Structural insight into the dual function of LbpB in mediating Neisserial pathogenesis
Figures
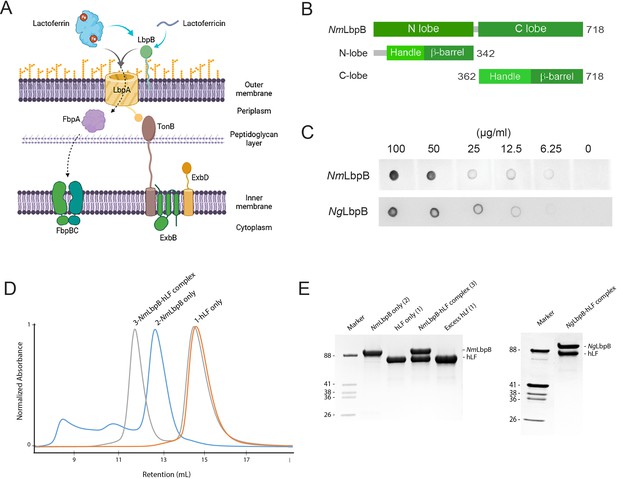
Lactoferrin-binding protein B (LbpB) forms a stable 1:1 complex with lactoferrin.
(A) The proposed role of Lbp system in iron acquisition from lactoferrin and protection from lactoferricin (Biorender). (B) Summary of LbpB constructs used in this study. (C) Solid-phase-binding assay of holo-lactoferrin (Lf) binding to N. meningitidis LbpB (NmLbpB; anti-Lf) and N. gonorrhoeae LbpB (NgLbpB; Lf-horse radish peroxidase [HRP]). (D) Formation of the NmLbpB–Lf complex over size-exclusion chromatography (SEC) from purified components. A leftward shift is observed for the complex compared to the individual components indicating the formation of the complex. (E) Sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS–PAGE) analysis of the NmLbpB–Lf complex formed from panel D, indicating the formation of the complex at a 1:1 ratio (lane 4). Similarly, the NgLbpB–Lf complex was formed by SEC from purified components which also formed a 1:1 complex as shown by SDS–PAGE analysis.
-
Figure 1—source data 1
LbpB forms a stable 1:1 complex with lactoferrin.
(A) The proposed role of lactoferrin-binding protein (Lbp) system in iron acquisition from lactoferrin and protection from lactoferricin (Biorender). (B) Summary of LbpB constructs used in this study. (C) Solid-phase-binding assay of holo-lactoferrin (Lf) binding to N. meningitidis LbpB (NmLbpB; anti-Lf) and N. gonorrhoeae LbpB (NgLbpB; Lf-horse radish peroxidase [HRP]). (The red arrow indicates the original blots that werecropped for this panel.) (D) Formation of the NmLbpB–Lf complex over size-exclusion chromatography (SEC) from purified components. A leftward shift is observed for the complex compared to the individual components indicating the formation of the complex. (E) Sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS–PAGE) analysis of the NmLbpB–Lf complex formed from panel D, indicating the formation of the complex at a 1:1 ratio (lane 4). Similarly, the NgLbpB–Lf complex was formed by SEC from purified components which alsoformed a 1:1 complex as shown by SDS–PAGE analysis. (The red arrow indicates the original SDS–PAGE gels that were cropped for this panel.)
- https://cdn.elifesciences.org/articles/71683/elife-71683-fig1-data1-v1.jpg
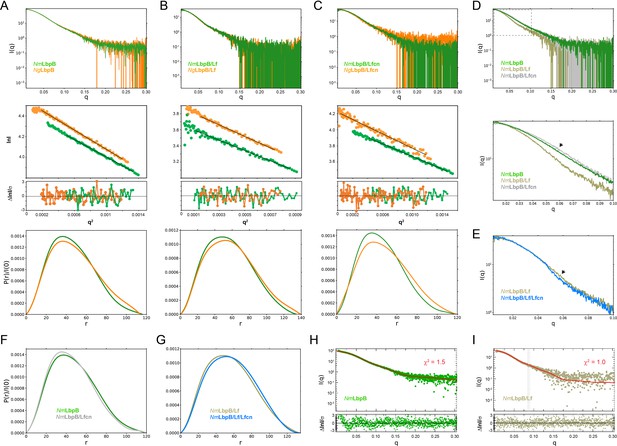
Solution structures of Nm/N. gonorrhoeae LbpB (NgLbpB) alone and in complexes using size-exclusion chromatography small-angle X-ray scattering (SEC-SAXS).
To compare the overall structures between the two species, SEC-SAXS analysis for both N. meningitidis LbpB (NmLbpB) (green) and NgLbpB (orange) was performed with resulting scattering profiles (top), Guinier plots (middle, offset for clarity), and P(r) plots (bottom) with closely matching Rg and Dmax values alone (A), in complex with lactoferrin (Lf) (B), and in complex with lactoferricin (Lfcn) (C). Since minimal differences were observed between the Nm and Ng samples, this supports the conclusion that LbpB has a conserved overall structure. (D) A superposition of scattering profiles for NmLbpB alone (green), in complex with Lf (olive), and in complex with Lfcn (grey). The bottom panel shows a zoomed view at lower q values to highlight the large change for the Lf complex and the small, but reproducible change observed for the Lfcn complex (green vs grey). Here, a significant difference in the shape of the curve was observed with Lf binding (green vs olive) confirming complex formation. And while small, a change upon Lfcn binding (green vs grey) could also be observed to confirm an overall structure change, albeit, much smaller given that the ligand is a peptide. (E) A zoomed view at lower q values comparing the NmLbpB–Lf complex in the absence (olive) and presence (blue) of Lfcn. Again, a reproducible small change is observed in the presence of Lfcn, confirming binding of the peptide. (F) P(r) plots for NmLbpB in the absence (green) and presence of Lfcn (grey) show that small structural changes occur upon Lfcn binding. (G) P(r) plots for NmLbpB–Lf in the absence (olive) and presence of Lfcn (blue), similarly showing that small structural changes occur upon Lfcn binding even in the presence of Lf. (H) A Crysol plot of the calculated scattering curve for the NmLbpB structure (red line) with the experimental scattering profile (green). (I) A Crysol plot of the calculated scattering curve for the NmLbpB–Lf complex structure (red line) with the experimental scattering profile (olive). Panels H and I provide confirmation that our X-ray crystal structure of NmLbpB–Lf agrees well with the ‘in-solution’ structures of NmLbpB alone and in complex Lf.
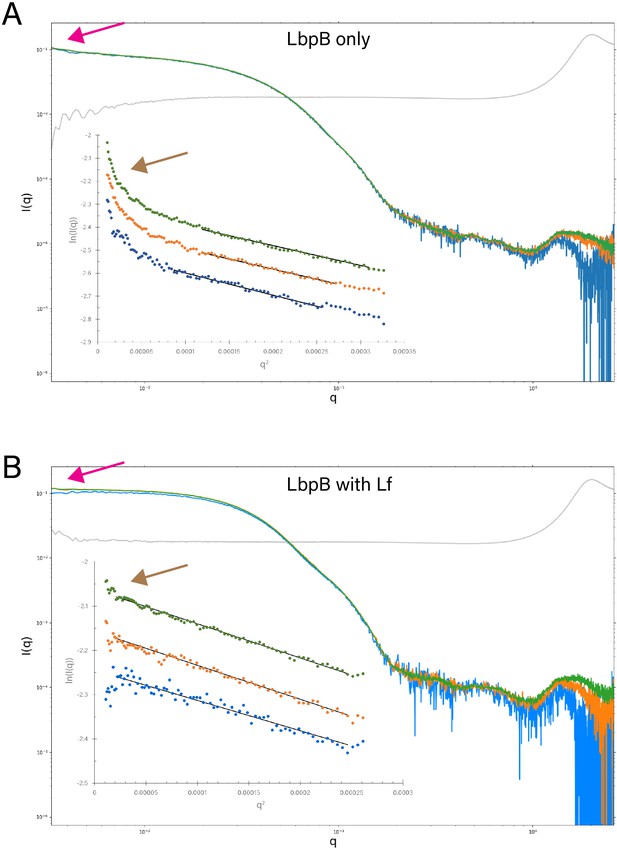
Static small-angle X-ray scattering (SAXS) analysis of lactoferrin-binding protein B (LbpB) alone and in complex with lactoferrin.
(A) A comparison of the static SAXS plots (log–log) of N. meningitidis LbpB (NmLbpB) at three different concentrations (0.85 mg/ml [blue], 1.7 mg/ml [orange], and 3.41 mg/ml [green]; blank is shown in grey). The inset shows respective Guinier plots at low q values (linear fits are shown as solid black lines); plots have been offset for clarity. (B) A comparison of the static SAXS plots (log–log) of NmLbpB in complex with lactoferrin (Lf) at three different concentrations (0.59 mg/ml [blue], 1.19 mg/ml [orange], and 2.38 mg/ml [green]; blank is shown in grey). The inset shows respective Guinier plots at low q values (linear fits are show as solid black lines); plots have been offset for clarity. For both panels, the magenta arrows highlight the intersection of the plots at the y-intercept, where LbpB with Lf is perpendicular, indicating a more stable sample; while LbpB only is not perpendicular, indicating a less stable sample. This is further confirmed when comparing the brown arrows showing the presence of Lf stabilizes the sample for SAXS analysis, whereas aggregation is observed at low q for the LbpB only sample.
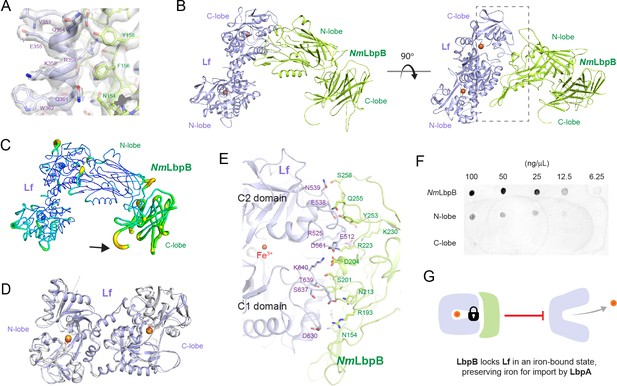
The 2.85 Å crystal structure of N. meningitidis LbpB (NmLbpB) in complex with human lactoferrin.
(A) Zoomed view at the interface between NmLbpB and lactoferrin (Lf) depicting the quality of the electron density shown as a grey isosurface (2FO–FC, 1.0σ). (B) Orthogonal views of the complex with NmLbpB in green, Lf in violet, and the iron atoms as red spheres. The N-lobe of NmLbpB interacts with only the C-lobe of Lf along an extended interface. (C) The C-lobe of NmLbpB has high B-factors with the large loops of this lobe not observed in our structure; the black arrow indicates the putative location of these loops. (D) An alignment of Lf from the complex with the structure of uncomplexed Lf (PDB ID 2BJJ) shows very little conformational changes upon binding NmLbpB (root-mean-square deviation (RMSD) of 1.3 Å). (E) A zoomed view of the binding interface shows extensive interactions along an elongated surface covering both the C1 and C2 domains of Lf (buried surface area 1760.8 Å2). (F) Solid-phase-binding assays show Lf binds both full-length and N-lobe NmLbpB, but not C-lobe only, supporting the observations in the complex structure. (G) Much like what has been proposed for the role of transferrin-binding protein (Tbp) B in the Tbp system, here we propose that lactoferrin-binding protein B (LbpB) also serves to bind and lock Lf in an iron-bound state for delivery to LbpA for iron import.
-
Figure 3—source data 1
The 2.85 Å crystal structure of N. meningitidis LbpB (NmLbpB) in complex with human lactoferrin.
(A) Zoomed view at the interface between NmLbpB and lactoferrin (Lf) depicting the quality of the electron density shown as a grey isosurface (2FO–FC , 1.0σ). (B) Orthogonal views of the complex with NmLbpB in green, Lf in violet, and the iron atoms as red spheres. The N-lobe of NmLbpB interacts with only the C-lobe of Lf along an extended interface. (C) The C-lobe of NmLbpB hashigh B-factors with the large loops of this lobe not observed in our structure; the black arrow indicates the putative location of these loops. (D) An alignment of Lf from the complex with the structure of uncomplexed Lf (PDB ID 2BJJ) shows very little conformational changes upon binding NmLbpB (RMSD of 1.3 Å). (E) A zoomed view of the binding interface shows extensive interactions along an elongated surface covering both the C1 and C2 domains of Lf (buried surface area 1760.8 Å 2 ). (F) Solid-phase-binding assays show Lf binds both full-length and N-lobe NmLbpB, but not C-lobe only, supporting the observations in the complex structure. (The red arrow indicates the original blot that was cropped for this panel.) (G) Much like what has been proposed for the role of transferrin-binding protein (Tbp) B in the Tbp system, here we propose that LbpB also serves to bind and lock Lf in an iron-bound state for delivery to LbpA for iron import.
- https://cdn.elifesciences.org/articles/71683/elife-71683-fig3-data1-v1.jpg
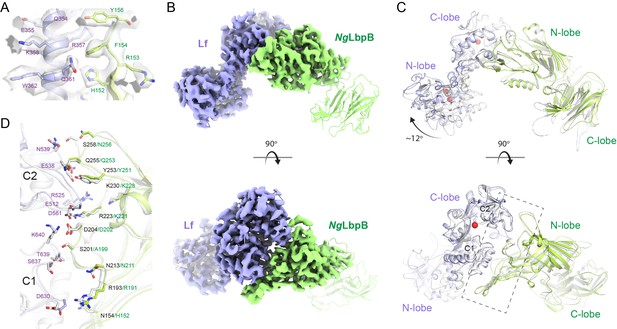
The 3.65 Å cryoEM structure of N. gonorrhoeae LbpB (NgLbpB) in complex with human lactoferrin.
(A) Zoomed view at the interface between NgLbpB and lactoferrin (Lf) depicting the quality of the density shown as a grey isosurface. (B) Orthogonal views of the full cryoEM map with NgLbpB in green and Lf in violet. (C) Orthogonal view of an alignment of the NgLbpB–Lf cryoEM structure (green/violet) with the N. meningitidis LbpB (NmLbpB)–Lf crystal structure (grey) (RMSD 1.4 Å along the interacting domains). (D) A zoomed view of the binding interface shows extensive interactions along an elongated surface covering both the C1 and C2 domains of Lf (buried surface area 1604 Å2).
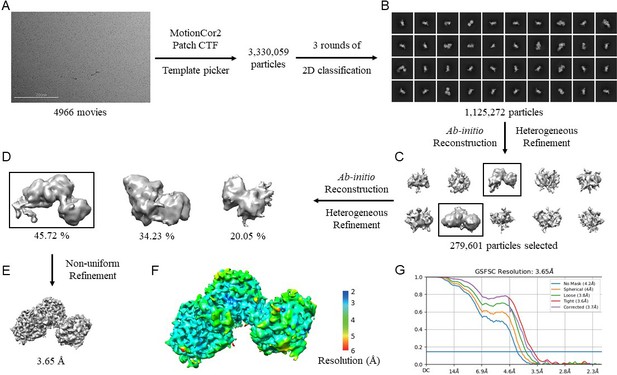
CryoEM data processing workflow for N. gonorrhoeae LbpB (NgLbpB)–Lf complex.
(A) Representative cryoEM micrograph from a total of 4966. Picked particles were subjected to iterative rounds of 2D classification. (B) Representative 2D class averages show different orientations of the particles. Two rounds of ab initio reconstruction followed by heterogeneous refinement produced 10 initial models in round 1 (C) and 3 initial models in rounds 2 (D). Boxed classes were selected for further processing. (E) Final 3D reconstruction map upon non-uniform refinement was obtained at 3.65 Å. (F) Local resolution of final cryoEM map. (G) Gold-standard Fourier shell correlation (GSFSC) curve of NgLbpB–Lf map. The horizontal blue line indicates 0.143 cutoff for resolution estimation.
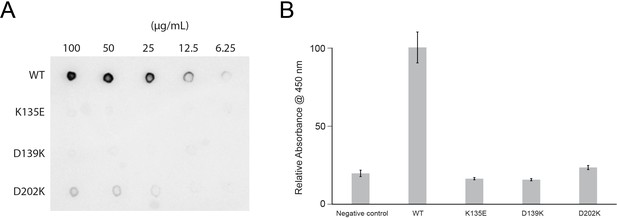
Lactoferrin binding to N. gonorrhoeae LbpB (NgLbpB) and NgLbpB mutants.
(A) Solid-phase-binding assays of dilutions of NgLbpB and mutants with horse radish peroxidase (HRP)-conjugated lactoferrin probe. (B) Enzyme-linked immunosorbent assays (ELISAs) of NgLbpB and mutants showing normalized absorbance. All experiments were done at least in triplicate.
-
Figure 4—figure supplement 2—source data 1
Lactoferrin binding to N. gonorrhoeae LbpB (NgLbpB) and NgLbpB mutants.
(A) Solid-phase-binding assays ofdilutions of NgLbpB and mutants with horse radish peroxidase (HRP)-conjugated lactoferrin probe. (The red arrow indicates the original blot that was cropped for this panel.) (B) Enzyme-linked immunosorbent assays (ELISAs) of NgLbpB andmutants showing normalized absorbance. All experiments were done at least in triplicate.
- https://cdn.elifesciences.org/articles/71683/elife-71683-fig4-figsupp2-data1-v1.jpg
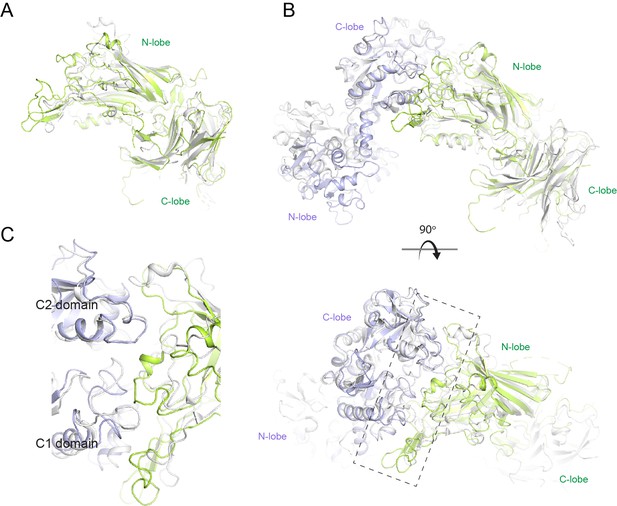
Structural comparison of N. meningitidis LbpB (NmLbpB) to NmTbpB.
(A) A superposition of the NmLbpB (green) and NmTbpB (grey) (PDB ID 3V8U) structures (RMSD of 2.0 Å). (B) Orthogonal views of a superposition of the NmLbpB–lactoferrin (Lf) (green/violet) and NmTbpB–transferrin (Tf) (grey) (PDB ID 3VE1) structures (overall RMSD of 4.0 Å; an RMSD of 1.4 Å was calculated for alignment of the interacting domains only). (C) A zoomed view from panel B of the dashed box along the binding interfaces.
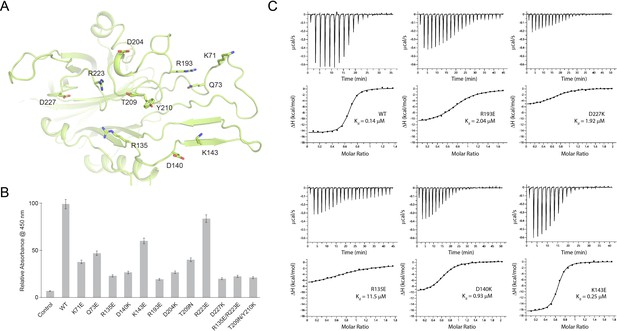
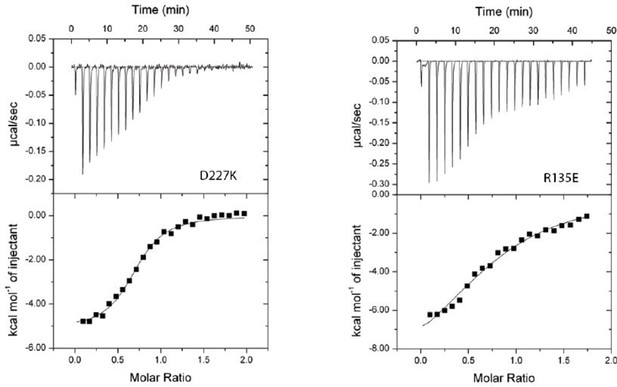
Probing the binding interface of N. meningitidis LbpB (NmLbpB) with lactoferrin.
(A) A zoomed view of the N-lobe of NmLbpB along the lactoferrin (Lf) interaction interface, highlighting primary residues involved in binding. (B) Enzyme-linked immunosorbent assays (ELISAs) to test the effects of structure-guided mutations of NmLbpB on Lf binding along the interaction interface. (C) Analysis of the binding parameters using isothermal titration calorimetry analysis of wild type and mutants of NmLbpB measuring the effects on Lf binding.
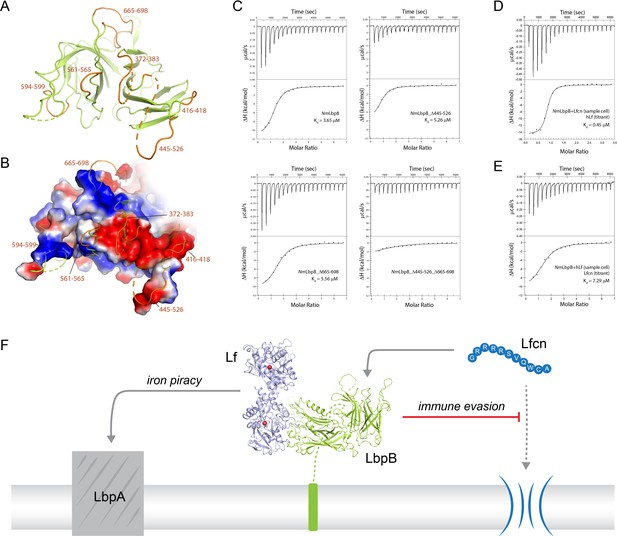
Probing the putative binding interface of N. meningitidis LbpB (NmLbpB) with lactoferricin.
(A) A zoomed view of the C-lobe of NmLbpB with the loops indicated in orange. (B) An electrostatic surface potential representation along the C-lobe of NmLbpB depicting the charged surfaces, including a strongly electronegative region (red). (C) Analysis of the binding parameters using isothermal titration calorimetry (ITC) analysis of wild type and loop-deletion mutants of NmLbpB measuring the effects on lactoferricin (Lfcn) binding. (D) ITC analysis of the NmLbpB–Lfcn complex titrated with lactoferrin (Lf), showing comparable binding to NmLbpB alone. (E) ITC analysis of the NmLbpB–(Lf) complex titrated with Lfcn, showing comparable binding to NmLbpB alone. (F) Model for the dual function of LbpB in mediating Neisserial pathogenesis by serving in both iron piracy and as an antimicrobial peptide (AMP) sink. While not shown, processing by NalP produces a soluble version of LbpB which can diffuse into the host environment to actively locate and neutralize AMP threats.
Additional files
-
Supplementary file 1
Supplementary Information.
(a) Summary of size-exclusion chromatography small-angle X-ray scattering (SEC-SAXS) parameters.
Lactoferrin (Lf) has a calculated molecular weight of 76.3 kDa, lactoferricin (Lfcn) 1.4 kDa, N. gonorrhoeae LbpB (NgLbpB) 78.4 kDa, and N. meningitidis LbpB (NmLbpB) 79.5 kDa. (b) Data collection and refinement statistics for the NmLbpB–Lf X-ray crystal structure. (c) Data collection and refinement statistics for the NgLbpB–Lf cryoEM structure. (d) Summary of the intermolecular interactions between NmLbpB and Lf. The information about interacting residues was obtained by QtPISA analysis. (e) Intermolecular interactions between NgLbpB and Lf. The information about interacting residues was obtained by QtPISA analysis. (f) Summary of ITC parameters for lactoferrin binding to NmLbpB mutants. These experiments were performed using a MicroCal iTC200 ITC calorimeter (Malvern Panalytical). (g) Summary of ITC parameters for lactoferricin binding to NmLbpB loop deletions. These experiments were performed using a Nano ITC calorimeter (TA Instruments). (h) Summary of ITC parameters for lactoferrin and lactoferricin binding to NmLbpB. These experiments were performed using a Nano ITC calorimeter (TA Instruments).
- https://cdn.elifesciences.org/articles/71683/elife-71683-supp1-v1.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/71683/elife-71683-transrepform1-v1.pdf
-
Source data 1
Source data for figures.
- https://cdn.elifesciences.org/articles/71683/elife-71683-supp2-v1.zip