap displaying the relative levels of expression for 73 potential diagnostic genes (rows) in each of the t-SNE clusters. Major groups identified in (A) are highlighted in the corresponding colors. Putative identities are shown in the x-axis. (C) Schematic showing the correspondence between sagittal (left) and coronal (right) sections at rostral (1) and caudal (2) positions within the septum (SEP), showing the lateral (LGE) and medial (MGE) ganglionic eminences for reference; the main proliferative area (ventricular zone) is highlighted in red. The compass on left panel indicates the rostrocaudal and dorsoventral axes. WiN, wiring neurons; PaN, pallial neurons; NbN, newborn neurons; IPC, intermediate progenitor cells; vMGE, ventral portion of the MGE; RG, radial glia; Divid, dividing cells; Eryt, erythrocytes; Vasc, vasculature; Mesen, mesenchymal cells; UI, unidentified; Neu, neurons; Oligo, oligodendrocytes; OPC, oligodendrocyte progenitor cells; Ast, astrocytes; Micro, microglia; Epend, ependymal cells.
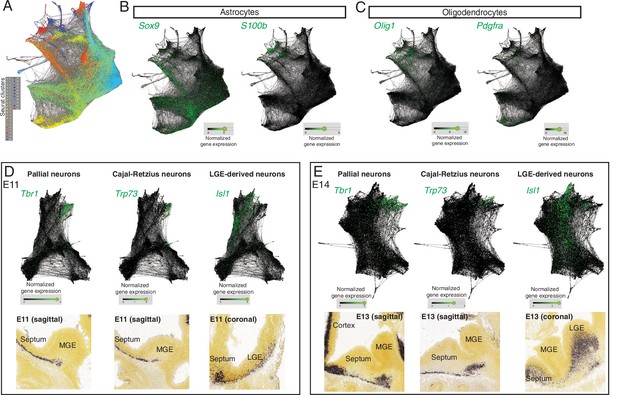
Additional cell identities in the SPRING plot.
(A) SPRING plot of all cells, as shown in Figure 1C, overlaid with the Seurat clusters identified in the t-SNE plot displayed in Figure 1—figure supplement 1A. (B, C) SPRING was used to highlight marker genes for progenitors (left) and differentiated cells (right) within the astrocyte (B) and oligodendrocyte (C) lineages. (D, E) Analysis of ‘protrusions’ within the neuronal portion of E11 (D) and E14 (B) SPRING plots shows enrichment in marker genes for newborn pallial neurons (Tbr1), Cajal–Retzius cells (Trp73) and potentially LGE-like (Isl1) newborn neurons in the septum, analogous across both stages; the presence of these neuronal types in the embryonic septum at similar stages is confirmed by in situ hybridization (bottom panels).
Image credit (bottom panels): Allen Institute – Allen Developing Mouse Brain Atlas.
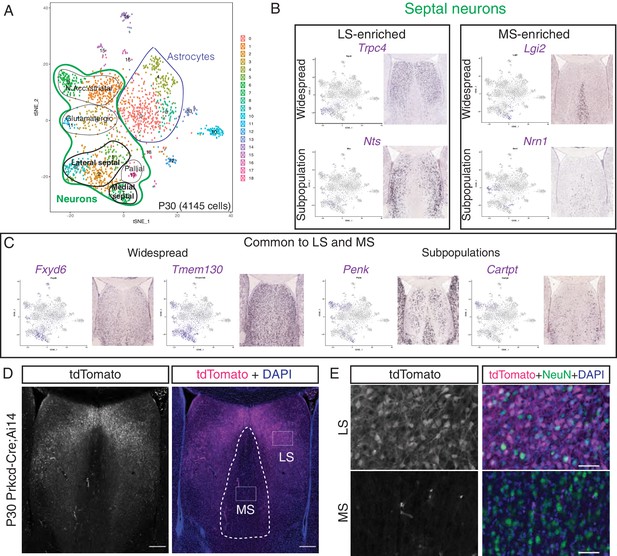
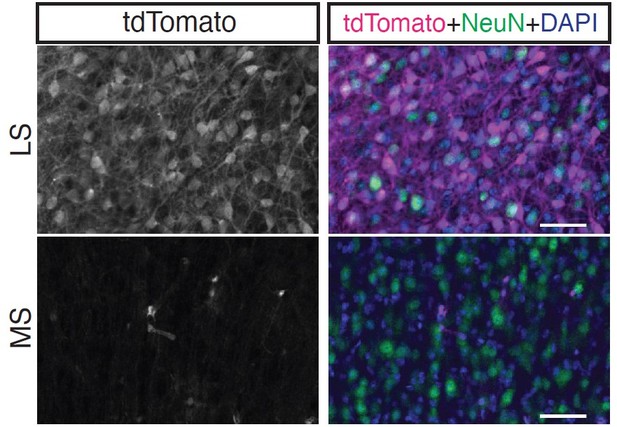
Analysis of postnatal day (P)30 dataset for discovery of novel septal neuron markers.
(A) t-SNE plot of P30 cells. Assigned identities are written over the corresponding clusters. Two major cluster groups contain astrocytes (blue), and neurons (green), of which a small proportion are either lateral or medial septal neurons (bold text). (B) Analysis of gene enrichment in different clusters (feature plots), validated with in situ hybridization images from the Allen Brain Atlas, led to the discovery of genes that are enriched in neurons of the lateral septum (LS) (left) or medial septum (MS) (right), either following widespread patterns (top) or representing a subpopulation within the corresponding septal nucleus (bottom). (C) Similarly, general septal markers common to LS and MS could be identified, either present in the majority of septal cells (left) or in a subset of them (right). (D) Coronal section through the septum of a P30 Prkcd-Cre;Ai14 mouse, where expression of tdTomato (gray; magenta in merge, counterstained with DAPI in blue), shows that cell labeling is restricted to the LS. Scale bars, 250 µm. (E) Insets from (D), showing overlap of immunostaining for tdTomato (gray; magenta in merge) and NeuN (green in merge) only in the LS.
Image credit for Panels B and C: Allen Mouse Brain Atlas.
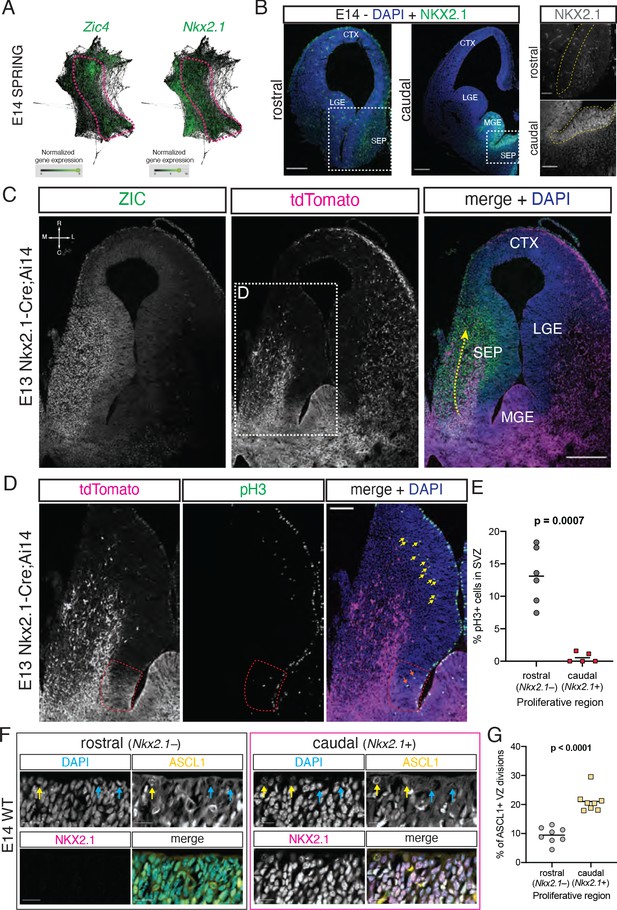
Neural progenitors within the septal eminence preferentially divide at the ventricular surface.
(A) SPRING plots at embryonic day (E)14 show that a set of cells at this stage (dashed magenta line) express both Zic4 (left) and Nkx2.1 (right). (B) Immunofluorescence staining for NKX2.1 (green; counterstained with DAPI, blue) at rostral and caudal locations of the septum on coronal sections of an E14 brain. Panels on the right display magnified insets (marked with dashed white squares on the corresponding overview images); the main proliferative area (ventricular zone) is highlighted by a yellow dashed line, showing NKX2.1-positive cells in the caudal, but not rostral, septal anlage. Scale bars: 250 µm (overview images); 100 µm (close-up images). (C) Horizontal section (compass on the top left of left panel indicates rostrocaudal and mediolateral axes) of the right hemisphere of an E13 Nkx2.1-Cre;Ai14 mouse brain. Immunofluorescence staining for ZIC proteins (left panel; green in merge) and the tdTomato fluorescent reporter (middle panel; magenta in merge), shown both as single channels and merged (right panel, with DAPI counterstaining in blue), show a subset of Nkx2.1-expressing septal progenitors in the septal eminence, as well as a caudal-to-rostral stream of migrating ZIC-positive neurons with a developmental history of Nkx2.1 expression (yellow dashed arrow). Scale bar, 250 µm. (D) Close-up of the area indicated by the dashed white line in (C); immunofluorescence staining for tdTomato (magenta in merge) and phosphorylated histone 3 (pH3, green in merge) shows difference in the number of subapically dividing cells in the rostral portion of the septum (yellow arrows) compared to the Nkx2.1-expressing (red dashed line) septal eminence (orange arrows). Scale bar, 100 µm. (E) Quantification of the proportion of dividing (pH3+) cells located in the subventricular zone of the rostral (gray dots) and caudal (red squares), that is, Nkx2.1– and Nkx2.1+, proliferative regions of the developing septum at E13. (F) Immunostaining for ASCL1 (yellow in merge) and NKX2.1 (magenta in merge), counterstained with DAPI (blue in merge), in rostral and caudal proliferative regions of the septum of an E14 mouse brain, highlighting ASCL1+ (yellow arrows on DAPI and ASCL1 panels) and ASCL1– (blue arrows on DAPI and ASCL1 panels) dividing cells at the apical surface. Scale bars, 20 µm. (G) Quantification of the proportion of ventricular surface divisions that are ASCL1+ in the rostral (gray dots) and caudal (yellow squares) proliferative regions of the developing septum at E14. All data points are represented; black bars represent the mean. Unpaired t-tests were performed; the p-values are indicated above the corresponding compared sets of data: bold typeface indicates statistically significant differences (p<0.05). CTX: cortex; LGE: lateral ganglionic eminence; MGE: medial ganglionic eminence; SEP: septum.
-
Figure 2—source data 1
Quantifications of septal eminence progenitors.
- https://cdn.elifesciences.org/articles/71545/elife-71545-fig2-data1-v2.xlsx
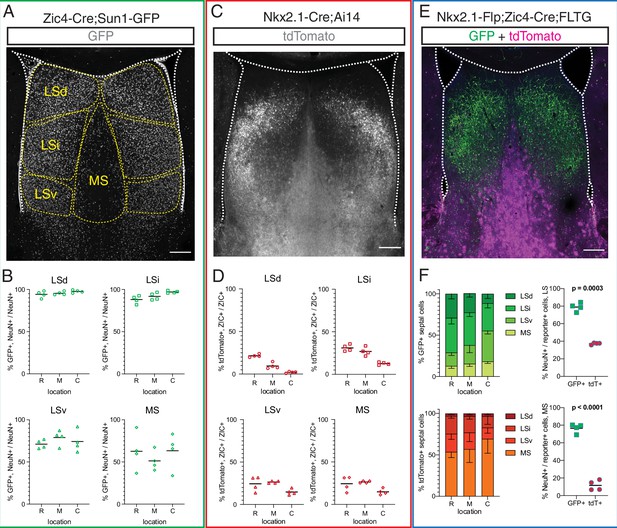
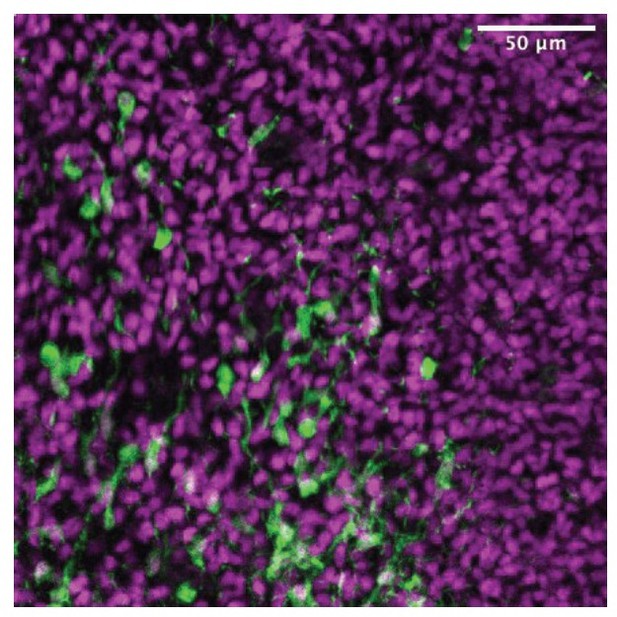
Fate mapping of septal eminence-derived neurons in the mature septum.
(A) Coronal section of the septum of a postnatal day (P)30 Zic4-Cre;Sun1-GFP mouse; immunofluorescence staining for the reporter GFP shows the location of Zic4-lineage cells. Yellow dashed lines indicate the location of medial septum (MS) and lateral septum (LS) nuclei as indicated. (B) Quantification of the proportion of neurons within the Zic4-lineage within the total NeuN+ neuronal population. (C) Coronal section of the septum of a P30 Nkx2.1-Cre;Ai14 mouse; immunofluorescence staining for the reporter tdTomato shows the location and morphology of Nkx2.1-lineage cells. (D) Quantification of the proportion of Nkx2.1-lineage cells within the ZIC+ population, as recognized by a pan-ZIC antibody. (E) Coronal section of the septum of a P30 Nkx2.1-Flp;Zic4-Cre;FLTG mouse; immunofluorescence staining for the reporters GFP (green) and tdTomato (magenta) shows the location and morphology cells within the Nkx2.1 lineage with (GFP+) or without (tdTomato+) a developmental history of Zic4 expression. (F) Left: proportion of the entire intersectional (GFP+, top) or subtractive (tdTomato+, bottom) populations allocated within each septal nucleus in the Nkx2.1-Flp;Zic4-Cre;FLTG line, as illustrated in (C); right: proportion of cells positive for the neuronal marker NeuN within the intersectional (GFP+, green squares) and subtractive (tdTomato+, red circles) populations in the entire LS (top) and in the MS (bottom). Scale bars (A, C, E), 250 µm. Quantifications in (B), (D), and (F) were obtained for each of the mouse lines above the corresponding set of graphs, across the dorsal, intermediate, and ventral nuclei of the LS (LSd, LSi, and LSv, respectively) and the MS, at rostral (R), medial (M), and caudal (C) locations along the rostrocaudal axis. Unpaired t-tests (right graphs in F) were performed; the p-values are indicated above the corresponding compared sets of data: bold typeface indicates statistically significant differences (p<0.05).
-
Figure 3—source data 1
Fate mapping of septal eminence derived cells.
- https://cdn.elifesciences.org/articles/71545/elife-71545-fig3-data1-v2.xlsx
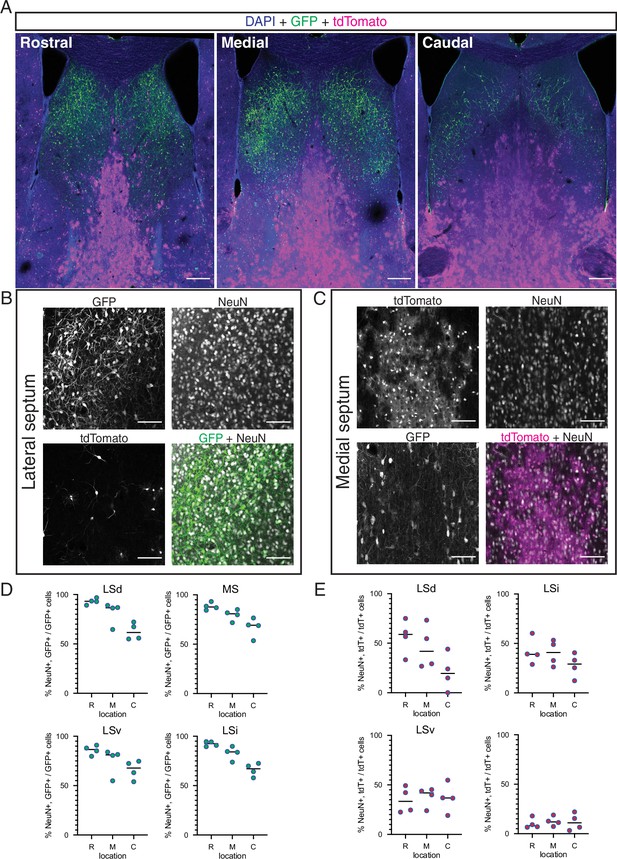
Neuronal identity within the intersectional and subtractive populations in the septum Nkx2.1-Flp;Zic4-Cre;FLTG mice.
(A) Overview of the septum along the rostrocaudal axis on coronal sections of a postnatal day (P)30 Nkx2.1-Flp;Zic4-Cre;FLTG mouse; immunofluorescence staining for the reporters GFP (green) and tdTomato (magenta) counterstained with DAPI (blue). (B, C) Confocal images of the lateral (B) and medial (C) septum of a P30 Nkx2.1-Flp;Zic4-Cre;FLTG mouse; immunostaining for GFP (green in merged image in B) and tdTomato (magenta in merged image in C) along with the neuronal marker NeuN (gray in merged images) allows the identification of neurons within the intersectional (B) and subtractive (C) populations in this genetic model. (D, E) Quantification of the proportion of labeled cells within the intersectional (GFP+, green circles; D) and subtractive (tdTomato+, red circles; E) positive for NeuN, across the dorsal, intermediate, and ventral nuclei of the lateral septum (LSd, LSi, and LSv, respectively) and the medial septum (MS), obtained at rostral (R), medial (M), and caudal (C) locations along the rostrocaudal axis. Scale bars, 250 µm (A), 100 µm (B, C).
-
Figure 3—figure supplement 1—source data 1
Percentage of neurons within fate-mapped populations.
- https://cdn.elifesciences.org/articles/71545/elife-71545-fig3-figsupp1-data1-v2.xlsx
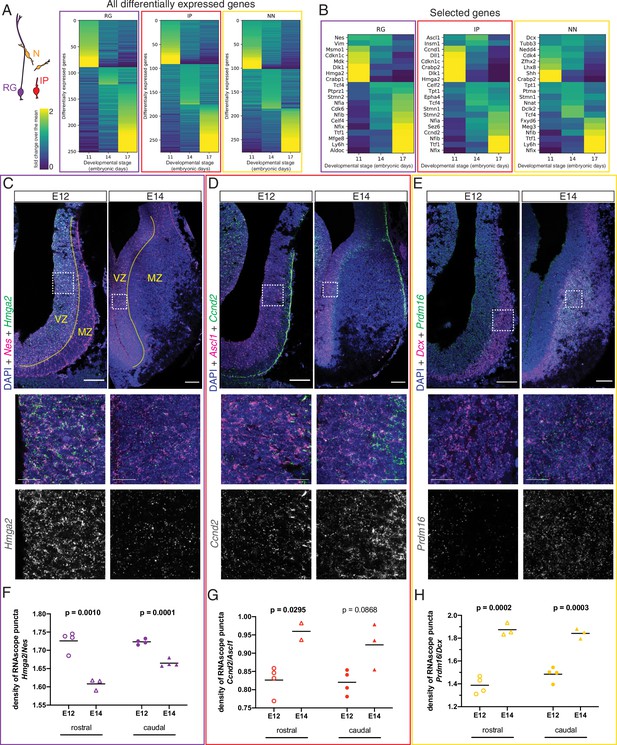
Different transcriptional programs are active during early and late septal neurogenesis.
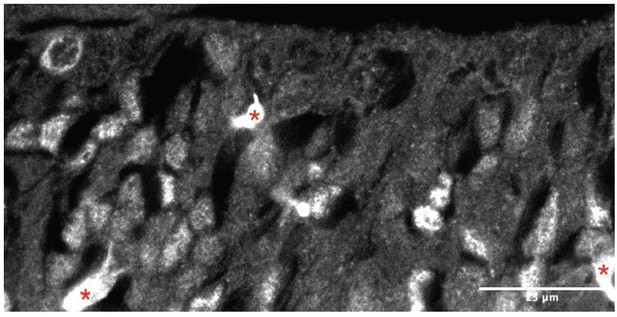
(A) Heatmaps illustrating all genes differentially enriched in the scRNA-Seq dataset across embryonic stages for the three cell types indicated in the cartoon: radial glia (RG, purple), intermediate progenitors (IP, red), and newborn neurons (NN, yellow). The Viridis color scale represents fold change over the mean, and applies to (A) and (B). (B) Heatmap showing the differential enrichment of a subset of selected genes, displayed as in (A). (C–E) Coronal brain sections showing the rostral septum at embryonic days (E)12 and 14, subjected to fluorescent single-molecule in situ hybridization for the genes Nes+ Hmga2 (C), Ascl1+ Ccnd2 (D), and Dcx+ Prdm16 (E). Cell-type marker mRNA puncta (Nes, Ascl1, and Dcx) are displayed in magenta, mRNA puncta for differentially enriched genes (Hmga2, Ccnd2, and Prdm16) in green, and DAPI counterstaining in blue. Yellow dashed lines in (C) mark the limit between ventricular zone (VZ) and mantle zone (MZ), as indicated. Dashed boxes indicate the location of the magnified 100 × 100 µm fields shown below, both as merged images (middle panels) and single channel for the corresponding differentially enriched genes (bottom panels). Scale bars: 100 µm (top panels), 25 µm (middle panels). (F–H) Quantification of density of mRNA puncta of differentially enriched genes, normalized to the density of cell-type marker mRNA; measurements were obtained from the rostral (empty symbols) and caudal (full symbols) portions of the septum at E12 (circles) and E14 (triangles). All data points are represented; black bars represent the mean. Unpaired t-tests were performed; p-values are indicated above the corresponding compared sets of data: those highlighted in bold indicate statistically significant differences (p<0.05).
-
Figure 4—source data 1
RNAscope puncta quantifications, Figure 4.
- https://cdn.elifesciences.org/articles/71545/elife-71545-fig4-data1-v2.xlsx
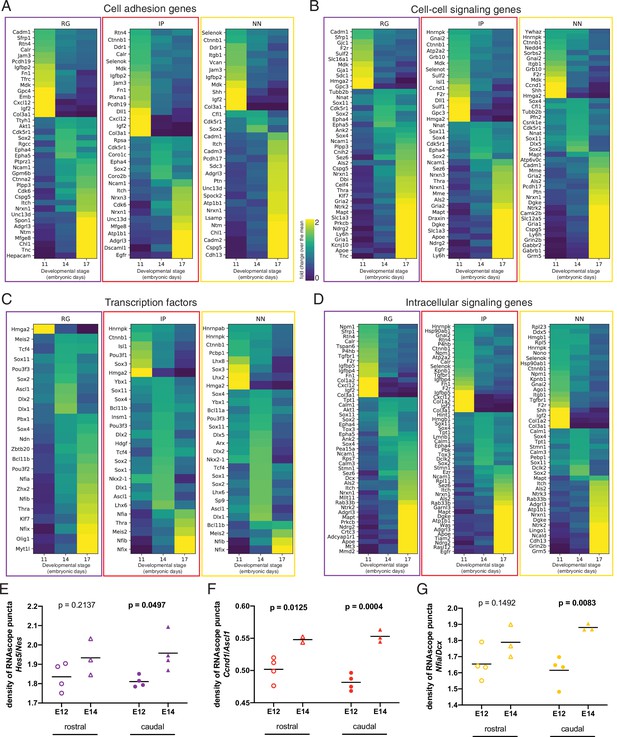
Additional categories and examples of genes differentially enriched across developmental stages.
(A–D) Heatmaps of differentially enriched classes of genes across embryonic stages for the three indicated cell types: radial glia (RG), intermediate progenitors (IP), and newborn neurons (NN), and sorted by biological function category: (A) cell adhesion genes; (B) cell-cell signaling genes; (C) transcription factors; (D) intracellular signaling. The Viridis color scale between (A) and (B) represents fold change over the mean, and applies to panels (A–D). (E, F) Quantification of mRNA puncta density of differentially enriched genes (E, Hes5; F, Ccnd1; G, Nfia), normalized to the density of cell-type marker mRNA (E, Nes; F, Ascl1; G, Dcx). Measurements were obtained from the rostral (empty symbols) and caudal (full symbols) portions of the septum at E12 (circles) and E14 (triangles). All data points are represented; black bars represent the mean. Unpaired t-tests were performed; p-values are indicated above the corresponding compared sets of data: those highlighted in bold indicate statistically significant differences (p<0.05).
-
Figure 4—figure supplement 1—source data 1
RNAscope puncta quantifications, Figure 4—figure supplement 1.
- https://cdn.elifesciences.org/articles/71545/elife-71545-fig4-figsupp1-data1-v2.xlsx
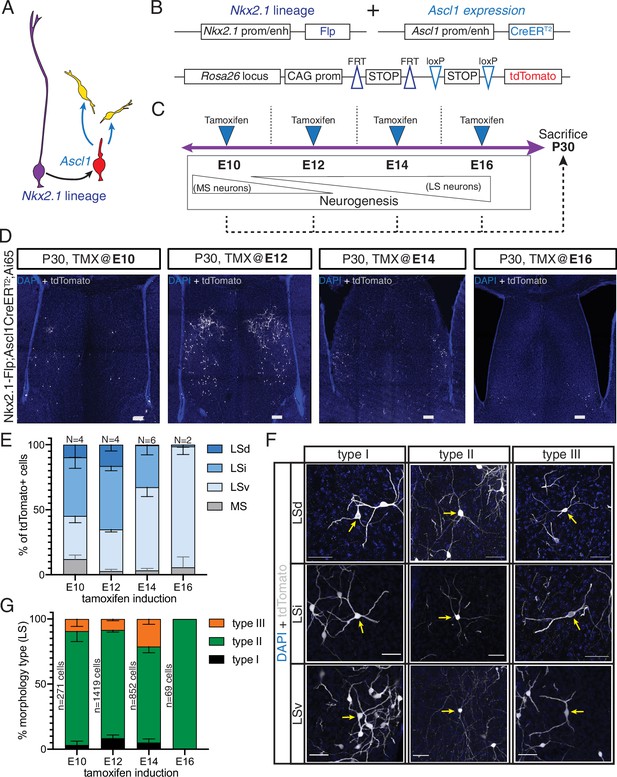
Generation of sequential temporal cohorts of neurons from the septal eminence.
(A) Schematic of neurogenesis in the septal eminence: radial glia (RG) (purple) within the Nkx2.1 lineage give rise to Ascl1-expressing transit-amplifying progenitors (red), which in turn divide to generate neurons. (B) Two driver lines, Nkx2.1-Flp and Ascl1-CreERT2, were crossed with an intersectional reporter line. The action of both Flp and Cre recombinases (i.e., within the Nkx2.1 lineage and in the presence of tamoxifen) leads to the expression of the fluorescent reporter tdTomato. (C) Experimental design: tamoxifen was administered to pregnant dams at embryonic day (E)10, E12, E14, or E16, covering the entire septal neurogenic period. Resulting litters were sacrificed and analyzed at postnatal day (P)30. (D) Representative images of coronal sections through the septum of P30 mice in which recombination was induced at the indicated stages. Cells derived from terminal progenitor divisions are labeled with tdTomato (gray; counterstained with DAPI, blue). Scale bars, 100 µm. (E) Quantification of the location of tdTomato+ cells in each septal nucleus as a percentage of total labeled cells (mean ± SD) within each temporal cohort. The number of biological replicates is indicated above each bar. (F) Representative images of neurons belonging to the three morphological subtypes within each LS nucleus, labeled with tdTomato (gray; counterstained with DAPI, blue). Arrows indicate the cell bodies of corresponding neuronal types. Scale bars, 50 µm. (G) Quantification of morphological neuron types as a percentage of the total number of classified cells (mean ± SD) within each temporal cohort. The total number of classified cells is indicated on the left side of the corresponding column; the number of biological replicates is the same as in (E) except for the E14 timepoint (N = 7).
-
Figure 5—source data 1
Location and morphology of septal eminence derived temporal cohorts.
- https://cdn.elifesciences.org/articles/71545/elife-71545-fig5-data1-v2.xlsx
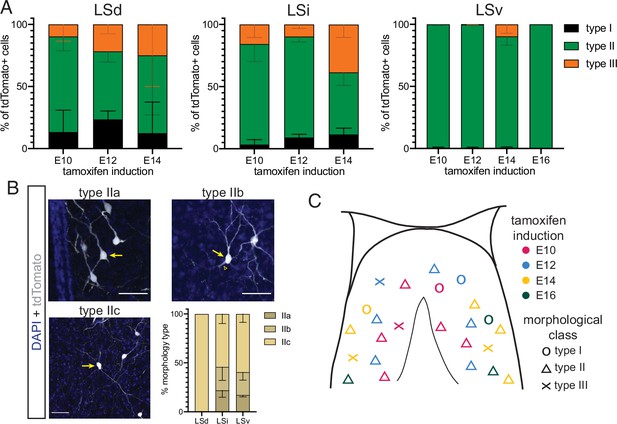
Detailed distribution of morphological cell types across temporal cohorts of septal eminence-derived neurons.
(A) Quantification of the distribution of the three main morphological neuron types in the dorsal (LSd, left), intermediate (LSi, middle), and ventral (LSv, right) nuclei of the lateral septum. Cells in each septal nucleus, represented as a percentage of total classified cells (mean ± SD) within each temporal cohort. (B) Proposed subdivision of morphological neuron type II into three subtypes: IIa (thick dendrites); type IIb (thick initial dendritic segments [arrowhead] that branch out into thinner processes); and type IIc (thin dendrites). Images show cells labeled with tdTomato (gray; counterstained with DAPI, blue); arrows indicate the somata of neurons within each proposed subtype. Quantification of each subtype (as % of total type II cells, mean ± SD) within each LS subdivision within the E14 temporal cohort. (C) Graphical summary of the distribution and morphological types of LS neurons derived from the septal eminence within each temporal cohort.
-
Figure 5—figure supplement 1—source data 1
Location and morphology of septal eminence derived temporal cohorts by septal nucleus; analysis of type II subtypes.
- https://cdn.elifesciences.org/articles/71545/elife-71545-fig5-figsupp1-data1-v2.xlsx
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | Mouse monoclonal anti-ASCL1 | BD Pharmingen | Cat# 556604;RRID:AB_396479 | (1:100) |
| Antibody | Chicken polyclonal anti-GFP | Aves | Cat# GFP-1020;RRID:AB_10000240 | (1:1000) |
| Antibody | Rat monoclonal anti-pH3 | Abcam | Cat # ab10543; RRID:AB_2295065 | (1:500) |
| Antibody | Mouse monoclonal anti-NeuN | Millipore | Cat# MAB377; RRID:AB_2298772 | (1:500) |
| Antibody | Rabbit polyclonal anti-NKX2.1 | Santa Cruz | Cat# sc-53136;RRID:AB_793529 | (1:250) |
| Antibody | Chicken polyclonal anti-RFP | Rockland | Cat# 600-901-379; RRID:AB_10704808 | (1:1000) |
| Antibody | Rabbit polyclonal anti-RFP | Rockland | Cat# 600-401-379; RRID:AB_2209751 | (1:1000) |
| Antibody | Rabbit polyclonal anti-ZIC | Segal Lab, DFCI;Borghesani et al., 2002 | n/a (gift) | (1:500) |
| Antibody | Goat polyclonal anti-chicken Alexa 488 | Thermo Fisher | Cat# A11039; RRID:AB_142924 | (1:1000) |
| Antibody | Goat polyclonal anti-chicken Alexa 546 | Thermo Fisher | Cat# A11040; RRID:AB_1500590 | (1:1000) |
| Antibody | Goat polyclonal anti-mouse Alexa 488 | Thermo Fisher | Cat# A11001; RRID:AB_2534069 | (1:1000) |
| Antibody | Goat polyclonal anti-rabbit Alexa 488 | Thermo Fisher | Cat# A11008; RRID:AB_143165 | (1:1000) |
| Antibody | Goat polyclonal anti-rabbit Alexa 546 | Thermo Fisher | Cat# A11010; RRID:AB_2534077 | (1:1000) |
| Antibody | Goat polyclonal anti-rat Alexa 647 | Thermo Fisher | Cat# A21247; RRID:AB_141778 | (1:1000) |
| Strain, strain background (Mus musculus) | B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J (‘Ai14’ in text) | Jackson Laboratory | Stock no. 007914; RRID:IMSR_JAX:007914 | (Gt(ROSA)26Sor) |
| Strain, strain background (M. musculus) | B6;129S-Gt(ROSA)26Sortm65.1(CAG-tdTomato)Hze/J (‘Ai65’ in text) | Jackson Laboratory | Stock no. 021875; RID:IMSR_JAX:021875 | (Gt(ROSA)26Sor) |
| Strain, strain background (M. musculus) | STOCK Ascl1tm1.1(Cre/ERT2)Jejo/J(‘Ascl1-CreERT2’ in text) | Jackson Laboratory | Stock no. 012882; RRID:IMSR_JAX:012882 | (Ascl1) |
| Strain, strain background (M. musculus) | CD-1 (‘wildtype; WT’ in text) | Charles River | Strain code 022; RRID:IMSR_CRL:022 | |
| Strain, strain background (M. musculus) | B6.Cg-Gt(ROSA)26Sortm1.3(CAG-tdTomato,-EGFP)Pjen/J (‘FLTG’ in text) | Jackson Laboratory | Stock no. 026932; RRID:IMSR_JAX:026932 | (Gt(ROSA)26Sor) |
| Strain, strain background (M. musculus) | C57BL/6J-Tg(Nkx2-1-cre)2Sand/J (‘Nkx2.1-Cre’ in text) | Jackson Laboratory | Stock no. 008661; RRID:IMSR_JAX:008661 | (Nkx2-1) |
| Strain, strain background (M. musculus) | Nkx2-1tm2.1(flpo)Zjh/J(‘Nkx2.1-Flp’ in text) | Jackson Laboratory | Stock no. 028577; RRID:IMSR_JAX:028577 | (Nkx2-1) |
| Strain, strain background (M. musculus) | STOCK Tg(Prkcd-glc-1/CFP,cre)EH124Gsat/Mmucd (‘Prkcd-Cre’ in text) | Jackson Laboratory | MMRRC:011559; RRID:MMRRC_011559-UCD | (Prkcd) |
| Strain, strain background (M. musculus) | B6;129-Gt(ROSA)26Sortm5(CAG-Sun1/sfGFP)Nat/J (‘Sun1-GFP’ in text) | Jackson Laboratory | Stock no. 021039;RRID:IMSR_JAX:021039 | (Gt(ROSA)26Sor) (Sun1) |
| Strain, strain background (M. musculus) | Zic4-iCre (‘Zic4-Cre’ in text) | Kessaris Lab, UCL (Rubin et al., 2010) | Animal code A611(gift) | (Zic4) |
| Other | Ascl1 RNAscope probe | ACD | Cat# 313291 | |
| Other | Ccnd1 RNAscope probe | ACD | Cat# 442671 | |
| Other | Ccnd2 RNAscope probe | ACD | Cat# 433211 | |
| Other | Dcx RNAscope probe | ACD | Cat# 478671 | |
| Other | Hes5 RNAscope probe | ACD | Cat# 400998 | |
| Other | Hmga2 RNAscope probe | ACD | Cat# 466641 | |
| Other | Nes RNAscope probe | ACD | Cat# 313161 | |
| Other | Nfia RNAscope probe | ACD | Cat# 586501 | |
| Other | Prdm16 RNAscope probe | ACD | Cat# 584281 | |
| Software, algorithm | Fiji 2.1.0/1.53c | Schindelin et al., 2012 | http://fiji.sc; RRID:SCR_002285 | |
| Software, algorithm | Prism 9 | GraphPad | RRID:SCR_002798 | |
| Software, algorithm | MATLAB (MATBOTS) | MathWorks | RRID:SCR_001622 |