Integrating multi-omics data reveals function and therapeutic potential of deubiquitinating enzymes
Figures
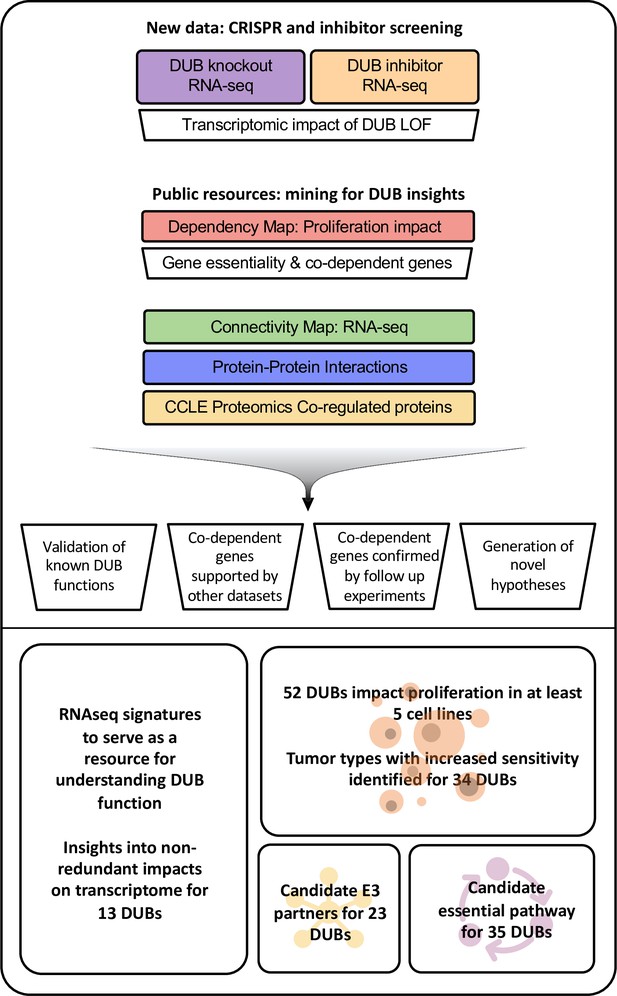
Approach to multi-omics analysis DUBs.
The functional impact of DUB loss was investigated via transcriptomic profiling of cells following CRISPR-Cas9 knockout (purple box); this was compared to signatures for genetic perturbations in the Broad Connectivity Map database of RNA-seq signatures (green). The impact of DUB knockout on cancer cell line proliferation was analyzed using the Broad Dependency Map (red) and compared to other gene knockouts in the dataset; this identified co-dependent genes (sets of genes whose knockout had similar effects on cell proliferation across cell lines). Multiple protein-protein interaction databases and co-expression (correlation in protein abundance across cell lines) in baseline proteomics in Cancer Cell Line Encyclopedia (CCLE; yellow) cell lines were used to provide support for physical or functional interactions among DUBs exhibiting co-dependency. We then used these analyses to explore the impact of DUB knockout on the transcriptome, determine the impact of DUBs on proliferation, and propose E3 ligase interactors and essential functions of DUBs. Acronyms used in figure: knockout (KO), loss of function (LOF), Cancer Cell Line Encyclopedia (CCLE).
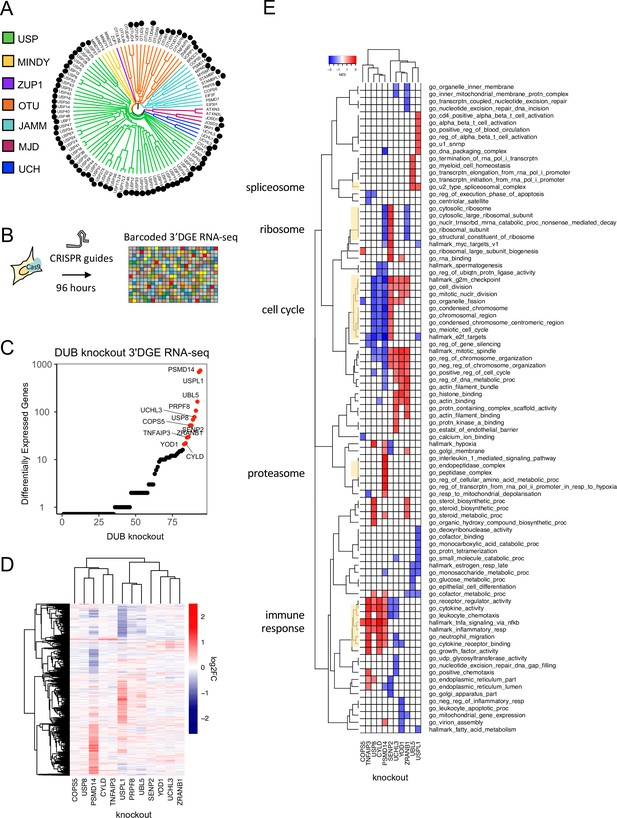
Measuring the impact of DUB CRISPR-Cas9 knockout on cell phenotypes.
(A) 81 DUBs covering the majority of the DUB phylogenetic tree were targeted in MDAMB231 cells using an arrayed CRISPR-Cas9 library (black circles (Figure 2 cont.) designate knockouts included in screen). (B) The impact of DUB loss on the transcriptome was profiled using high-throughput 3’ DGE RNA-seq 96 hr after transfection with CRISPR-Cas9. (C) The impact of individual DUB knockouts on the transcriptome of MDAMB231 cells, 4 days post CRISPR-Cas9 guide transfection as quantified by the number of differentially expressed genes (adjusted p-value <0.05). Guide RNA transfections were performed in triplicate and transcriptional responses then averaged. Knockouts that resulted in more than 20 differentially expressed (DE) genes are colored red. (D) Hierarchical clustering of log2FC values for significantly differentially expressed (DE) genes (adjusted p-value <0.05) for the knockouts colored red in (C). (E) Gene set enrichment analysis results for DUB knockouts that resulted in at least 20 DE genes. Gene sets that were significantly enriched (FDR <0.05) and in the top five up- or down-regulated gene sets in at least one condition are shown. PRPF8 knockout did not result in any significantly enriched pathways so it is not displayed.

Assessment of CRISPR-Cas9 guide target downregulation.
(A) Western blots showing select CRISPR-Cas9 guide target downregulation relative to a non-targeting control guide in MDAMB231 cells 96 hr post transfection. For each CRISPR-Cas9 knockout, four (cont.) crRNA guides for the target DUB were pooled together. (B) Change in CRISPR-Cas9 guide target mRNA abundance (log2 fold change vs. log 10 p-value, p-value <0.05 colored red) in MDAMB231 cells 96 hr post transfection.
-
Figure 2—figure supplement 1—source data 1
Original image from western blots in Figure 2—figure supplement 1 (images for target genes).
- https://cdn.elifesciences.org/articles/72879/elife-72879-fig2-figsupp1-data1-v1.png
-
Figure 2—figure supplement 1—source data 2
Original image from western blots in Figure 2—figure supplement 1 (images for loading controls).
- https://cdn.elifesciences.org/articles/72879/elife-72879-fig2-figsupp1-data2-v1.png
-
Figure 2—figure supplement 1—source data 3
Original image from western blots in Figure 2—figure supplement 1 (images for target genes, darker exposure).
- https://cdn.elifesciences.org/articles/72879/elife-72879-fig2-figsupp1-data3-v1.png
-
Figure 2—figure supplement 1—source data 4
Original image from western blots in Figure 2—figure supplement 1 (images for GAPDH for USP1 KO).
- https://cdn.elifesciences.org/articles/72879/elife-72879-fig2-figsupp1-data4-v1.png
-
Figure 2—figure supplement 1—source data 5
Original image from western blots in Figure 2—figure supplement 1 (images for USP1 for USP1 KO).
- https://cdn.elifesciences.org/articles/72879/elife-72879-fig2-figsupp1-data5-v1.png
-
Figure 2—figure supplement 1—source data 6
Labeled, uncropped blots for Figure 2—figure supplement 1 (USP7 KO).
- https://cdn.elifesciences.org/articles/72879/elife-72879-fig2-figsupp1-data6-v1.png
-
Figure 2—figure supplement 1—source data 7
Labeled, uncropped blots for Figure 2—figure supplement 1 (USP8 KO).
- https://cdn.elifesciences.org/articles/72879/elife-72879-fig2-figsupp1-data7-v1.png
-
Figure 2—figure supplement 1—source data 8
Labeled, uncropped blots for Figure 2—figure supplement 1 (USP10 KO).
- https://cdn.elifesciences.org/articles/72879/elife-72879-fig2-figsupp1-data8-v1.png
-
Figure 2—figure supplement 1—source data 9
Labeled, uncropped blots for Figure 2—figure supplement 1 (USP1 KO).
- https://cdn.elifesciences.org/articles/72879/elife-72879-fig2-figsupp1-data9-v1.png
-
Figure 2—figure supplement 1—source data 10
Labeled, uncropped blots for Figure 2—figure supplement 1 (USP11 KO).
- https://cdn.elifesciences.org/articles/72879/elife-72879-fig2-figsupp1-data10-v1.png
-
Figure 2—figure supplement 1—source data 11
Labeled, uncropped blots for Figure 2—figure supplement 1 (UCHL5 KO).
- https://cdn.elifesciences.org/articles/72879/elife-72879-fig2-figsupp1-data11-v1.png
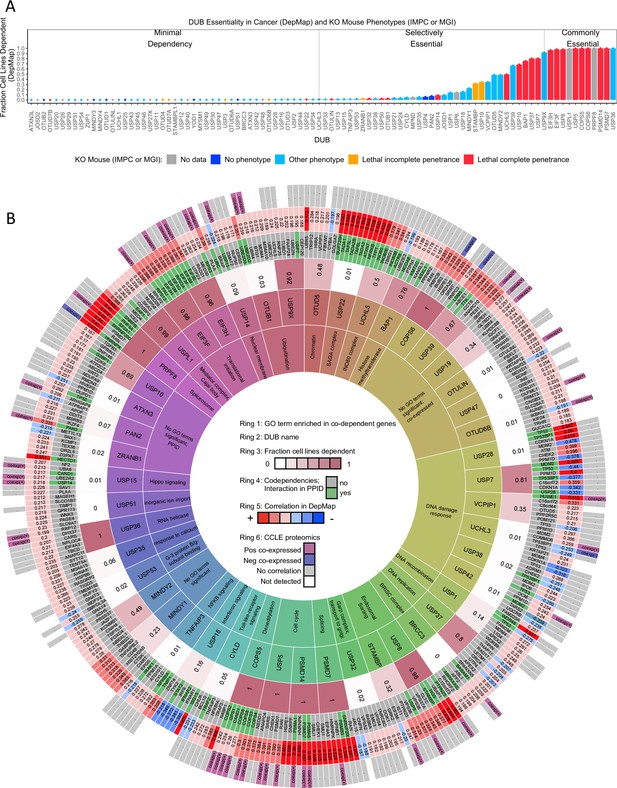
DUB essentiality and codependency relationships in DepMap.
(A) The fraction of cancer cell lines present in the DepMap that are strongly dependent on each DUB (using recommended threshold CERES <–0.5). Bars are colored coded based on knockout mouse phenotype data from the IMPC and MGI datasets. (B) The strongest co-dependent genes for each DUB (n=7 but similar results were obtained for n=5–10). For visualization purposes, only DUBs that either had a significantly enriched GO term, a co-dependent gene supported by PPID or CCLE proteomics co-expression, or scored as essential in at least 20% of DepMap cell lines are displayed (see Supplementary files 6 and 7 for complete codependency results). The inner ring (ring 1) contains the top GO term for the co-dependent genes for each DUB (highly similar GO terms are grouped to aid in viewing the data, see Supplementary file 5 for all GO results). The second ring contains the DUB gene name. The third ring contains the fraction of cell lines strongly dependent on the DUB. The fourth ring contains the co-dependent gene name (green if DUB – co-dependent gene pair exists in a protein-protein interaction database). The fifth ring contains the Pearson correlation value for the DUB-co-dependent gene pair (red represents a positive correlation and blue represents a negative correlation). The sixth ring designates which co-dependent genes had similar transcriptomic profiles in CMap or were co-expressed in baseline proteomics with the respective DUB.
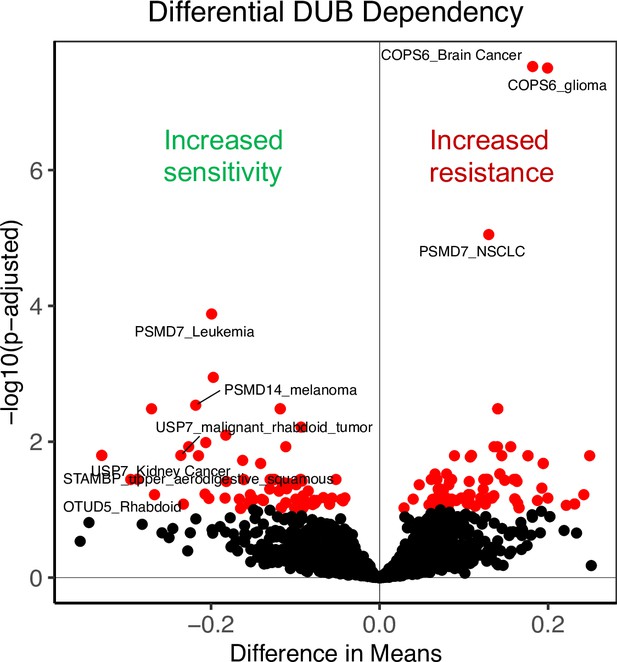
Differential DUB dependency by cancer type.
Two-sided t-tests were conducted between a selected cancer type and all other cell lines for each DUB knockout (plotted as difference in mean dependency score vs adjusted p-value, adjusted p-value <0.1 colored red). Negative difference in means represents a cancer type with increased dependence on that knockout (stronger impact on proliferation) and a positive difference in means represents a cancer type with decreased dependence on that knockout (weaker impact on proliferation).

Assessing the association between DUB abundance and dependency.
(A) Correlations between the impact of DUB knockout on proliferation (CERES dependency score) and DUB copy number, mRNA abundance, and protein abundance for each DUB. A negative correlation occurs when the DUB abundance is higher in the more sensitive cells and a positive correlation occurs when the DUB abundance is lower in the more sensitive cells. (B) The fraction of each tumor type in the DepMap with copy number loss of the eleven DUBs with correlations between copy number and sensitivity to DUB knockout greater than 0.2. (C) The fraction of all cell lines with copy number loss of the eleven DUBs with correlations between copy number and sensitivity to DUB knockout greater than 0.2.
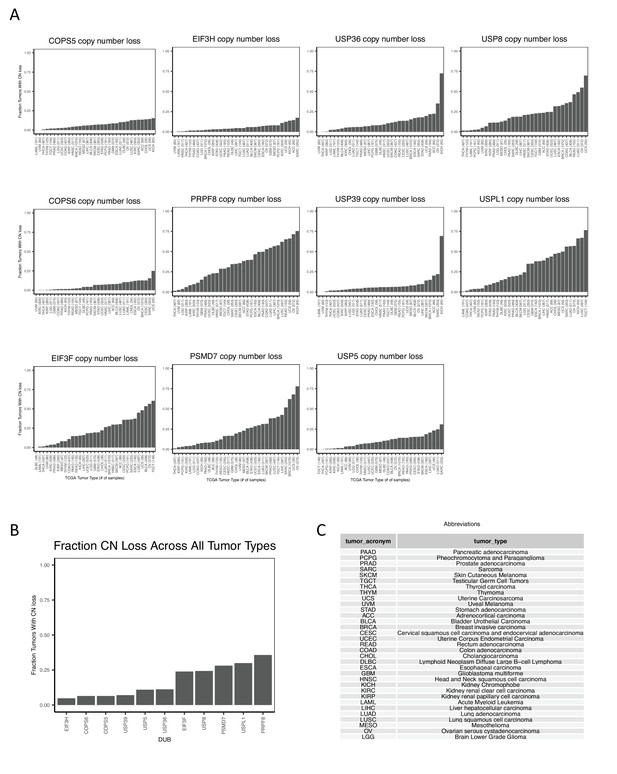
Copy number loss of individual DUBs in various tumor types.
(A) The fraction of each tumor type in TCGA PanCancer Atlas studies with copy number loss of individual DUBs. The eleven DUBs with correlations between copy number and sensitivity to DUB knockout greater than 0.2 in the DepMap are displayed. (B) The fraction of all tumor types with copy number loss of the 11 DUBs with correlations between copy number and sensitivity to DUB knockout greater than 0.2. (C) Abbreviations for TCGA tumor types.
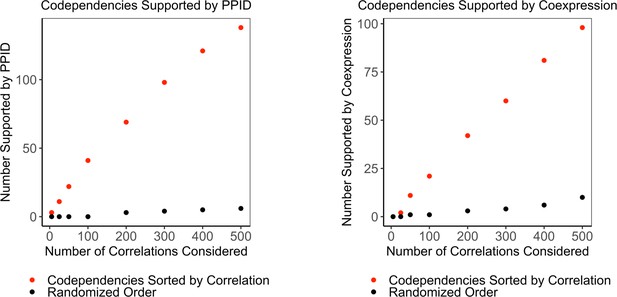
Enrichment of interactors and co-expressed genes in co-dependent genes.
(A) The number of co-dependent genes sorted by correlation value supported by protein-protein interactions (red) compared to the number supported when co-dependent genes are randomly shuffled (black). (B) The number of co-dependent genes sorted by correlation value supported by co-expression (red) compared to the number supported when co-dependent genes are randomly shuffled (black).

Enriched GO terms that are common across analyses.
(A) The significant GO terms from the DepMap co-dependent gene analysis were compared to GO terms enriched in the interactors for each DUB and the co-expressed proteins in the Cancer Cell Line Encyclopedia proteomics dataset. (B) The most significant GO term from DepMap analysis (smallest FDR <0.05) for each DUB as well as the GO term from DepMap analysis with the largest sum of -log(p-value) across datasets (if different). DUBs that were not present in the dataset are distinguished with a black dot.
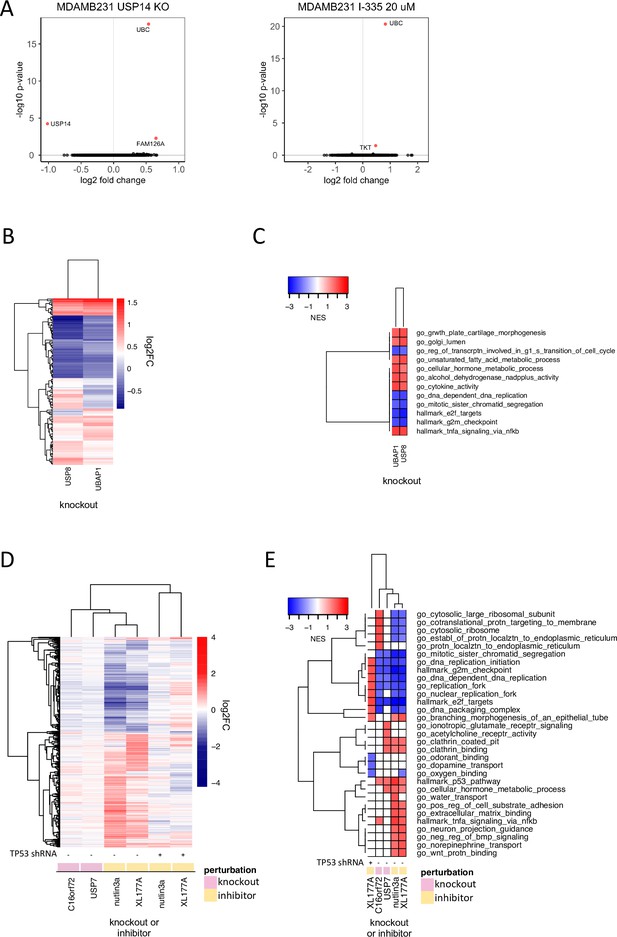
Discriminating the functions of well-studied DUBs.
(A) Changes in gene expression (log2 fold change vs. log 10 adjusted p-value, adjusted p-value <0.05 colored red) in MDAMB231 cells 96 hr following USP14 knockout by CRISPR-Cas9 (left) or 24 hr after treatment with the USP14 inhibitor I-335 at 20 µM (right). (B) Hierarchical clustering of significantly differentially expressed genes (adjusted p-value <0.05) 96 hr following knockout of USP8 or UBAP1 in MDAMB231 cells. (C) Gene sets significantly (FDR <0.05) enriched in MDAMB231 cells 96 hr after knockout of USP8 or UBAP1. The top five upregulated and top five downregulated gene sets for each condition are shown. (D) Hierarchical clustering of significantly differentially expressed genes (adjusted p-value <0.05) 96 hr after knockout of USP7 or C16orf72 in wild-type MCF7 cells or following a 24 hr treatment with 5 µM nutlin3a or 1 µM XL-177A in wildtype or p53 knockdown MCF7 cells. (E) Gene sets significantly enriched (FDR <0.05) for the conditions shown in (D). The top five up- and down-regulated gene sets for each condition are shown.
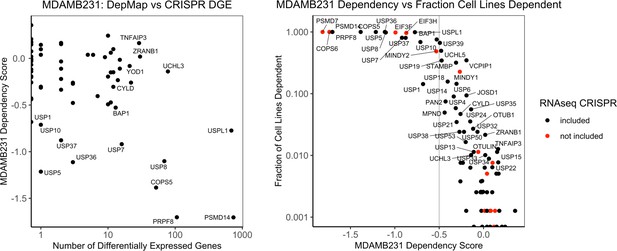
Comparison of transcriptional phenotypes and dependency in DepMap.
(A) The number of differentially expressed genes in the MDAMB231 RNAseq screen for each DUB vs the impact of each DUB knock on the proliferation of MDAMB231 cells in the DepMap (CERES dependency score). (B) The impact of DUB knock on the proliferation of MDAMB231 in the DepMap (CERES dependency score) vs the fraction of cell lines dependent on each DUB (CERES dependency score <–0.5).

Additional analysis of knockouts of endosomal sorting proteins as well as characterization of small molecules targeting MDM2 and USP7.
(A) Hierarchical clustering of significantly differentially expressed genes (adjusted p-value <0.05) 96 hr following knockout of USP8, UBAP1, HGS, and PTPN23 in MDAMB231 cells. (B) Gene sets significantly enriched (FDR <0.05) in MDAMB231 cells 96 hr after knockout of USP8, UBAP1, HGS, (cont.) and PTPN23. The top five upregulated and top five downregulated gene sets for each condition are shown. (C) Quantification of immunofluorescence measuring p21 levels following 24 hr treatments with XL177A or nutlin3a at the concentrations indicated in MCF7 wt and p53 knock down cells.
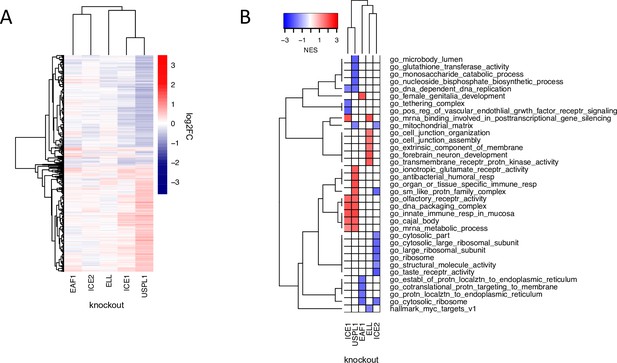
The role of USPL1 role in the Little Elongation Complex.
(A) Hierarchical clustering of log2FC values of differentially expressed genes (adjusted p-value <0.05) 96 h following knockout of USPL1, ICE1, ELL, ICE2, and EAF1 in MDAMB231. (B) Gene sets significantly enriched (FDR <0.05) for the conditions shown in (A). The top five up- and down-regulated gene sets for each condition are shown.
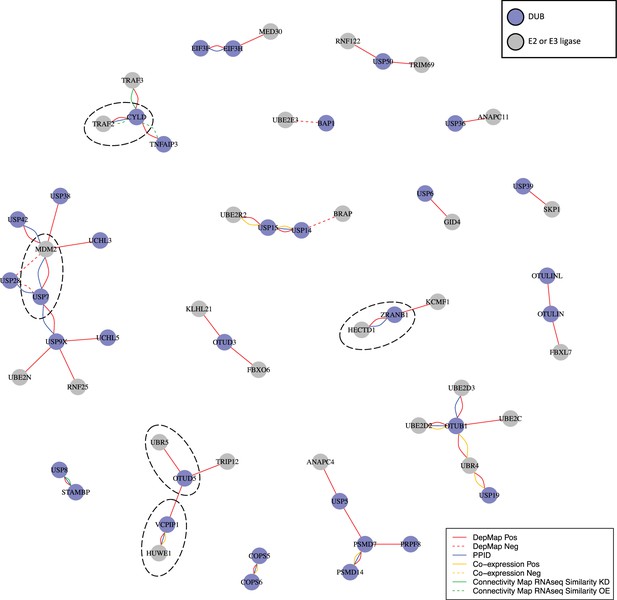
DUB E3 ligase network.
: Ubiquitin or ubiquitin-like transferases whose co-dependency relationships correlated with DUBs in the DepMap. DUBs are colored blue and ubiquitin transferases are colored grey. Red lines represent correlations in the top seven co-dependent genes. Green lines represent similarity by CMap (tau similarity score >90). Yellow lines represent co-expression in proteomics (FDR <0.01 and |z-score|>2). Blue lines represent interaction in protein-protein interaction databases.
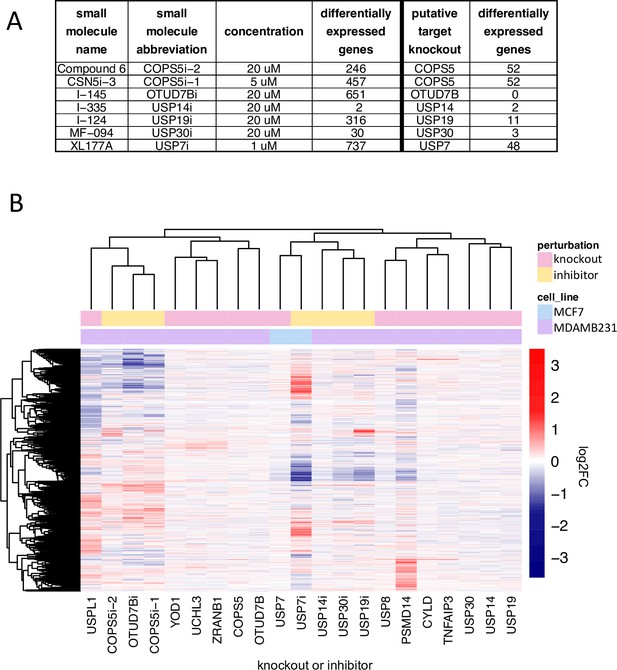
Comparison of DUB knockout and inhibition.
(A) The number of significantly differentially expressed genes (adjusted p-value <0.05) as a result of small molecule DUB inhibition (24 hr treatment) and knockout of the putative target (96 hours after transfection with guide). (B) Hierarchical clustering of log2FC values for significantly differentially expressed genes (adjusted p-value <0.05) for small molecule inhibitors of DUBs, the knockout of the putative DUB targets of the small molecules, and the DUB knockout hits that resulted in more than 20 differentially expressed genes. inhibition with XL177A, the DUB inhibitor and knockout were strongly correlated, but the inhibitor resulted in substantially more DE genes, and (iii) in five cases (inhibition of COPS5 with Compound 6 or CSN5i-3, inhibition of OTUD7B with I-145, inhibition of USP19 with I-124, and inhibition of USP30 with MF-094) the inhibitor and knockout had dissimilar signatures (a and b). CRISPR-Cas9 mediated knockout or small molecule inhibition of USP14 with I-335 resulted in only two DE genes, and the most strongly perturbed DE gene was the same in both instances: UBC. From these data, we conclude that I-335 is a potent and selective inhibitor of USP14.
Tables
Data and public resources used to infer the functions of DUB genes.
| Resource | Description | Key Insights | Terms / abbreviations |
|---|---|---|---|
| DGE RNA-seq | High-throughput RNA-seq data collected for this study following knockout or inhibition of individual DUBs using CRISPR-Cas 9. 81 DUB knockouts (found in a commercial library) and 7 small molecules were characterized in biological replicate. | Transcriptional signatures acquired from cells in which individual DUBs are inactivated. This provides insight into potential gene function. Similarity between signatures of gene knockout and small molecules provides insight into small molecule selectivity. | 3’DGE-seq: 3’ Digital Gene Expression, a type of high-throughput RNA-seq. Differentially Expressed (DE) gene: Gene with different level of expression across two samples as defined by a false discovery rate (FDR) adjusted P<0.05. |
| Connectivity Map (CMap) | A Broad Institute database of post-perturbation RNA-seq signatures generated from multiple cell lines following knockdown (RNAi or CRISPR-Cas 9), gene over-expression, or treatment of cells with small molecule drugs. Signatures in CMAP resource are comprised of 978 landmark genes measured using a Luminex bead-based assay. The expression of 11,350 genes is then inferred. Data are available for ~3000 genes and ~5000 small molecules. | Enables identification of genes that, when silenced with RNAi, or overexpressed, have similar transcriptional effects as a query L1000 or DGE-Seq signature. The effects of gene knockout/over-expression can be compared to the effects of drugs for mechanism of action studies. | Query mRNA profile: the transcriptomic signature that is used to query CMap and retrieve similar signatures. Tau score: a parameter that quantifies similarity between the query mRNA profile and CMap signatures (tau similarity is computed by counting the number of pairwise mismatches between two ranked lists). CMap recommends a threshold value for tau similarity scores of >90. |
| Dependency Map (DepMap) | A Broad Institute database of gene essentiality scored in >700 cancer cell lines based on genome-wide pooled CRISPR-Cas9 knockout screens. Dropout of specific Guide RNAs is used as a measure of essentiality. | Enables identification of genes that are essential for cell proliferation or survival in specific cell lines. Patterns of essentiality across cell line panels (the DepMap score) can be computed to identify genes potentially having related biological functions. | DepMap score – a measure of cell line dropout rate in a pooled genome-wide CRISPR screen (a gene with a dependency score <–0.5 is considered an essential gene in that cell line) Co-Dependency. Genes with similar DepMap scores are said to be co-dependent. In our study, we analyzed the top seven co-dependent genes, but similar data were obtained when more or fewer co-dependent genes were considered. |
| Cancer Cell Line Encyclopedia (CCLE) Proteomics | A Broad Institute database of baseline shotgun proteomics collected from CCLE cell lines. Data from ~375 cell lines and 12,000 proteins per line are available. | Protein co-expression across cell line panels provides insight into functional interactions: proteins in the same complex are often co-expressed to a significant degree across CCLE cell clines. | Co-expressed genes: correlation in protein abundance across cell lines with FDR <0.01 and |z-score|>2. These thresholds are set based on previous publications (Nusinow et al., 2020). |
| BioGRID | Protein-protein interaction database compiling interaction data from multiple sources. Protein interactions are measured using multiple physical assays including affinity capture MS, affinity capture western blotting, and assembly of reconstituted complexes from purified recombinant subunits in vitro. | Discovery of protein-protein interactions using a variety of methods that focus on physical interaction. | PPID: protein-protein interaction database. |
| IntAct | Protein-protein interaction database that compiles interaction data from multiple sources. | Discovery of protein-protein interactions using a variety of methods that focus on physical interaction. | PPID: protein-protein interaction database. |
| Pathway Commons | Protein-protein interaction database compiling interaction data from multiple sources. | Discovery of protein-protein interactions using a variety of methods that focus on physical interaction. | PPID: protein-protein interaction database. |
| NURSA | Physical interactions among proteins, particularly those involved in transcription. | Discovery of proteins that interact directly as determined by affinity capture mass spectrometry; approximately ~3000 IP assays are currently included. | PPID: protein-protein interaction database. NURSA: Nuclear Receptor Signaling Atlas |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (human) | DUB CRISPR-Cas9 screening library: Dharmacon EDIT-R crRNA Library - Human Deubiquitinating Enzymes | Dharmacon (Horizon Discovery) | GC-004700 Lot 17,107 | |
| Genetic reagent (human) | Dharmacon Edit-R tracrRNA | Dharmacon (Horizon Discovery) | U-002005–05 | |
| Transfected construct | Dharmafect 4 | Dharmacon (Horizon Discovery) | T-2004–02 | |
| Cell line (human) | MDAMB231 | ATCC | CRM-HTB-26 | |
| Cell line (human) | MCF7 | ATCC | HTB-22 | |
| Antibody | Flag-tag (L5) antibody (rat monoclonal) | Thermo Fischer | MA1-142 | 1:1,000 dilution |
| Antibody | USP7 antibody (rabbit monoclonal) | Cell Signaling | 4,833 | 1:1,000 dilution |
| Antibody | USP8 antibody (mouse monoclonal) | Santa Cruz Biotechnology | sc-376130 | 1:1,000 dilution |
| Antibody | USP10 antibody (rabbit monoclonal) | Cell Signaling | 8,501 | 1:1,000 dilution |
| Antibody | USP1 antibody (rabbit monoclonal) | Cell Signaling | D37B4 | 1:1,000 dilution |
| Antibody | USP11 antibody (rabbit monoclonal) | abcam | ab109232 | 1:1,000 dilution |
| Antibody | UCHL5 antibody (mouse monoclonal) | Santa Cruz Biotechnology | sc-271002 | 1:1,000 dilution |
Additional files
-
Supplementary file 1
cRNAs used for CRISPR-Cas9 knockout studies.
- https://cdn.elifesciences.org/articles/72879/elife-72879-supp1-v1.xlsx
-
Supplementary file 2
Full results for gene set enrichment analysis for CRISPR-Cas9 knockout studies.
- https://cdn.elifesciences.org/articles/72879/elife-72879-supp2-v1.txt
-
Supplementary file 3
Full results for Connectivity Map analysis for CRISPR-Cas9 knockout studies.
- https://cdn.elifesciences.org/articles/72879/elife-72879-supp3-v1.tsv
-
Supplementary file 4
Full t-test results for lineage sensitivity analysis of DUBs in the DepMap.
- https://cdn.elifesciences.org/articles/72879/elife-72879-supp4-v1.tsv
-
Supplementary file 5
Dataset integration results used to generate Figure 3.
- https://cdn.elifesciences.org/articles/72879/elife-72879-supp5-v1.tsv
-
Supplementary file 6
Full dataset integration results for integration of DepMap, CMap results, CCLE proteomics, and PPIDs.
- https://cdn.elifesciences.org/articles/72879/elife-72879-supp6-v1.tsv
-
Supplementary file 7
Column descriptors to facilitate interpretation of Supplementary file 6.
- https://cdn.elifesciences.org/articles/72879/elife-72879-supp7-v1.xlsx
-
Supplementary file 8
Identifiers for small molecules used in RNA-seq screen.
- https://cdn.elifesciences.org/articles/72879/elife-72879-supp8-v1.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/72879/elife-72879-transrepform1-v1.docx