CREB5 reprograms FOXA1 nuclear interactions to promote resistance to androgen receptor-targeting therapies
Figures
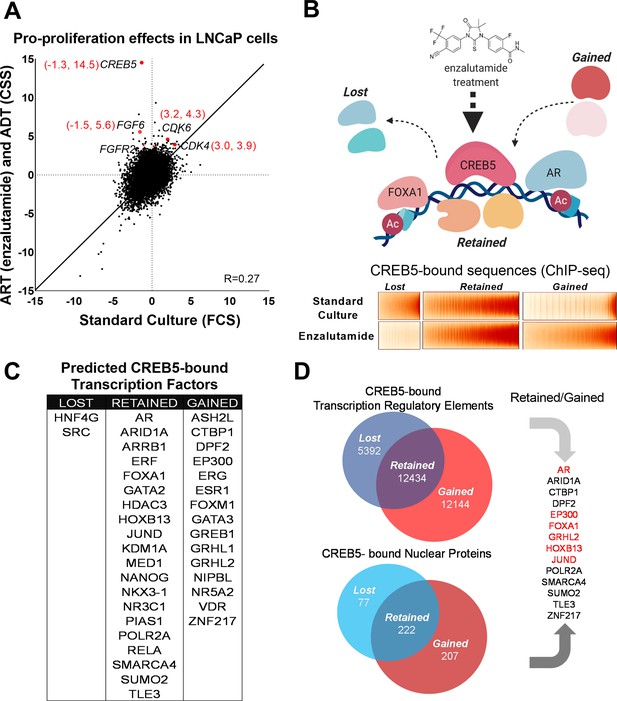
CREB5 overexpression and nuclear interactions that are reprogrammed upon androgen receptor-targeted therapy (ART) treatments.
(A) Analysis of enzalutamide resistance genes in LNCaP cells based on a genome-scale screen, including 17,255 open reading frames (ORFs). Z-scores are displayed for the experimental arms conducted in either standard culture (FCS, x-axis) or treatment (enzalutamide +CSS, y-axis) conditions. CREB5 and other enzalutamide -specific hits (Z > 3) and their proliferation scores are highlighted in red. (B) A model that depicts changes in chromatin -associated interactions of CREB5 that occur post enzalutamide treatment. Bottom,: CREB5 ChIP-seq data is presented in accordance to three categories of CREB5 binding behavior. Categories are grouped by significant changes by enzalutamide treatments. (C) GIGGLE analyses predicts transcription factors that are CREB5-bound based on the ChIP-seq experiments as categorized in B. (D) Rapid immunoprecipitation and mass spectroscopy of endogenous proteins (RIME) experiments were performed to identify CREB5 interaction profiles in control or enzalutamide -treated cultures. The common proteins identified by both RIME and GIGGLE are highlighted for the retained and gained groups.
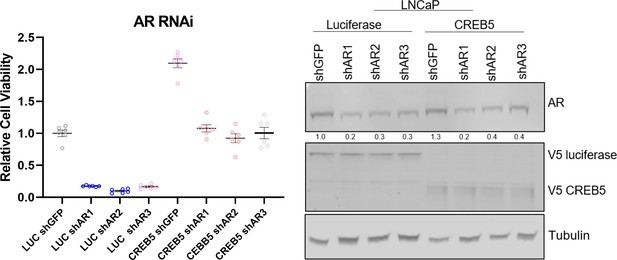
LNCaP cells overexpressing CREB5 or luciferase negative control (LUC) were transduced with lentivirus with shRNAs that target a GFP sequence or three distinct regions of androgen receptor (AR).
Cell viability was determined with standard error of the mean displayed (left). The degree of AR suppression is shown in immunoblots (right). V5 indicates expression of CREB5 or luciferase and tubulin is a loading control.
-
Figure 1—figure supplement 1—source data 1
Immunoblots were used to detect expression of V5-tagged CREB5 or luciferase in the indicated samples for Figure 1—figure supplement 1.
- https://cdn.elifesciences.org/articles/73223/elife-73223-fig1-figsupp1-data1-v2.tif
-
Figure 1—figure supplement 1—source data 2
The area highlighted was used to develop the figure for V5-tagged CREB5 or luciferase in the indicated samples for Figure 1—figure supplement 1.
- https://cdn.elifesciences.org/articles/73223/elife-73223-fig1-figsupp1-data2-v2.tif
-
Figure 1—figure supplement 1—source data 3
Immunoblots were used to detect expression of androgen receptor (AR) in the indicated samples for Figure 1—figure supplement 1.
- https://cdn.elifesciences.org/articles/73223/elife-73223-fig1-figsupp1-data3-v2.tif
-
Figure 1—figure supplement 1—source data 4
The area highlighted was used to develop the figure for androgen receptor (AR) in the indicated samples for Figure 1—figure supplement 1.
- https://cdn.elifesciences.org/articles/73223/elife-73223-fig1-figsupp1-data4-v2.tif
-
Figure 1—figure supplement 1—source data 5
Immunoblots were used to detect expression of tubulin in the indicated samples for Figure 1—figure supplement 1.
- https://cdn.elifesciences.org/articles/73223/elife-73223-fig1-figsupp1-data5-v2.tif
-
Figure 1—figure supplement 1—source data 6
The area highlighted was used to develop the figure for tubulin in the indicated samples for Figure 1—figure supplement 1.
- https://cdn.elifesciences.org/articles/73223/elife-73223-fig1-figsupp1-data6-v2.tif
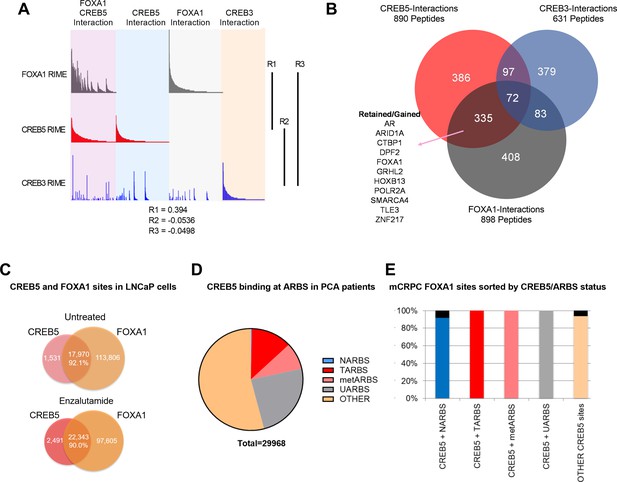
CREB5 and FOXA1 share chromatin-associated functions in metastatic castration-resistant prostate cancer (mCRPC) based on binding sequences and rapid immunoprecipitation and mass spectroscopy of endogenous proteins (RIME) interaction profiles.
(A) RIME analysis depicting the interaction profiles of FOXA1 (greay), CREB5 (red), and CREB3 (blue). Proteins that interact with FOXA1 and CREB5 are also shown. The Pearson correlation coefficients (R) are shownn. (B) Venn diagram depicting unique peptide interactions that are either independent or shared between CREB5 (red), CREB3 (blue), and FOXA1 (greay). Peptides identified to be induced by enzalutamide are highlighted as Retained/Gained. (C) ChIP-seq experiments were used to examine CREB5 and FOXA1 interactions in LNCaP cells with or without enzalutamide treatments. The Venn diagram depicts total binding sites in each condition and the overlapping sites and percentage of shared transcription regulatory elements. (D) CREB5 -bound sites are analyzed and represented as AR binding sites (ARBS) observed in clinical samples. This includes ARBS exclusive in normal (NARBS), tumor (TARBS), mCRPC (metARBS), all tissues (UARBS), as well as all non -ARBS (OTHER). E. CREB5 -bound ARBs are further classified and depicted as % of FOXA1 sites observed in mCRPC (y-axis). The colors represent the overall percentage of FOXA1 sites while the black represents non -FOXA1 sites.
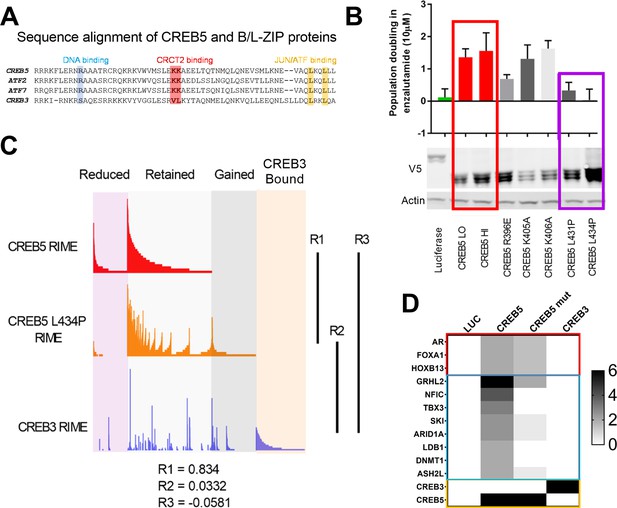
A loss of resistant CREB5 mutant was identified and determines transcription co-regulators associated with androgen receptor-targeted therapy (ART)-resistant proliferation.
(A) Alignment of CREB5 sequence with ATF2, ATF7, and CREB3, highlighting the DNA binding domains (blue), CRCT2 binding domains (red), and JUN/ATF binding domains (yellow). (B) Population doubling (y-axis) of LNCaP cells overexpressing wild-type CREB5 variants (red), CREB5 JUN/FOS-binding mutants (purple), and a luciferase negative control (green) in 10 μM enzalutamide. V5 expression represents V5-tagged CREB5 protein levels. Actin is a loading control. (C) Rapid immunoprecipitation and mass spectroscopy of endogenous proteins (RIME) analysis depicting the interaction profiles of wild-type CREB5 (red), CREB5 L434P (orange), and CREB3 (blue). CREB5 interactions that were reduced, retained, or gained upon enzalutamide treatments are depicted. The Pearson correlation coefficients (R) are shown. (D) A heatmap depicts the RIME interactions of luciferase control, wild-type CREB5, L434P CREB5, and CREB3. Several canonical AR co-factors (AR, FOXA1, HOXB13) interact with both CREB5 and CREB5 L434P and are shown.
-
Figure 3—source data 1
Immunoblots were used to detect expression of V5-tagged CREB5 or luciferase in the indicated samples for Figure 3B.
- https://cdn.elifesciences.org/articles/73223/elife-73223-fig3-data1-v2.tif
-
Figure 3—source data 2
The area highlighted was used to develop the figure for V5-tagged CREB5 or luciferase in the indicated samples for Figure 3B.
- https://cdn.elifesciences.org/articles/73223/elife-73223-fig3-data2-v2.tif
-
Figure 3—source data 3
Immunoblots were used to detect expression of actin in the indicated samples for Figure 3B.
- https://cdn.elifesciences.org/articles/73223/elife-73223-fig3-data3-v2.tif
-
Figure 3—source data 4
The area highlighted was used to develop the figure for actin in the indicated samples for Figure 3B.
- https://cdn.elifesciences.org/articles/73223/elife-73223-fig3-data4-v2.tif
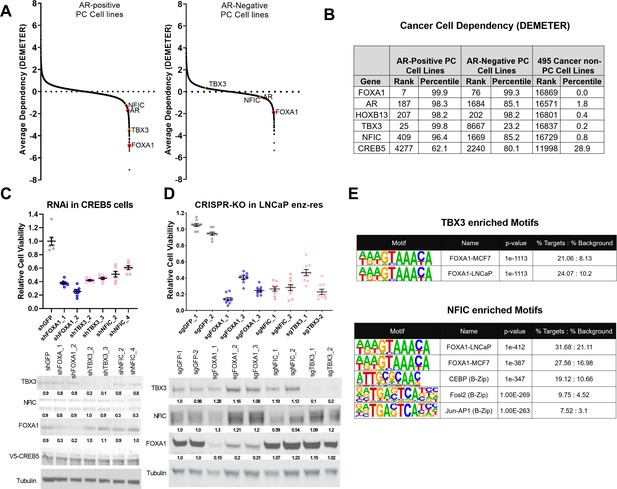
TBX3 and NFIC are key regulators in prostate cancer cells including those that are enzalutamide resistant.
(A) Analysis of genome-scale RNAi screening data ranking the average dependency of 16,869 genes (x-axis) in androgen receptor (AR)-positive (Lleft) and AR-negative (Rrightt.) prostate cancer cell lines (Project Achilles 2.20.1). Average DEMETER score (y-axis) indicates the dependency correlations of FOXA1 and CREB5-interacting proteins. A negative DEMTER score indicates gene dependency in these specific PC cell lines. (B) Average ranks and percentiles based on DEMETER dependency scores are shown for selected genes in AR-positive, AR-negative, and non-PC cell lines. (C). shRNA was utilized to deplete experimental (NFIC, TBX3), negative (GFP) or positive controls (FOXA1) genes in LNCaP cells overexpressing CREB5. The overall cell numbers are depicted post -perturbation. A representative immunoblots depicts depletion of proteins from the proliferation experiments. Tubulin was used as a loading control. The relative depletion is quantified based on the average of all experiments after normalizing to tubulin. (D) CRISPR-Cas9 was utilized to deplete experimental (NFIC, TBX3), negative (GFP) or positive controls (FOXA1) genes in LNCaP cells that spontaneously developed resistance to enzalutamide. The overall cell numbers are depicted post -perturbation. A representative immunoblots depicts depletion of proteins from (C, upper panel) in proliferation experiments. Tubulin was used as a loading control. The relative depletion is quantified based on the average of all experiments after normalizing to tubulin. (E) ChIP-seq data from NFIC and TBX3 was analyzed to predict interaction with CREB5 or FOXA1 motifs. Enriched motifs, the targeted cell lines, and significance levels are depicted.
-
Figure 4—source data 1
Immunoblots were used to detect expression of TBX3 and FOXA1 in the indicated samples for Figure 4C.
- https://cdn.elifesciences.org/articles/73223/elife-73223-fig4-data1-v2.zip
-
Figure 4—source data 2
The area highlighted was used to develop the figure for TBX3 and FOXA1 in the indicated samples for Figure 4C.
- https://cdn.elifesciences.org/articles/73223/elife-73223-fig4-data2-v2.zip
-
Figure 4—source data 3
Immunoblots were used to detect expression of tubulin in the indicated samples for Figure 4C.
- https://cdn.elifesciences.org/articles/73223/elife-73223-fig4-data3-v2.zip
-
Figure 4—source data 4
The area highlighted was used to develop the figure for tubulin in the indicated samples for Figure 4C.
- https://cdn.elifesciences.org/articles/73223/elife-73223-fig4-data4-v2.zip
-
Figure 4—source data 5
Immunoblots were used to detect expression of NFIC in the indicated sample for Figure 4C.
- https://cdn.elifesciences.org/articles/73223/elife-73223-fig4-data5-v2.tif
-
Figure 4—source data 6
The area highlighted was used to develop the figure for NFIC in the indicated samples for Figure 4C.
- https://cdn.elifesciences.org/articles/73223/elife-73223-fig4-data6-v2.tif
-
Figure 4—source data 7
Immunoblots were used to detect expression of V5-tagged CREB5 or luciferase in the indicated samples for Figure 4C.
- https://cdn.elifesciences.org/articles/73223/elife-73223-fig4-data7-v2.tif
-
Figure 4—source data 8
The area highlighted was used to develop the figure for V5-tagged CREB5 or luciferase in the indicated samples for Figure 4C.
- https://cdn.elifesciences.org/articles/73223/elife-73223-fig4-data8-v2.tif
-
Figure 4—source data 9
Immunoblots were used to detect expression of FOXA1 and TBX3 in the indicated samples for Figure 4D.
- https://cdn.elifesciences.org/articles/73223/elife-73223-fig4-data9-v2.zip
-
Figure 4—source data 10
The area highlighted was used to develop the figure for FOXA1 and TBX3 in the indicated samples for Figure 4D.
- https://cdn.elifesciences.org/articles/73223/elife-73223-fig4-data10-v2.zip
-
Figure 4—source data 11
Immunoblots were used to detect expression of tubulin in the indicated samples for Figure 4D.
- https://cdn.elifesciences.org/articles/73223/elife-73223-fig4-data11-v2.zip
-
Figure 4—source data 12
The area highlighted was used to develop the figure for tubulin in the indicated samples for Figure 4D.
- https://cdn.elifesciences.org/articles/73223/elife-73223-fig4-data12-v2.zip
-
Figure 4—source data 13
Immunoblots were used to detect expression of NFIC in the indicated samples for Figure 4D.
- https://cdn.elifesciences.org/articles/73223/elife-73223-fig4-data13-v2.zip
-
Figure 4—source data 14
The area highlighted was used to develop the figure for NFIC in the indicated samples for Figure 4D.
- https://cdn.elifesciences.org/articles/73223/elife-73223-fig4-data14-v2.zip
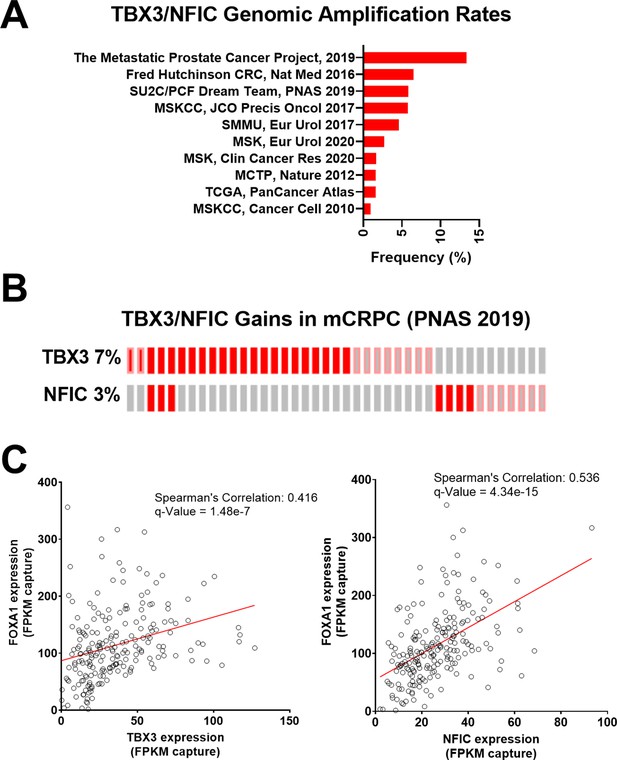
TBX3 and NFIC are amplified in prostate cancer cells.
(A) The genomic amplification rates of TBX3 and NFIC are examined in various prostate cancer studies. (B) In one metastatic castration-resistant prostate cancer (mCRPC) study, the rates of TBX3, NFIC, and FOXA1 gains are depicted. (C and D). The expression of TBX and NFIC are compared in one mCRPC study in which the regression line, Spearman’s correlation coefficients, and q-values are depicted.
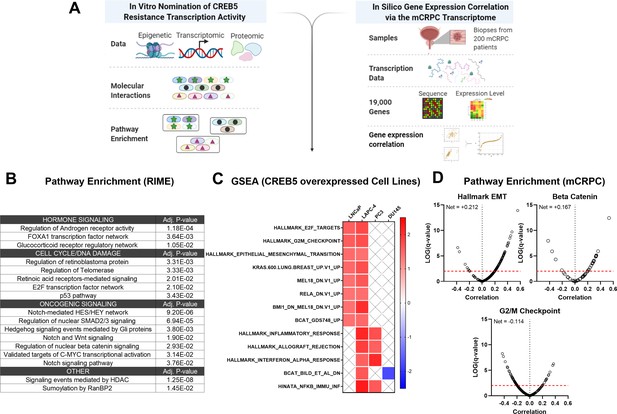
Integrative analysis of CREB5 activity.
(A) A workflow of the informatics analysis of CREB5 using in vitro and metastatic castration-resistant prostate cancer (mCRPC) data. (B) Spectrum of shared CREB5 and FOXA1 protein interactions identified by rapid immunoprecipitation and mass spectroscopy of endogenous proteins (RIME) are analyzed. The enriched pathways and statistical significance are presented for specific pathways. (C) Gene Set Enrichment Analysis (GSEA) analysis of RNA-seq data from CREB5 or luciferase overexpressing androgen receptor (AR)-positive (LNCaP and LAPC-4) and AR-negative (PC3, DU145) prostate cancer cells. (D) Based on RNA-seq from clinical mCRPC, Spearman’s correlation coefficients compare CREB5 expression with EMT, betaβ-catenin, and G2/M signaling. Correlation coefficient values (Rho, σ, x-axis) for CREB5 against each gene, as represented by a single dot, and the statistical significance (negative log of p-value, y-axis) areis displayed. Pp-vValue is marked (red dotted line).
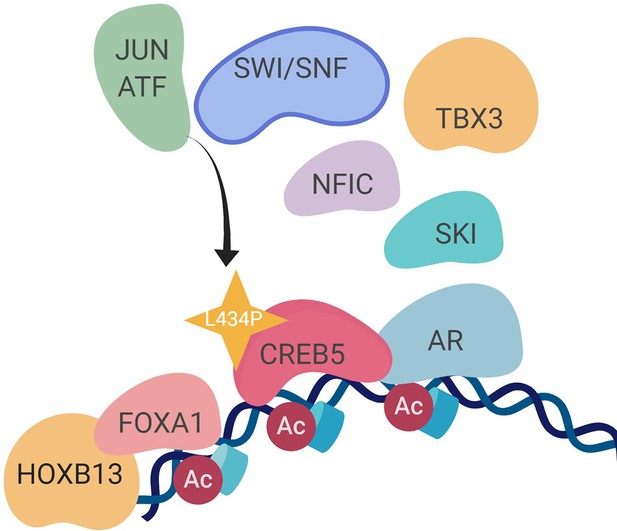
A molecular model of the CREB5 complex and transcription promoting androgen receptor-targeted therapy (ART) resistance.
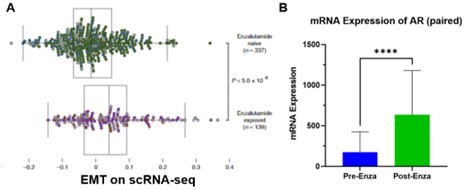
A.
Figure adapted from He et al. (He et al., Nat Med, 2021). Single cells from a mCRPC patient pre- and post-treatment were examined based on single cell RNA-seq approach and the most enriched pathway was EMT after enzalutamide treatment. B. AR expression levels were examined in the sample tumor cells, which increased with statistical significance (Student’s t-test).
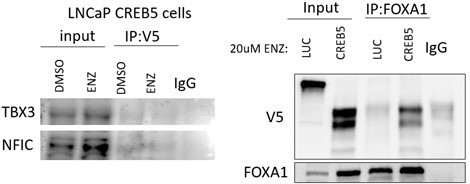
Left.
V5-tagged CREB5 was targeted for immunoprecipitation in cell lysates in vehicle control (DMSO) or enzalutamide treated (ENZ) LNCaP cells. TBX3 and NFIC were detected in the total lysate (input) and precipitates (IP) using immunoblots. IgG was used as a negative precipitation control. Right. FOXA1 was targeted in enzalutamide treated cells in a Co-IP experiment. CREB5 and FOXA1 were detected in the total lysate (input) and precipitates (IP) using immunoblots. IgG was used as a negative precipitation control.
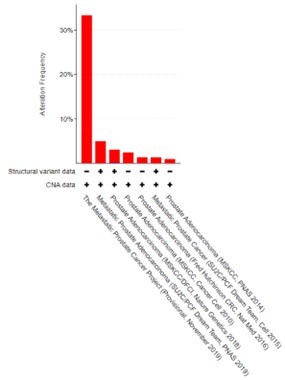
CREB5 amplifications (red) and deletions (blue) are examined and displayed across prostate cancer cohorts as of 12.2.2021.
In total, there were variable amplification rates and we only detected one homozygous deletion in a primary prostate tumor in the 2018 study that included 680 primary prostate cancer samples and 333 mCRPC. Adapted from cBioportal.
Tables
Results from RIME, transcript association analyses, Dependency analyses are presented for TBX3, NFIC, DNMT1 and LDB1.
| Unique Peptide Counts, CREBS RIME | Unique Peptide Counts, FOXA1 RIME | mCRPC Transcript Correlation with FOXA1 | Dependency in AR-positive PC, Percentile | Dependency in AR-negative PC, Percentile | Overall Dependency, Percentile | |
|---|---|---|---|---|---|---|
| TBX3 | 3.00 | 6.50 | 0.54 | 99.78 | 23.27 | 0.19 |
| NFIC | 4.00 | 4.00 | 0.42 | 96.38 | 85.22 | 0.82 |
| DNMT1 | 2.00 | 0.50 | -0.09 | 28.25 | 60.21 | 63.38 |
| LBD1 | 2.00 | 1.50 | 0.07 | 14.42 | 65.56 | 85.23 |
Transcription data from three independent studies were examined.
Pearson correlations were performed to examine the associations between CREB5 and AR as well as AR and FOXA1. The cohort features are described along with the correlation value and p-value. The p-values were are marked as significant (**) or not significant (n.s.) based on type I error levels at less than 0.05.
| CREB5, AR | AR, FOXA1 | |||
|---|---|---|---|---|
| R | p-val | R | p-val | |
| PCF/SU2C (n=266, mCRPC only) (Abida et al., Proc Natl Acad Sci USA, 2019) | -0.11 | 0.0612 (n.s.) | 0.41 | 3.70E-12** |
| TCGA PRAD (n=488, Primary only) | 0.08 | 0.0707 (n.s.) | 0.37 | 2.29E-17** |
| MSK (n=128, primary and mCRPC) (Taylor et al., Cancer Cell, 2010) | 0.08 | 0.34 (n.s.) | 0.4 | 2.92E-06** |
Additional files
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/73223/elife-73223-transrepform1-v2.pdf
-
Supplementary file 1
Tables.
Table 1. Z-scores for each gene from the genome-scale screen are presented and ranked.Table 2. Unique peptide counts are displayed for the RIME experiment based on each condition.Table 3. Unique peptide counts are displayed for the RIME experiment based on each condition.Table 4. Unique peptide counts are displayed for the RIME experiment based on each condition.
- https://cdn.elifesciences.org/articles/73223/elife-73223-supp1-v2.xlsx





