Branched ubiquitin chain binding and deubiquitination by UCH37 facilitate proteasome clearance of stress-induced inclusions
Figures
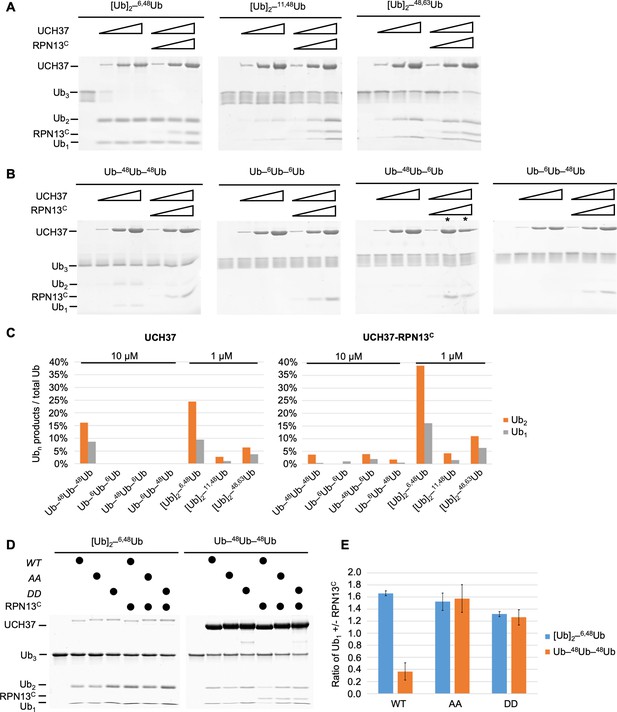
UCH37–RPN13C preferentially cleaves branched polyUb chains.
Branched (A) or linear (B) Ub3 substrates (5 μM) with the indicated Ub–Ub linkages were incubated for 1 hr at 37°C with 1, 5, or 10 μM His-TEV-UCH37, with or without the addition of equimolar RPN13C. Reactions were analyzed by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS–PAGE) and Coomassie staining. (C) Quantification of the Ub2 and Ub1 products from (A) and (B). For linear Ub3 substrates, results from incubations with 10 μM enzyme are shown; for branched Ub3 substrates, results from 1 μM enzyme are shown. (D) Wild-type (WT), M148A F149A (AA), or M148D F149D (DD) UCH37, alone or with the addition of equimolar of RPN13C, were incubated with Ub3 substrates for 2 hr at 37°C. Left, reactions contained 0.5 μM enzymes and 10 μM branched substrates. Right, reactions contained 10 μM enzymes and 5 μM linear Ub3. (E) Quantification from (D), plotted as the ratio of Ub1 produced in the presence over absence of RPN13C. Mean ± standard deviation (SD) from two independent replicates are shown.
-
Figure 1—source data 1
Uncropped gels in Figure 1.
- https://cdn.elifesciences.org/articles/72798/elife-72798-fig1-data1-v2.docx

UCH37–RPN13C preferentially cleaves branched polyUb chains.
(A) Notation used to describe architectures of polyUb chains used in this paper (adopted from Varadan et al., 2005). Note that we use the term ‘linear’ to refer to nonbranched Ub chains, which include but are not exclusively M1-linked Ub chains. (B) UCH37 exclusively cleaves the K48 linkage in a branched chain. Ub3 substrates were incubated with the indicated DUB (i.e., UCH37–RPN13C complex or OTUB1) and the reaction products were detected using either the K6 linkage-specific anti-Ub affimer (Michel et al., 2017) or K48 linkage-specific anti-Ub antibody. In Lane 5, OTUB1 was added after UCH37–RPN13C had completely hydrolyzed the K6/K48-branched Ub3, thereby producing K6-linked Ub2 that was refractory to OTUB1. (C) Crystal structure of the UCH37–RPN13C–Ub complex (PDB code: 4WLR). Part of the active site crossover loop (ASCL) is disordered and is represented as a dotted line. UCH37 ASCL residues M148 F149 contact RPN13C and are shown in gray. UCH37 residues E34, W36, and I216 contact the hydrophobic patch on Ub and are shown in orange. UCH37 catalytic-site residue C88 is shown in red. (D) Quantification from Figure 1D, plotted as the ratio of Ub2 in the presence over absence of RPN13C. Mean ± standard deviation (SD) from two independent replicates are shown.
-
Figure 1—figure supplement 1—source data 1
Raw western data in supplement 1B.
- https://cdn.elifesciences.org/articles/72798/elife-72798-fig1-figsupp1-data1-v2.docx
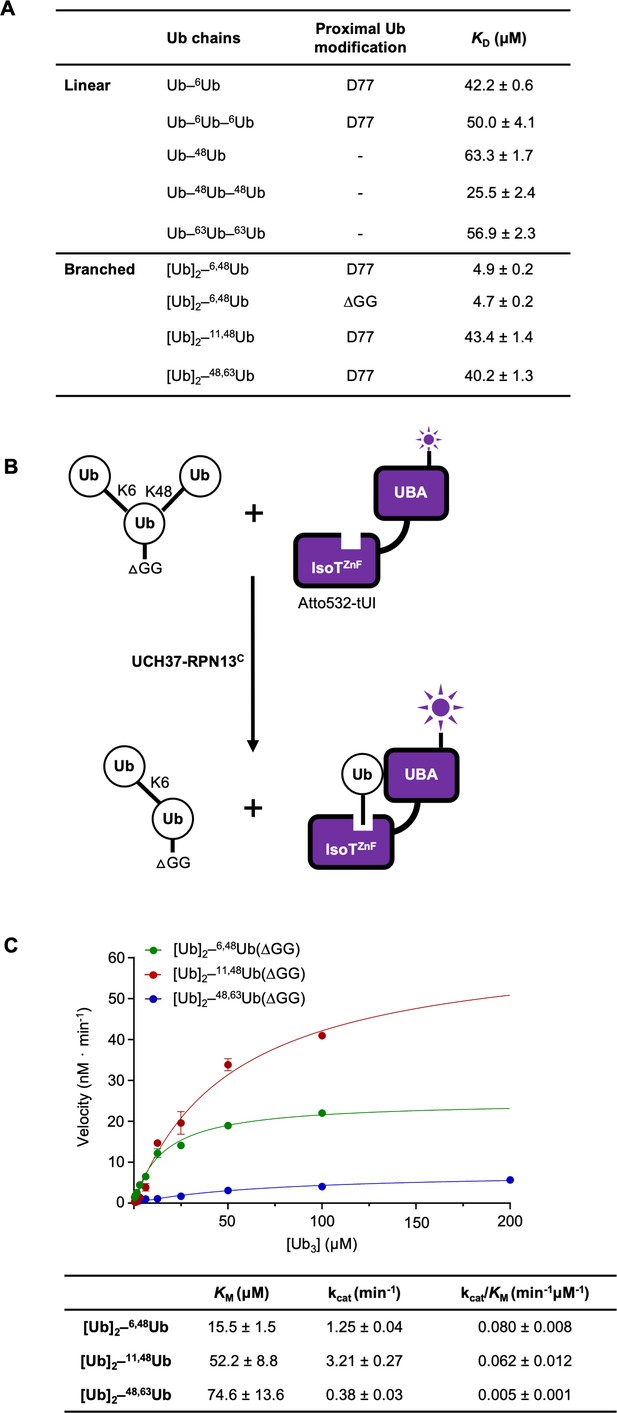
UCH37–RPN13C preferentially binds and deubiquitinates K6/K48-branched Ub3.
(A) Binding affinities between His-TEV-UCH37(C88S)–RPN13C and various polyUb chains were measured by microscale thermophoresis. Binding isotherms ( Figure 2—figure supplement 1) were fit with a single-site-binding model; best-fit KD values are shown with standard errors. (B) Schematic of the free Ub sensor-based deubiquitination assay. (C) Michaelis–Menten kinetics of branched Ub3 hydrolysis by NS–UCH37–RPN13C. The table shows best-fit KM, kcat, and kcat/KM values with standard errors from two independent replicates.
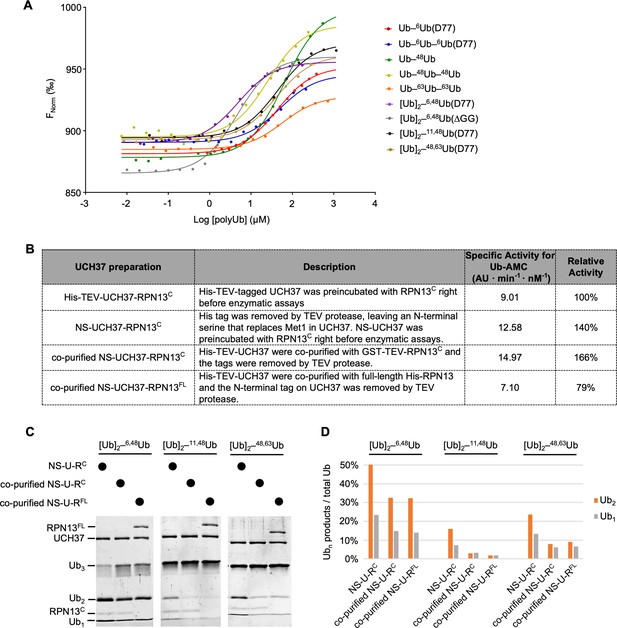
UCH37–RPN13C preferentially binds and deubiquitinates K6/K48-branched Ub3.
(A) Ub chain-binding isotherms of His-TEV-UCH37(C88S)–RPN13C were determined by microscale thermophoresis; fluorescent labeling used RED-tris-NTA that binds to the His-tag. Lines show the fitted curves using a single-site-binding model.(B) Each UCH37 preparation was evaluated by Ub-AMC hydrolysis. Specific activities were determined with 1 nM enzyme and 8 µM Ub-AMC; AU, arbitrary fluorescence unit. (C) Gel-based deubiquitinating enzyme (DUB) assays were performed to compare debranching efficiencies by the UCH37 preparations described in (B). Enzymes (1 µM) were incubated with 5 µM substrate as indicated for 2 hr at 37 °C. Reaction products were quantified and plotted in (D). U–RC and U–RFL represent UCH37–RPN13C and UCH37–RPN13FL, respectively.
-
Figure 2—figure supplement 1—source data 1
Uncropped gels in supplement 1C.
- https://cdn.elifesciences.org/articles/72798/elife-72798-fig2-figsupp1-data1-v2.docx
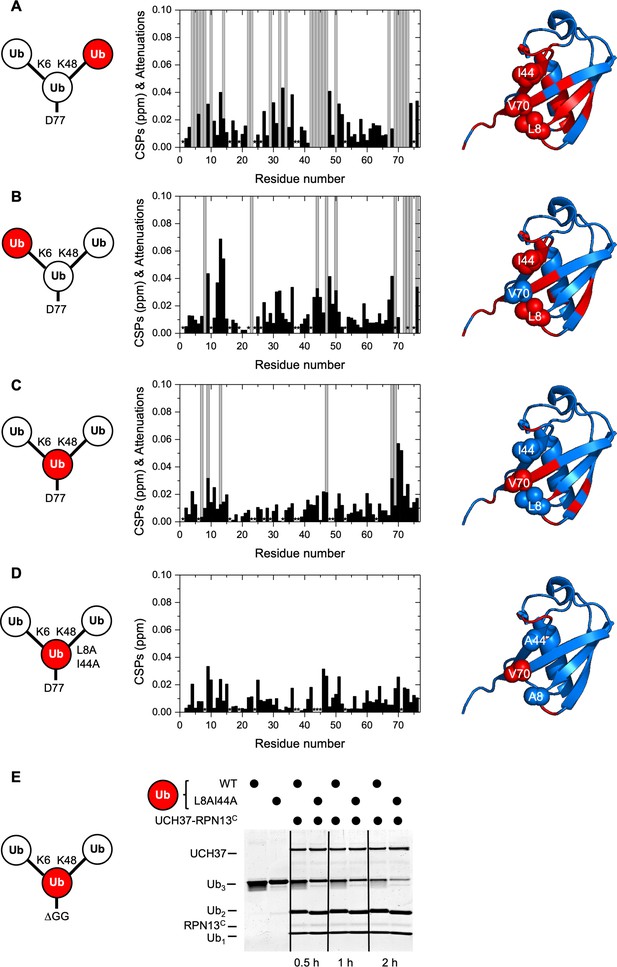
UCH37–RPN13C contacts the hydrophobic patches on both distal ubiquitin (Ub) units in a branched Ub3 chain.
Residue-specific perturbations of backbone amide NMR signals in the (A) K48-linked distal Ub, (B) K6-linked distal Ub, (C) the proximal Ub, and (D) mutated proximal Ub(L8A,I44A) in branched K6/K48-linked Ub3 caused by the addition of 1.2 molar equivalents of copurified UCH37(C88A)–RPN13C. The NMR spectra are shown in Figure 3—figure supplement 1. Black bars represent chemical shift perturbations (CSPs, in ppm), gray bars mark residues exhibiting strong signal attenuations. Residues that were not observed or could not be unambiguously assigned/quantified due to signal overlap are marked with asterisks. Residues with strong signal attenuations or CSP >0.025 ppm are mapped (red) on the 3D structure of Ub (PDB code: 1UBQ); the hydrophobic patch residues are shown in sphere representation. (E) 1 µM UCH37–RPN13C was incubated with 5 µM substrate as indicated at 37 °C. Reaction products were analyzed by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS–PAGE) and Coomassie staining.
-
Figure 3—source data 1
Uncropped gel in Figure 3E.
- https://cdn.elifesciences.org/articles/72798/elife-72798-fig3-data1-v2.docx
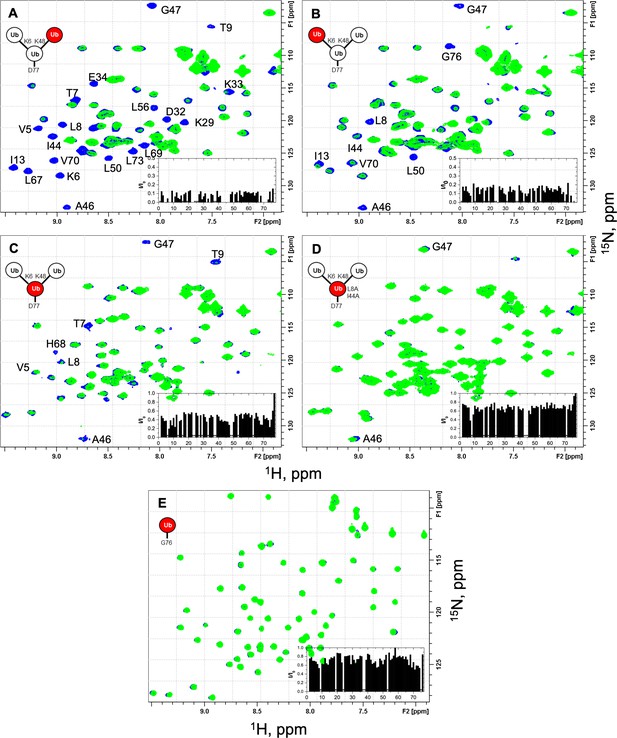
NMR spectra show unit-specific Ub3 binding to UCH37–RPN13C.
Overlays of 1H–15N and 1H–15N SOFAST-HMQC spectra (at 23 °C) of the individual 15N-labeled ubiquitin (Ub) units in the branched K6/K48-linked Ub3 and of monomeric Ub before (blue) and after (green) the addition of a 1.2 molar equivalent of UCH37(C88S)–RPN13C. Shown are fragments of the ‘fingerprint’ region of the spectra of (A) K48-linked distal Ub, (B) K6-linked distal Ub, (C) proximal Ub D77, and (D) proximal Ub D77 (L8A,I44A), as well as (E) monomeric WT Ub. Residues exhibiting significant signal attenuations as well as hydrophobic patch residues are indicated. The insets depict the ratio (I/I0) of signal peak intensities after (I) and before (I0) the addition of UCH37(C88S)–RPN13C as a function of residue number. The spectra of unbound protein were recorded with 64 (A, B), 128 (C, D), or 32 scans (E), those of the complex were recorded with 2048 (A, B), 512 (C), and 256 (D, E) scans to compensate for the overall signal decrease as a result of binding. The overall attenuation of peak signal intensities (I/I0) caused by the addition of UCH37(C88S)–RPN13C was 0.09 ± 0.03 for K48-linked distal Ub and 0.14 ± 0.04 for K6-linked distal Ub (completely attenuated signals excluded), 0.43 ± 0.09 for the proximal Ub and 0.67 ± 0.07 for the proximal Ub with the L8A,I44A mutation (signals from flexible G76 and D77 excluded), and 0.73 ± 0.10 for monoUb. Residues that exhibited complete signal attenuation and those with I/I0< mean − 2 × SD (in A, B) or I/I0< mean − 1.5 × SD (in C) are marked with gray bars in Figure 3A–C.
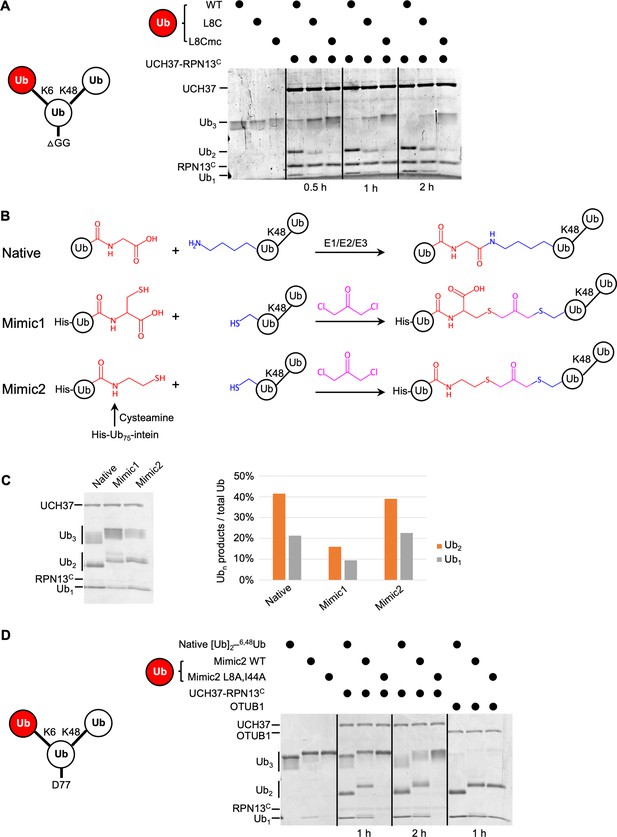
The hydrophobic patch on the K6-linked distal ubiquitin (Ub) is required for debranching.
(A) 1 µM enzyme was incubated with 5 µM of the indicated substrate at 37 °C; at the times indicated, aliquots were taken and analyzed by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS–PAGE) and Coomassie staining. (B) Schematic showing assembly strategies and structures of branched K6/K48–Ub3 and crosslinked mimics. (C) Comparison of native K6/K48–Ub3 with mimics 1 and 2 in gel-based DUB assays using NS–UCH37–RPN13C as described in (A). Quantification of Ub2 and Ub1 products from the gel are plotted as shown. (D) Native K6/K48–Ub3 or Mimic2 Ub3 containing either wild-type or L8A,I44A mutant Ub crosslinked to Ub–48Ub(K6C) were analyzed by gel-based DUB assays as described in (A).
-
Figure 4—source data 1
Uncropped gels in Figure 4.
- https://cdn.elifesciences.org/articles/72798/elife-72798-fig4-data1-v2.docx
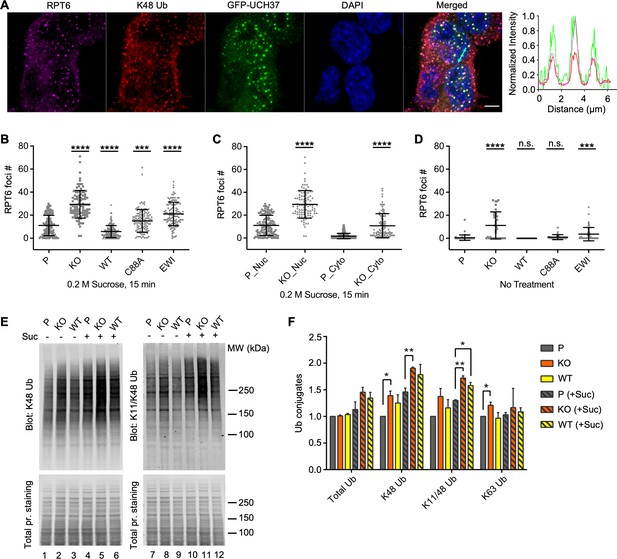
UCH37 activity regulates proteasome condensates upon osmotic stress.
(A) HCT116 cells expressing Flag-GFP-UCH37(C88A) were treated with 0.2 M sucrose for 15 min, then fixed and immunostained with RPT6 and K48-specific antibodies. The line profile represents the magenta (RPT6), red (K48), and green (GFP) channel intensities along the arrow shown in cyan (merged panel). Scale bar, 5 μm. (B–D) RPT6 foci numbers in each cell were quantified and are shown as mean ± standard deviation (SD); n > 100 cells were measured for each cell type. Unpaired t-tests were performed between each cell type and P: ***, p < 0.001; ****, p < 0.0001; n.s., not significant. (E) Whole-cell lysates were collected from P, KO, and WT cells with or without 30 min 0.2 M sucrose treatment, and then analyzed by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS–PAGE) and immunblotting with the indicated antibodies. (F) Quantification of Ub conjugates from whole-cell lysates immunoblotted with FK2 antibody (for total Ub conjugates) or with linkage-specific anti-Ub antibodies. Representative blots are shown in (E). Anti-Ub signals relative to those in P were plotted after normalization using the signal intensities from total protein stain. Mean ± SD from two independent replicates are plotted. Unpaired t-tests were performed between each cell type and P: *, p < 0.05; **, p < 0.01.
-
Figure 5—source data 1
Raw western data in Figure 5E, F.
- https://cdn.elifesciences.org/articles/72798/elife-72798-fig5-data1-v2.docx
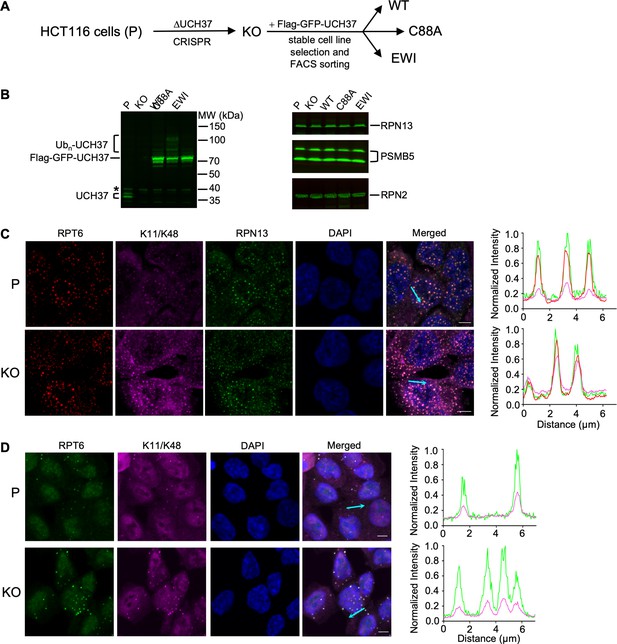
UCH37 regulates branched Ub chain-containing inclusions induced by proteolytic stresses.
(A) Schematic of cell line constructions. UCH37 knockout (KO) was derived from parental HCT116 cells (P) by CRISPR. KO cells were then infected with retrovirus encoding wild-type, C88A, or EWI versions of Flag-GFP-UCH37, followed by stable cell line selection to create WT, C88A, and EWI polyclonal cell lines. (B) Whole-cell lysates from cells as described in (A) were analyzed by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS–PAGE) and immunoblotting; * denotes a nonspecific band. Note that endogenous UCH37 has two isoforms, and UCH37(C88A) is oligoubiquitinated. (C, D) Cells are treated with 0.2 M sucrose for 15 min (C) or 0.5 mM sodium arsenite for 90 min (D), fixed and immunostained with the indicated antibodies. The line profiles represent the magenta, red, and green channel intensities along the arrow shown in cyan (merged panel). Scale bar, 5 μm.
-
Figure 5—figure supplement 1—source data 1
Raw western data in supplement 1B.
- https://cdn.elifesciences.org/articles/72798/elife-72798-fig5-figsupp1-data1-v2.docx
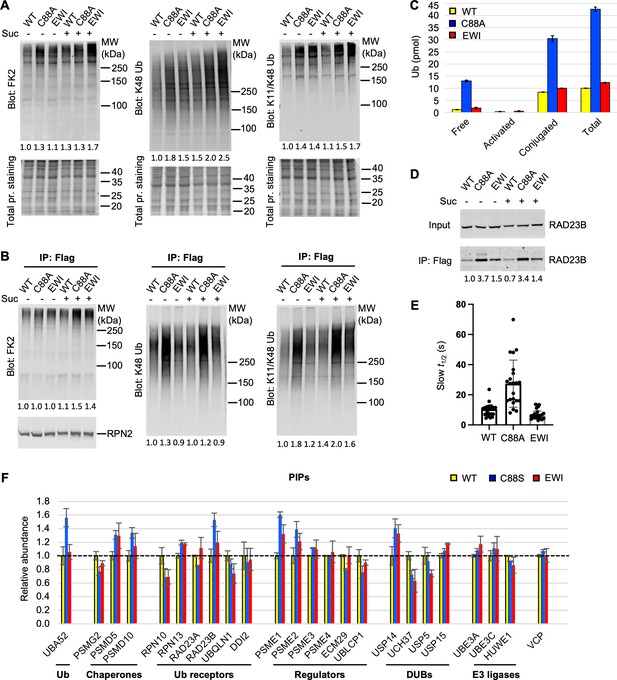
UCH37(C88A)-containing proteasomes accumulate polyUb species and RAD23B.
(A) Soluble cell lysates were collected from WT, C88A, and EWI cells with or without 30 min 0.2 M sucrose treatment, analyzed by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS–PAGE) and immunblotting with indicated antibodies. Numbers indicate Ub signals relative to those in untreated WT cells after normalization against total protein stain signals. (B) UCH37-containing proteasomes were immunoprecipitated and analyzed as described in (A). Numbers below the lanes indicate Ub signals relative to those in untreated WT cells after normalization against RPN2 signals. (C) Different types of Ub species associated with UCH37-containing proteasomes immunoprecipitated from HEK293 cells were quantified by a free Ub sensor-based assay (Choi et al., 2019). Shown are mean ± SD from two independent replicates. (D) RAD23B accumulation detected on UCH37(C88A)-containing proteasomes as described in (A) and (B). (E) Fluorescence recovery after photobleaching (FRAP) analysis of GFP-UCH37 after 0.2 M sucrose treatment demonstrates that C88A-containing proteasomes are less mobile. At least 20 foci from 7 or 8 cells were analyzed from each cell line. After fitting the FRAP curve with two exponentials, the slow t1/2 of each focus were plotted with mean and SD indicated. (F) UCH37-containing proteasomes from WT, C88S, and EWI cells were isolated with immobilized anti-GFP nanobodies, trypsin digested, and the peptides analyzed by tandem mass tag (TMT) mass spectrometry. Protein abundances were measured from two independent pulldowns, each analyzed by mass spectrometry in triplicate, and plotted as mean ± coefficient of variation (CV) after normalizing against signals from WT samples.
-
Figure 6—source data 1
Raw western data in Figure 6A, B, D.
- https://cdn.elifesciences.org/articles/72798/elife-72798-fig6-data1-v2.docx
-
Figure 6—source data 2
Excel file of fluorescence recovery measurements for each sucrose-induced focus.
- https://cdn.elifesciences.org/articles/72798/elife-72798-fig6-data2-v2.xlsx
-
Figure 6—source data 3
Excel file of proteomic analyses of UCH37-associated proteasomes by tandem mass tag (TMT).
- https://cdn.elifesciences.org/articles/72798/elife-72798-fig6-data3-v2.xlsx

UCH37(C88A) and UCH37(C88S)-containing proteasomes accumulate polyUb species.
(A) Analysis of UCH37-containing proteasomes from HEK293 cells. Flp-In T-REx 293 cell lines were constructed to inducibly express different versions of Flag-HA-tagged UCH37. After Dox induction for 48 hr, anti-Flag immunoprecipitates from the cells were analyzed by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS–PAGE) and immunoblotting with the indicated antibodies. (B) Anti-HA immunoprecipitates from cells expressing different versions of HA-tagged UCH37 were analyzed as described in (A). * denotes a non-specific band (IgG heavy chain). (C) Anti-Flag immunoprecipitates from cells inducibly express UCH37 C88A or C88S were analyzed as described in (A). (D) Microscale thermophoresis were performed to determine binding affinities between UCH37(C88A), or UCH37(C88S), and K6/K48-branched Ub3, as described in Figure 2A. (E) Microscale thermophoresis was performed to determine binding affinities between full-length RAD23B and Ub3 chains of different topologies. Shown are best-fit KD values with standard errors from a single-site-binding model. (F) Tandem mass tag (TMT) mass spectrometry analysis of UCH37-containing proteasomes as described in Figure 6F.
-
Figure 6—figure supplement 1—source data 1
Raw western data in supplement 1A, B, C.
- https://cdn.elifesciences.org/articles/72798/elife-72798-fig6-figsupp1-data1-v2.docx
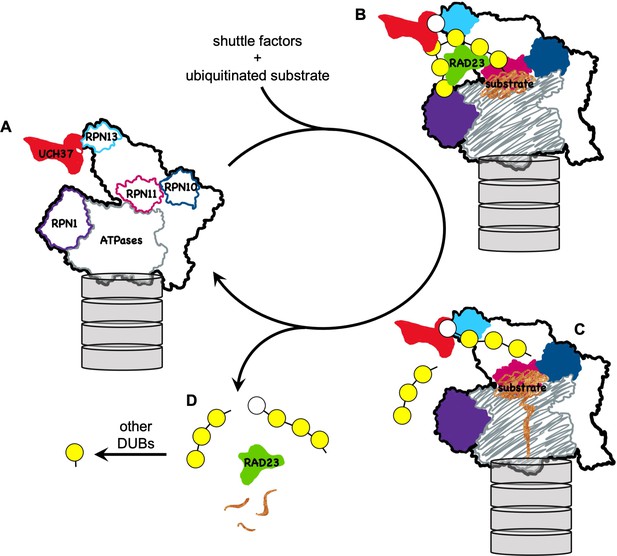
A model for how UCH37 promotes substrate processing and recycling of the proteasome through debranching of polyUb chains.
(A) A schematic of singly capped 26S proteasome in s1 state (PDB code: 4CR2) with locations indicated for intrinsic Ub receptors (RPN1, RPN10, and RPN13) and associated DUBs (RPN11 and UCH37); for clarity, USP14 is not shown. (B) RAD23 delivers a substrate (brown) modified by polyUb with a single branch point. UCH37 binds both distal Ubs at the branch point. K48-linked Ubs are shown in yellow whereas a non-K48-linked Ub is shown in white. (C) A substrate-engaged proteasome (PDB code: 4CR4) where substrate starts to be translocated and polyUb is removed en bloc by RPN11. Whether debranching by UCH37 occurs before or after RPN11 action is unknown. (D) UCH37 cleavage of the K48 linkage at the branch point facilitates release of free (poly)Ub chains and the shuttle factors.
Additional files
-
Supplementary file 1
Sequence alignment for UCH37 proteins used in this work.
The active site C88 is highlighted in yellow and EWI residues are indicated in orange.
- https://cdn.elifesciences.org/articles/72798/elife-72798-supp1-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/72798/elife-72798-transrepform1-v2.docx





