Characterization of the endogenous DAF-12 ligand and its use as an anthelmintic agent in Strongyloides stercoralis
Figures
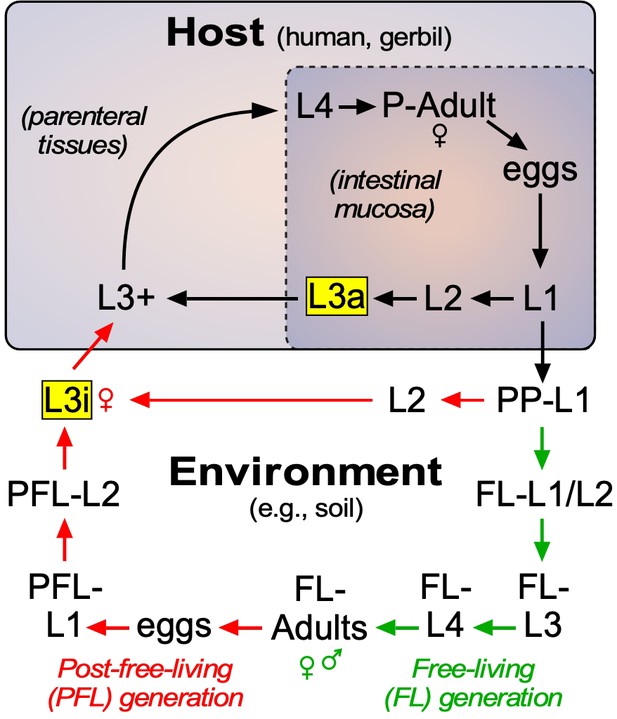
Lifecycle of S. stercoralis.
Similar to other nematodes, S. stercoralis hatch from eggs and undergo four larval (L) molts to become adults, either in the environment or in the host. The parasite has two infectious stages (highlighted in yellow), one that exists in the environment as L3i and one that exists in the mucosa of the host intestine as an autoinfective L3a. In the postparasitic (PP) environment, larvae can undergo two developmental fates. Under host-like temperature conditions, the PP-larvae arrest their development directly as infectious third-stage larvae. However, under more temperate conditions, the postparasitic L1 (PP-L1) develop through one free-living (FL) generation (green arrows), followed by a post-free-living (PFL) generation (red arrows) that invariably arrests as L3i. Pharmacologically activating the nuclear receptor DAF-12 has been shown to prevent L3i arrest in the PFL generation (Albarqi et al., 2016; Wang et al., 2009). In this study, we characterized the role of the endogenous DAF-12 ligand at each of these developmental stages. See text for details.
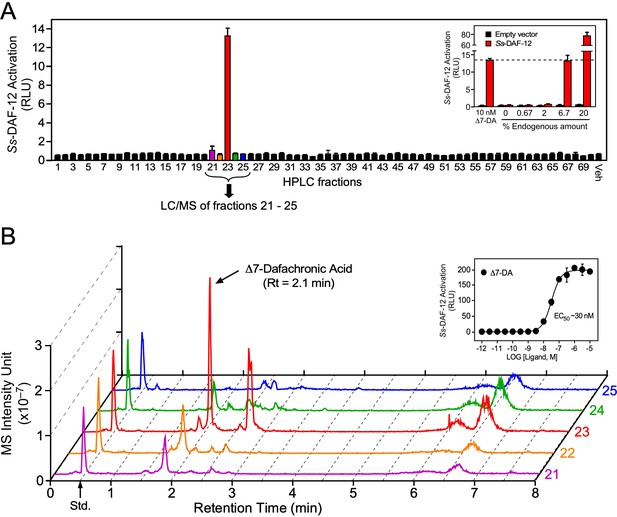
Identification of Δ7-dafachronic acid as the endogenous DAF-12 ligand in S. stercoralis.
(A) Purification of the endogenous S. stercoralis ligand for Ss-DAF-12. Lipids from free-living L3 worms were extracted and fractionated as described in Figure 2—figure supplement 1. The resulting lipid fractions were then tested in a Ss-DAF-12 cell-based reporter assay. Inset: Dose response of the endogenous activity in fraction 23. RLUs, relative light units. Data are presented as the mean ± standard deviation (SD) of technical triplicates. (B) Δ7-DA is specifically present in the active lipid fraction. High-performance liquid chromatography (HPLC) fractions 21–25 were analyzed by ultra-performance liquid chromatography coupled with mass spectrometry (UPLC–MS). CDCA-2H4 (100 nM) was added to each fraction as an internal standard and has a retention time (RT) of 0.5 min (arrow). Inset: Dose response of Δ7-DA in the Ss-DAF-12 reporter assay. Data are presented as the mean ± SD of technical triplicates and were repeated three times. See also Figure 2—figure supplements 1 and 2.
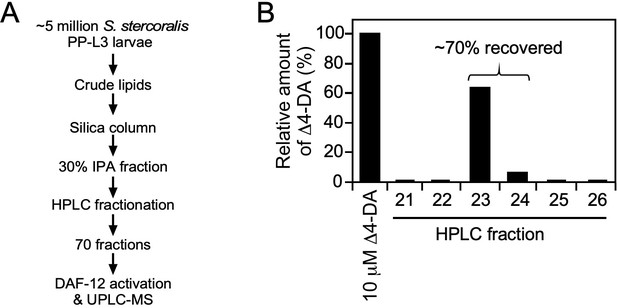
Strategy for activity-based, DAF-12 ligand purification in S. stercoralis.
(A) DAF-12 ligand purification scheme. (B) Determination of purification efficacy. To estimate the efficiency of DA purification, 10 μM of Δ4-DA was added as a standard to ~2 million C. elegans daf9daf12 worms, a mutant strain that lacks endogenous DA. Following extraction, the lipid fraction was further purified by high-performance liquid chromatography (HPLC). The amount of Δ4-DA in each of the 70 collected HPLC fractions was analyzed by liquid chromatography (LC)/mass spectrometry (MS) in negative selective ion monitoring (SIM) mode at m/z 413. Shown are the only fractions containing detectable Δ4-DA. Up to 70% of the Δ4-DA was recovered, the majority of which was in fraction 23. The experiment was repeated twice.
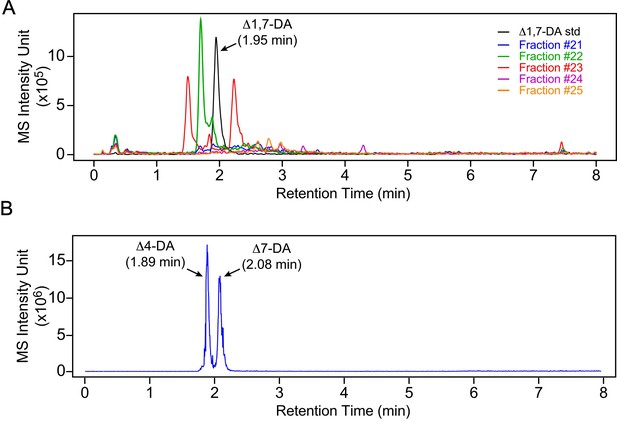
Δ4-DA and Δ1,7-DA are not present in free-living L3 parasites.
(A) Δ1,7-DA is undetectable in FL-L3 larvae. The active DAF-12 fractions shown in Figure 2 were analyzed by ultra-performance liquid chromatography coupled with mass spectrometry (UPLC–MS) in negative selective ion monitoring (SIM) mode at m/z 411 and compared to the Δ1,7-DA standard. Δ1,7-DA has a distinct molecular mass that permits it to be separated from other DA congeners with this method. If Δ1,7-DA was present, it would have been detected in fraction 23. (B) Δ4-DA is undetectable L3 parasites. Δ4-DA and Δ7-DA standards were separated and detected by UPLC–MS in negative SIM mode at m/z 413. Although Δ4-DA has the same molecular mass as Δ7-DA, the two compounds can be completely separated by retention times. Notably, the retention time of the peak in the active fraction shown in Figure 2 only matches Δ7-DA.
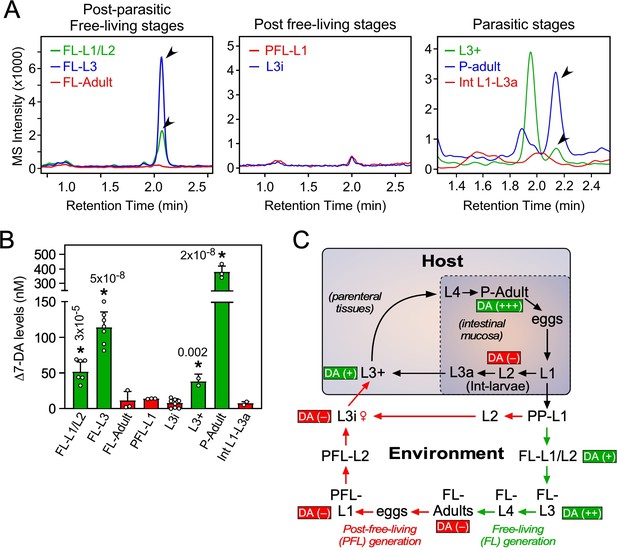
Profiling of Δ7-DA in developmental stages of S. stercoralis.
(A) Detection of Δ7-DA during the lifecycle of S. stercoralis. Lipid extracts from the indicated stages of S. stercoralis were analyzed by derivatizing Δ7-DA to Δ7-DA-picolylamine, which then was detected by ultra-performance liquid chromatography coupled with mass spectrometry (UPLC–MS) in positive multiple reaction monitoring (MRM) mode with m/z transition 505 → 487. Parasitic stages were recovered from hyperinfected gerbils. Arrowheads show Δ7-DA peaks. Note that in the extracts from L3+ larvae, the peak with the faster retention time (1.95 min) is an unknown metabolite that is only found at this stage. (B) Δ7-DA levels were determined in the stages shown in (A) by comparison to a known standard. Data are presented as the mean ± standard deviation (SD) (n = 2–8); *p values (shown in the figure) were determined by Student’s t-test compared to L3i larvae. (C) Schematic summary of Δ7-DA levels in the development stages of S. stercoralis. Δ7-DA is absent in larvae developing to infectious stages (i.e., L3i and L3a) and is present in larvae undergoing reproductive development.
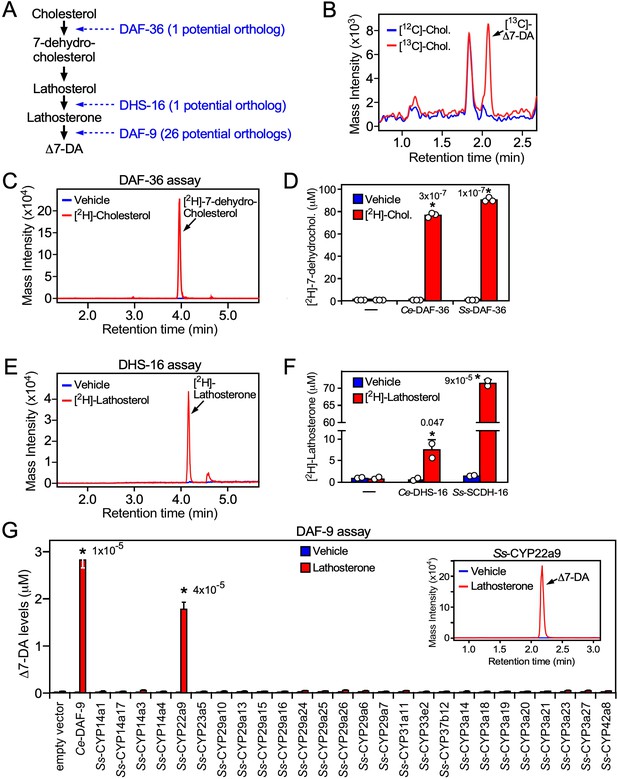
Characterization of the Δ7-DA biosynthetic pathway in S. stercoralis.
(A) Diagram of Δ7-DA biosynthetic pathway in C. elegans. In blue are the known C. elegans enzymes followed in parentheses by the number of candidate orthologs found in S. stercoralis. (B) Δ7-DA is synthesized de novo in S. stercoralis. Extracts from FL-L3 worms cultured from PP-L1s in the presence or absence of [13C]-cholesterol were assayed by ultra-performance liquid chromatography coupled with mass spectrometry (UPLC–MS) for incorporation of [13C] into Δ7-DA. (C, D) Ss-DAF-36 catalyzes the synthesis of 7-dehydrocholesterol in the first step of Δ7-DA biosynthesis. Sf9 microsomes expressing Ss-DAF-36 were incubated with vehicle or 100 μM [2H]-cholesterol and assayed for the synthesis of [2H]-7-dehydrocholesterol by UPLC–MS chromatography (C), and the amount quantitated relative to that produced by the C. elegans ortholog, Ce-DAF-36 (D). (E, F) Ss-SCDH-16 catalyzes the of synthesis of lathosterone in the penultimate step of Δ7-DA biosynthesis. Sf9 microsomes expressing Ss-SCDH-16 were incubated with vehicle or 100 μM [2H]-lathosterol and assayed for the synthesis of [2H]-lathosterone by UPLC–MS chromatography (E), and the amount quantitated relative to that produced by the C. elegans ortholog, Ce-DHS-16 (F). (G) Ss-CYP22a9 catalyzes the synthesis of Δ7-DA from lathosterone. Sf9 cells expressing one of each of the 26 S. stercoralis cytochrome P450 homologs were incubated with vehicle or 10 μM lathosterone and assayed for the production of Δ7-DA by UPLC–MS as in Figure 2. Ce-DAF-9 is shown as a positive control. Inset, chromatogram from the reaction with Ss-CYP22a9. Data are presented as the mean ± standard deviation (SD) (n = 2–3); *p values (shown in the figure) were determined by Student’s t-test compared to vehicle. See also Figure 4—figure supplement 1.
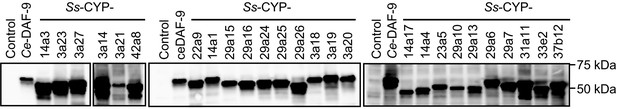
Expression of the 26 S. stercoralis cytochrome P450 enzymes in insect Sf9 cells.
P450 enzymes were fused to C-terminal HA tags and detected by immunoblot using an anti-HA antibody. The C. elegans DAF-9 enzyme is shown as a positive control. The experiment was repeated three times. For full gel blot images, see Figure 4—figure supplement 1—source data 1.
-
Figure 4—figure supplement 1—source data 1
Full gel images for the expression of Ss-CYPs.
- https://cdn.elifesciences.org/articles/73535/elife-73535-fig4-figsupp1-data1-v1.pdf
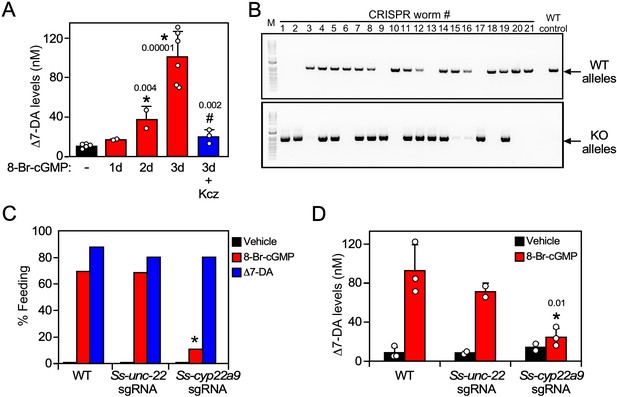
Ss-CYP22a9 is required for L3i activation and Δ7-DA synthesis in S. stercoralis.
(A) Inhibition of cytochrome P450 activity blocks Δ7-DA synthesis in parasites. L3i (1000 worms/group) were treated with 0.5 mM 8-Br-cGMP in the presence or absence of 25 μM ketoconazole (Kcz). Data are presented as the mean ± standard deviation (SD) (n = 2–6); p values (shown in the figure) were determined by t-test compared to vehicle (*) or 3-day treatment with 8-Br-cGMP (#). (B, C) Ss-CYP22a9 is required for cGMP-induced L3i activation. The Ss-cyp22a9 gene was disrupted by CRISPR/Cas9-mediated, homology-directed repair as shown in Figure 5—figure supplement 1C. The resulting F1 generation of L3i worms expressing the positive selection marker (GFP) were sorted manually, subjected to single worm genotyping (B) and assayed for feeding behavior (C). Disruption of the Ss-unc-22 gene was used as a control. *p = 1 × 10−12 compared to wild-type (WT) and p = 1 × 10−7 compared to Ss-unc-22 by Fisher’s exact test (n = 40–200 from three independent experiments). For full gel blot images, see Figure 5—source data 1. (D) Δ7-DA synthesis is abolished in Ss-cyp22a9 knockout parasites. Ss-cyp22a9 or Ss-unc-22 (as a control) genes were disrupted by CRISPR using the same sgRNA plasmids in (B, C) and F1 generation L3i worms were assayed for Δ7-DA levels as in Figure 3 after treatment with 0.5 mM 8-Br-cGMP. Data represent the mean ± SD (n = 2–3); *p < 0.03 by t-test compared to WT or Ss-unc-22 worms treated with 8-Br-cGMP. See also Figure 5—figure supplement 1.
-
Figure 5—source data 1
Full gel images for single worm genotyping.
- https://cdn.elifesciences.org/articles/73535/elife-73535-fig5-data1-v1.pdf
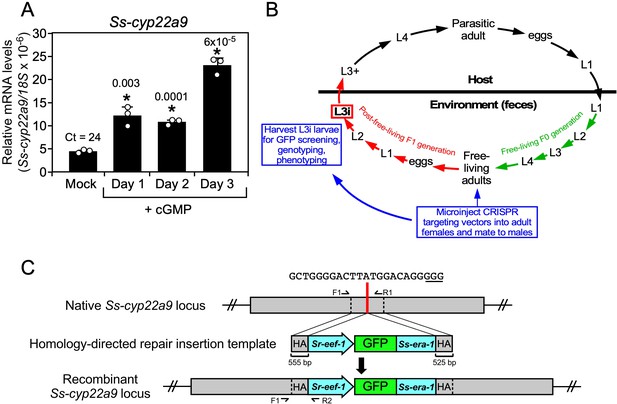
In vivo characterization of the DAF-9 homolog (Ss-CYP22a9) in S. stercoralis.
(A) Ss-cyp22a9 mRNA expression is increased by cGMP in L3i larvae. L3i larvae were treated as in Figure 5 and Ss-cyp22a9 mRNA levels were measured by qPCR and compared to 18S rRNA levels. Results expressed as means ± standard deviation (SD) from technical triplicates. *p values (shown in the figure) determined by t-test. Duplicate experiments were performed with similar results. Ct value shown for mock treated. (B) CRISPR knockout strategy for targeting Ss-cyp22a9 in S. stercoralis: free-living adult females were microinjected with CRISPR targeting vectors (for either nonhomologous end joining [NHEJ] or homologous directed repair [HDR] gene editing) and allowed to mate. F1 progeny were grown to the L3i stage and GFP-positive worms were phenotyped and genotyped. (C) Homology-directed repair targeting vector for Ss-cyp22a9 CRISPR. Red line indicates position of the insertion repair template in the P450 domain of the Ss-cyp22a9 gene (SSTP_0001032100). The guide sequence and PAM site (underlined) are shown. The recombinant locus expresses a GFP marker from the Sr-eef-1 promoter fused to the 3′-UTR of the Ss-era-1 gene. F and R, forward and reverse primers, respectively; HA, homology arms. WT and recombinant alleles produce 1052 and 823 bp products with F1/R1 and F1/R2, respectively.
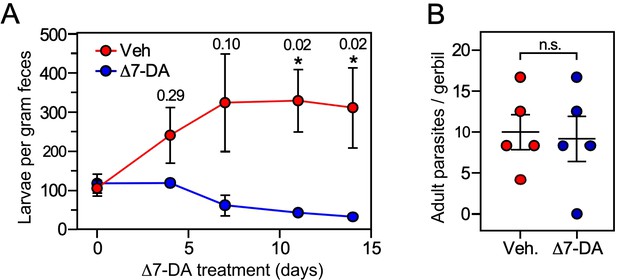
Δ7-DA suppresses output of fecal larvae in latent, uncomplicated strongyloidiasis.
(A) Δ7-DA treatment reduces fecal larvae by >90% in gerbils infected with S. stercoralis. (B) Adult parasite burden in infected gerbils measured at 14-day post-treatment. Data are plotted as the mean ± standard error (SE) (n = 5); q values (shown in the figure) were determined by Mann–Whitney U test compared to vehicle (*statistically significant; n.s., not significant). See also Figure 6—figure supplement 1 for individual data points in (A).
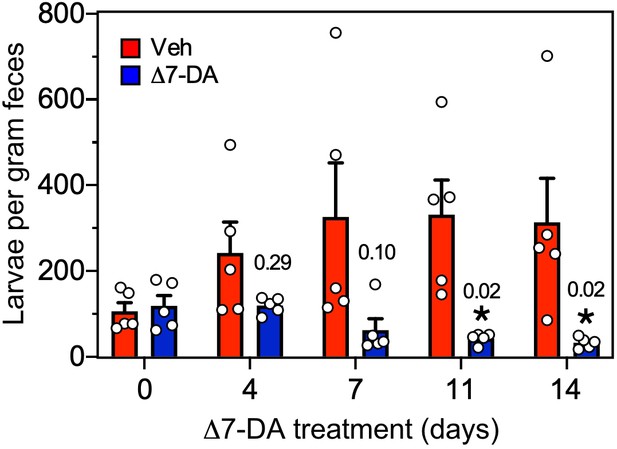
Δ7-DA suppresses output of fecal larvae in latent, uncomplicated strongyloidiasis.
Δ7-DA treatment reduces fecal larvae by >90% in gerbils infected with S. stercoralis. This is a replot of Figure 6A showing individual data points. Data are plotted as the mean ± standard error (SE) (n = 5); q values (shown in the figure) were determined by Mann–Whitney U test compared to vehicle (*statistically significant).
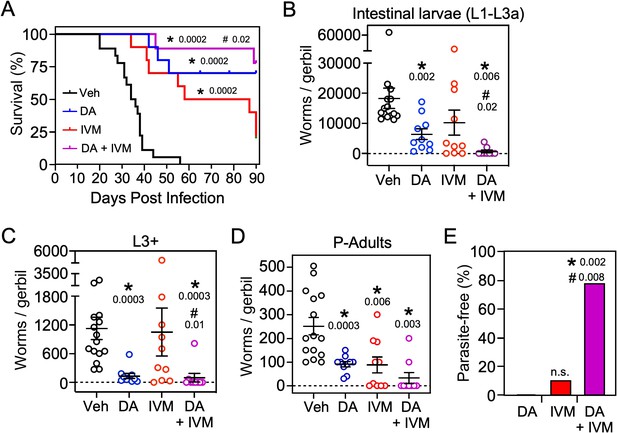
Δ7-DA and ivermectin act cooperatively to treat disseminated strongyloidiasis hyperinfection.
(A) Kaplan–Meier survival curves of hyperinfected gerbils treated with vehicle (Veh), Δ7-DA, and/or ivermectin (IVM). Sample sizes: n = 18 (Veh), 10 (DA), 10 (IVM), 9 (DA+ IVM); q values (shown in the figure) were determined by log-ranked test compared to treatment with vehicle (*) or ivermectin alone (#). (B–D) Parasite burden at various lifecycle stages from the hyperinfected gerbil treatment groups shown in (A). At the time of death, live intestinal L1-L3a (B), L3+ (C), and adult (D) parasites were counted. Sample sizes: n = 15 (Veh), 10 (DA), 10 (IVM), 9 (DA+ IVM); q values (shown in the figure) were determined by Mann–Whitney U test compared to treatment with vehicle (*) or ivermectin alone (#). (E) Cotreatment with Δ7-DA and ivermectin eradicates parasites in hyperinfected gerbils. Animals from (A) with no detectable parasites in any part of the body after 90 days of treatment were scored as parasite-free. Notably, all the animals in the cotreatment group that survived hyperinfection (shown in (A)) were found to be parasite-free. Sample sizes: n = 10 (DA), 10 (IVM), 9 (DA+ IVM); q values (shown in the figure) were determined by Fisher’s exact test compared to treatment with vehicle (*) or ivermectin alone (#); n.s., not significant. DA, 50 μM Δ7-DA administered in drinking water; IVM, 300 μg/kg ivermectin by i.p. injection. See also Figure 7—figure supplement 1.
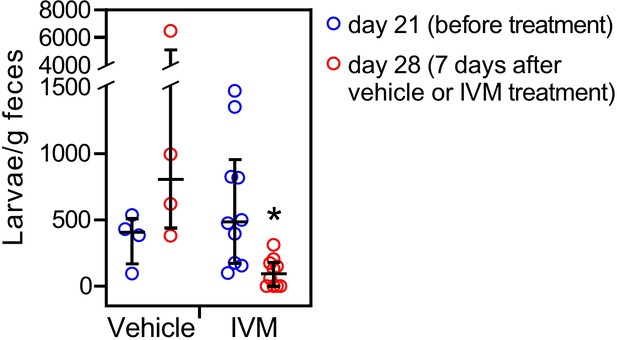
Ivermectin (IVM) mimics the treatment of a human S. stercoralis hyperinfection in gerbil models.
Vehicle or IVM (300 mg/kg) was injected i.p. into S. stercoralis hyperinfected gerbils at 21-day postinfection and fecal larval output was measured 7 days later. IVM treatment results in ~40% decrease in fecal larval output, but does not permanently eliminate parasites. n = 4 for vehicle, 10 for IVM treatment groups. *q = 0.02 comparing to the day 21 data, by Mann–Whitney–Wilcoxon test corrected with FDR method. Error bars represent the median ± interquartile.

Strategy for using DAF-12-based therapeutics to treat strongyloidiasis.
Administration of DAF-12 ligands like Δ7-DA disrupts the lifecycle of nematode parasites by preventing the development of the infective L3i and L3a worms, where DAF-12 is normally unliganded. Pharmacologic activation of DAF-12 with Δ7-DA prevents both environmental infection and autoinfection, which are essential features of the latent and hyperinfection forms of strongyloidiasis. In contrast, ivermectin kills only the actively developing stages and therefore is unable to target developmentally-quiescent infective L3i and L3a larvae. For this reason, the combination of the DAF-12 ligand and ivermectin achieves a synergistic, double blockade of the lifecycle.
Tables
Detection methods for the compounds in this study.
| Steroids | Retention time(min) | MS detection mode | Parent ion (m/z) | Product ion (m/z) |
|---|---|---|---|---|
| Δ7-DA | 2.1 | Negative SIM | 413 | N/A |
| Δ4-DA | 1.9 | Negative SIM | 413 | N/A |
| Δ1,7-DA | 2.0 | Negative SIM | 411 | N/A |
| Δ7-DA-PA | 2.1 | Positive MRM | 505 | 487 |
| [13C]-Δ7-DA-PA | 2.1 | Positive MRM | 508 | 490 |
| [2H]–7-Dehydrocholesterol | 3.9 | Positive MRM | 374 | 109 |
| [2H]-Lathosterone | 4.1 | Positive MRM | 392 | 109 |
-
MRM, multiple reaction monitoring; SIM, selective ion monitoring.
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene(S. stercoralis) | Ss_cyp22a9 | Wormbase | SSTP_0001032100 | |
| Gene(S. stercoralis) | Ss-daf-36 | Wormbase | SSTP_0000037900 | |
| Gene(S. stercoralis) | Ss-scdh-16 | Wormbase | SSTP_0001031100 | |
| Cell line (Cercopithecus aethiops) | COS-7 | ATCC | Cat# CRL-1651; RRID:CVCL_0224 | |
| Cell line (Spodoptera frugiperda) | Sf9 | ATCC | Cat# CRL-1711; RRID:CVCL_0549 | |
| Strain, strain background(Meriones unguiculatus, male) | Mongolian gerbil | Charles Rivers | Strain code: 243 | |
| Strain, strain background(Canis familiaris, male) | Dog | Oak Hill Genetics | N/A | |
| Antibody | anti-HA tag antibody [HA.C5] (Mouse monoclonal) | Abcam | Cat# ab18181; RRID:AB_444303 | WB (1:2000) |
| Recombinant DNA reagent | pGL4.53(plasmid) | Promega | E5011 | |
| Recombinant DNA reagent | pCMX-CeSs-DAF-12(plasmid) | Wang et al., 2009 (PMID:19497877) | N/A | |
| Recombinant DNA reagent | pNL3.1-DAF-12RE-lit-1(plasmid) | This paper | N/A | DAF-12 reporter |
| Recombinant DNA reagent | pFastBac-Dual-hOR(plasmid) | Motola et al., 2006 (PMID:16529801) | N/A | |
| Recombinant DNA reagent | pFastBac-Dual-hOR-CYPs(plasmids) | This paper | N/A | Express hOR and CYPs in Sf9 cells |
| Recombinant DNA reagent | pFastBac-Dual-hOR-DAF-36s(plasmids) | This paper | N/A | Express hOR and DAF-36s in Sf9 cells |
| Recombinant DNA reagent | pFastBac-Dual-hOR-DHS-16s(plasmids) | This paper | N/A | Express hOR and DHS-16s in Sf9 cells |
| Recombinant DNA reagent | pML60-Ss-unc-22(plasmid) | Gang et al., 2017 (PMID:29016680) | N/A | |
| Recombinant DNA reagent | pML60-Ss-cyp22a9(plasmid) | This paper | N/A | Guide RNA plasmid for Ss-cyp22a9 |
| Recombinant DNA reagent | pPV540(plasmid) | Lok, 2019 (PMID:31379923) | N/A | |
| Commercial assay or kit | Nano-Glo Dual-luciferase kits | Promega | N1620 | |
| Commercial assay or kit | NADPH regeneration system | Promega | V9510 | |
| Commercial assay or kit | Microsome Isolation Kit | Abcam | ab206995 | |
| Chemical compound, drug | Ketoconazole | Sigma | K1003 | |
| Chemical compound, drug | 8-Bromo-cGMP | Tocris | 1089 | |
| Chemical compound, drug | Methylprednisolone acetate | Zeotis | DEPO-MEDRO | 20 mg/ml |
| Chemical compound, drug | Ivermectin | Merial Limited | Ivermec | 1% solution |
| Chemical compound, drug | Chenodeoxycholic acid-2H4 | Sigma | 614,122 | |
| Chemical compound, drug | Triphenylphosphine | Sigma | T84409 | |
| Chemical compound, drug | 2,2′-Dipyridyl disulfide | Sigma | D5767 | |
| Chemical compound, drug | 2-Picolylamine | Sigma | A65204 | |
| Chemical compound, drug | Cholesterol-13C3 | Cambridge Isotope | CLM-9139 | |
| Chemical compound, drug | Cholesterol-2H7 | Avanti Polar Lipids | 700041 P | |
| Chemical compound, drug | Lathosterol-2H7 | Avanti Polar Lipids | 700,056 | |
| Chemical compound, drug | 7-Dehydrocholesterol-2H7 | Avanti Polar Lipids | 700116P | |
| Chemical compound, drug | Lathosterone | Steraloids | C7500-000 | |
| Chemical compound, drug | Alexa Fluor 594 fluorescent dye | Thermo Fisher | A33082 |
Sample preparation for Δ7-DA quantification in S. stercoralis.
| Parasite sample | Amount | Lysis method | Standard preparation |
|---|---|---|---|
| FL-L1/L2 | 200,000 | Sonication | 100 nM Δ7-DA compound spiked in 200,000 FL-L1/L2 |
| FL-L3 | 50,000 | Sonication | 100 nM Δ7-DA compound spiked in 50,000 FL-L3 |
| FL-adult | 5000 | Sonication | 100 nM Δ7-DA compound spiked in 5000 FL-adults |
| PFL-L1 | 200,000 | Sonication | 100 nM Δ7-DA compound spiked in 200,000 PFL-L1 |
| L3i | 50,000 | Sonication | 100 nM Δ7-DA compound spiked in 50,000 L3i |
| L3+ | 1000 | Proteinase K | 100 nM Δ7-DA compound spiked in 1000 L3+ |
| P-adult | 500 | Proteinase K | 100 nM Δ7-DA compound spiked in 500 FL-adults |
| Intestinal L1-L3a | 5000 | Proteinase K | 100 nM Δ7-DA compound spiked in 5000 Int-larvae |
| cGMP-treated L3i | 1000 | Proteinase K | 100 nM Δ7-DA compound spiked in 1000 L3i |
Oligo sequences used in this study.
| Sequence | Description | |
|---|---|---|
| Forward | GGCATCACCATACAAAACAG | Ss-cyp22a9 wild-type allele genotyping |
| Reverse | TTTGTATGAGGAGGGTTGTG | |
| Forward | GGCATCACCATACAAAACAG | Ss-cyp22a9 KO allele genotyping |
| Reverse | CATCACATTCATCAAAAGTCCACT | |
| Forward | TCCTGGCCAGTGCTAATGTTATT | Ss-cyp22a9 qPCR |
| Reverse | CTATTTGGACGGGATGAGAAGACT | |
| Forward | TGGTGCATGGCCGTTCTTA | Ss-18SRNA qPCR |
| Reverse | CTCGCTCGTTATCGGAATCAA | |
| Forward | GCTGGGGACTTATGGACAGGgttttagagctagaaatagcaag | sgRNA expression plasmid |
| Reverse | /5phos/CATTGTATTGGATGGCAATC | targeting Ss-cyp22a9 |





